Abstract
The variety of chemically diverse pharmacologically-active compounds administered to patients is large and seemingly forever growing, and, with every new drug released and administered, there is always the potential of an allergic reaction. The most commonly occurring allergic responses to drugs are the type I, or immediate hypersensitivity reactions mediated by IgE antibodies. These reactions may affect a single organ, such as the nasopharynx (allergic rhinitis), eyes (conjunctivitis), mucosa of mouth/throat/tongue (angioedema), bronchopulmonary tissue (asthma), gastrointestinal tract (gastroenteritis) and skin (urticaria, eczema), or multiple organs (anaphylaxis), causing symptoms ranging from minor itching and inflammation to death. It seems that almost every drug is capable of causing an immediate reaction and it is unusual to find a drug that has not provoked an anaphylactic response in at least one patient. These facts alone indicate the extraordinary breadth of recognition of IgE antibodies for drugs ranging from relatively simple structures, for example, aspirin, to complex molecules, such as the macrolide antibiotics composed of a large macrocyclic ring with attached deoxy sugars. This wide recognition profile is borne out at the molecular level by results of quantitative immunochemical studies where hapten inhibition investigations have identified structural determinants complementary to IgE antibodies in the sera of allergic subjects. Allergenic determinants have been identified on a variety of drugs including neuromuscular blockers, penicillins, cephalosporins, opioids, thiopentone, sulfonamides, trimethoprim, quinolones, chlorhexidine and the non-steroidal anti-inflammatory drug aspirin. It is already clear that IgE can distinguish fine structural differences on a wide variety of molecules, determinants may be at least as small as an amino group or encompass the whole molecule, and individual drugs may demonstrate allergenic heterogeneity.
1. Introduction: IgE Antibody Recognition of Drugs
From the foundation years of the science of immunology, a prime requirement for a substance to exhibit immunogenicity, that is to act as an antigen, was said to be the inherent property of a certain minimum size. For a molecule to induce an antibody response it was generally believed, and often stated, that a molecular mass of at least ~ 5 to 10 kDa was necessary, that proteins in particular were good immunogens and ‘small’ covalent compounds such as most drugs and other chemicals (usually <1 kDa) were non-antigenic in their free state [1]. Under certain circumstances some exceptions to these requirements were noted. Following Landsteiner's early demonstrations that simple chemicals such as acyl chlorides and acid anhydrides could be made antigenic by coupling these haptens to a suitable carrier protein (for example albumins) [2], it was found that employment of certain adjuvants and immunization schedules sometimes elicited good antibody responses to so-called ‘small’ peptides (<5 kDa) and linkage of a wide variety of other simple chemicals and drugs to a macromolecular carrier produced immunogenic hapten-carrier complexes capable of inducing clear humoral immune responses [3]. Despite this perceived need for drugs to be presented in haptenated macromolecular form to induce an antibody response, specific IgE responses occur to a number of different unreactive drugs lacking both suitable functional groups and properties to account for the formation of drug-carrier antigens. In addition, IgE antibody responses, sometimes manifesting as life-threatening anaphylaxis, are known to occur in some patients upon first exposure to the drug [4,5]. Here we examine IgE antibody responses in immediate, type I hypersensitivities to a range of different drugs together with findings so far on the specificities of the antibody combining site-drug allergenic determinants interactions.
2. Type I Immediate Hypersensitivity: IgE Antibodies, Mast Cells, Mediators and Clinical Reactions
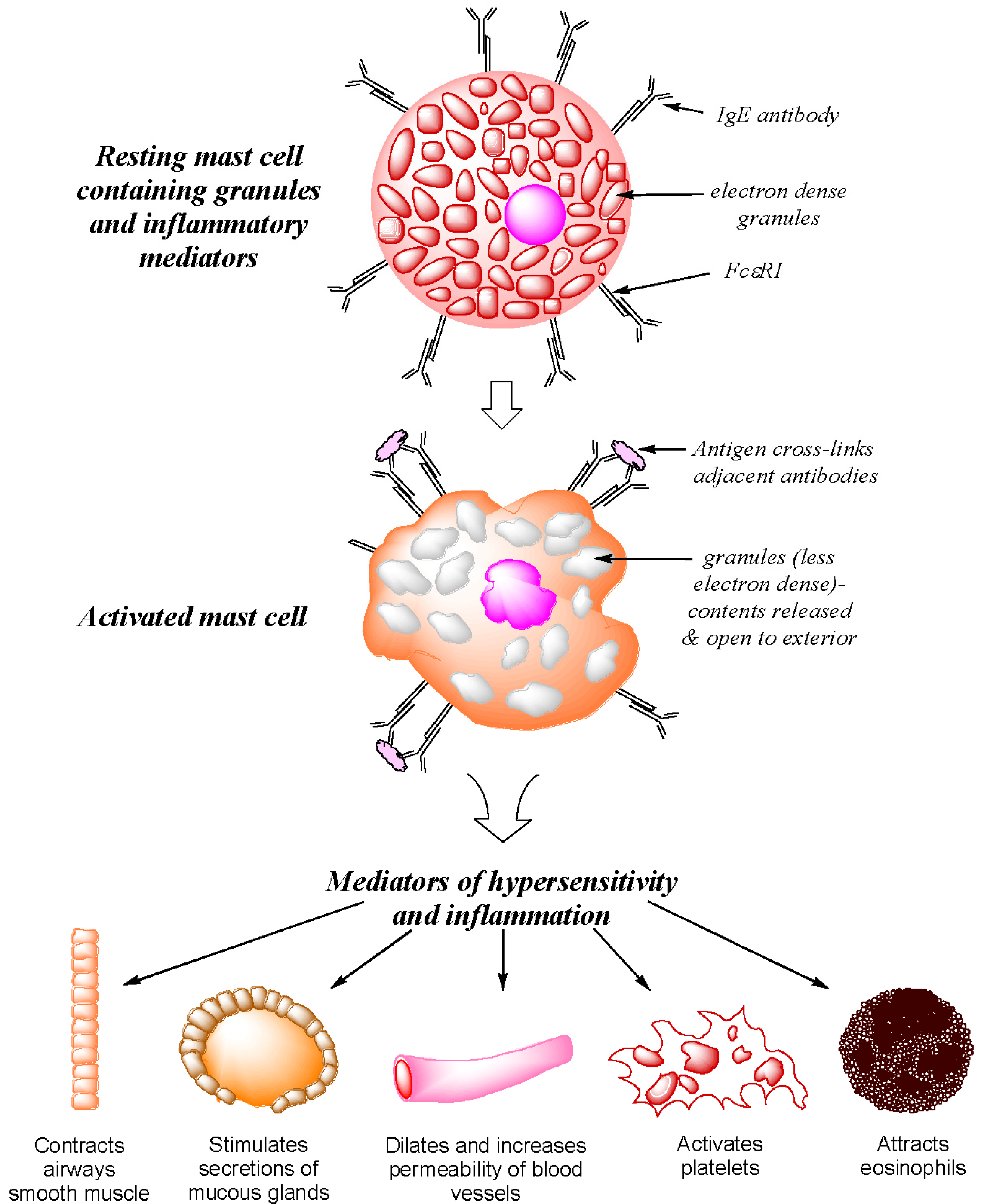
Before considering the recognition of individual drugs in immediate allergic reactions, it should be remembered that in addition to the humoral interaction of IgE with its complementary allergen, the signs and symptoms of a type I hypersensitivity are a direct consequence of cell-mediated processes involving a three component interaction between allergen, antibody and cell. IgE antibodies mediate the immediate (type I) allergic response by interacting with their complementary allergens and with mast cells and basophils. The antibodies bind strongly (~Ka 10−10 M) via the Cε3 domain linker regionto the high affinity FcεRI receptor abundantly expressed on mast cell and basophil surfaces forming a long-lasting IgE–FcεRI complex that dissociates slowly. Interaction of the combining sites of the cell-bound bivalent antibodies with the complementary determinants of the provoking allergen in an allergic subject effects cross-linkage of adjacent antibodies, aggregation of the FcεRI receptors and the triggering of a rapid release of preformed mediators from the secretory granules of the cell [5,6,7] (Figure 1). The released preformed mediators including histamine, platelet activating factor (PAF), heparin, neutrophil, eosinophil and monocyte chemotactic factors, serotonin and the enzymes tryptase, chymase and carboxypeptidase produce the early signs and symptoms seen in a type I hypersensitivity response, namely, vasodilation, edema, bronchospasm and pruritus. In an on-going reaction, newly synthesized mediators are also released. These comprise leukotrienes, prostaglandin D2 (PGD2), a host of pro- and anti-inflammatory cytokines and chemokines, and growth and stimulating factors including interleukins, tumor necrosis factor (TNF), granulocyte-macrophage colony stimulating factor (GM-CSF) and monocyte chemotactic protein-1 (MCP-1) [8].
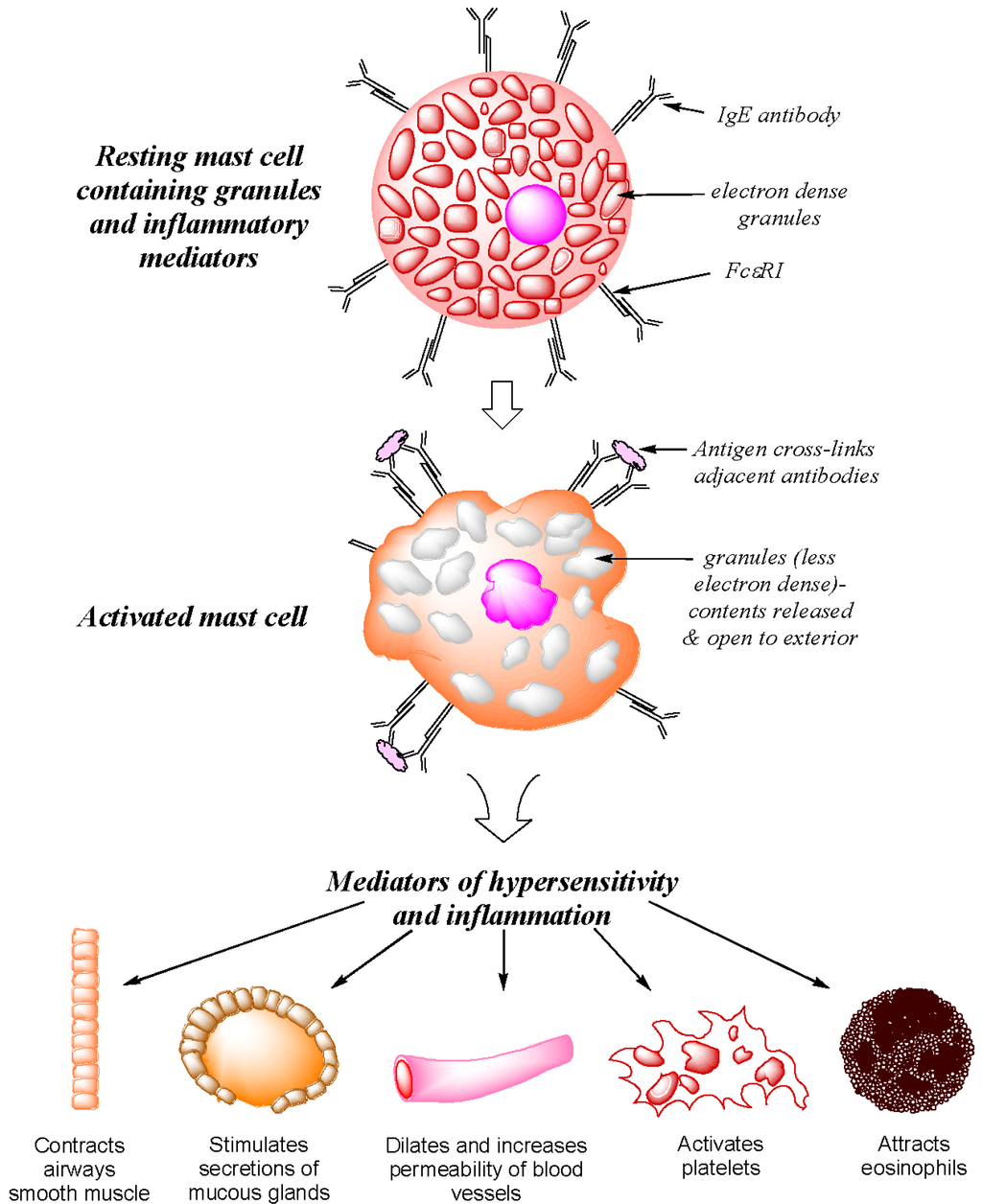
Figure 1.
Diagrammatic representation of allergen-induced mediator release from sensitized mast cells and their main physiological and pharmacological effects on body tissues.
Figure 1.
Diagrammatic representation of allergen-induced mediator release from sensitized mast cells and their main physiological and pharmacological effects on body tissues.
At the clinical level, the host of mediators released in an immediate, type I response evokes a rapid reaction, usually within 30–60 min, but the response may sometimes appear within a few minutes and be extremely dramatic as occurs in anaphylaxis [5,9]. In an anaphylactic reaction, multiple organs and tissues may be affected but IgE-mediated immediate reactions can affect a single organ such as bronchopulmonary tissue (as in asthma), the nasopharynx (allergic rhinitis), mucosa of mouth/throat/tongue (angioedema), the gastrointestinal tract (gastroenteritis), eyes (conjunctivitis) or skin (urticaria, eczema). In addition to anaphylaxis which may be severe enough to cause death, urticaria and angioedema which often occur together, are reactions commonly caused by drugs. Urticaria or hives, manifests as raised, pruritic and often transient erythematous plaques (Figure 2). Angioedema is a vascular reaction producing swelling of the face, often around the mouth and eyes (Figure 3), as well as the mouth mucosa, throat, tongue and genitalia and sometimes other regions of the body including the hands. Swelling is a result of increased permeability and fluid leakage producing edema of the subcutaneous and submucosal tissues [5].
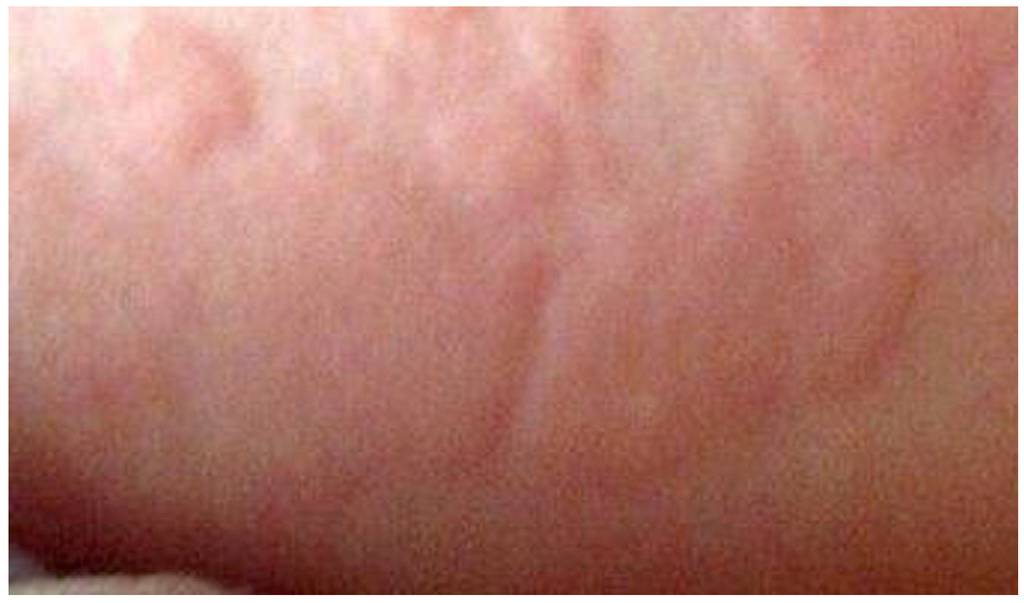
Figure 2.
Urticaria or hives showing pale red, raised, pruritic plaques. Photograph courtesy of Dr Sheryl Van Nunen.
Figure 2.
Urticaria or hives showing pale red, raised, pruritic plaques. Photograph courtesy of Dr Sheryl Van Nunen.
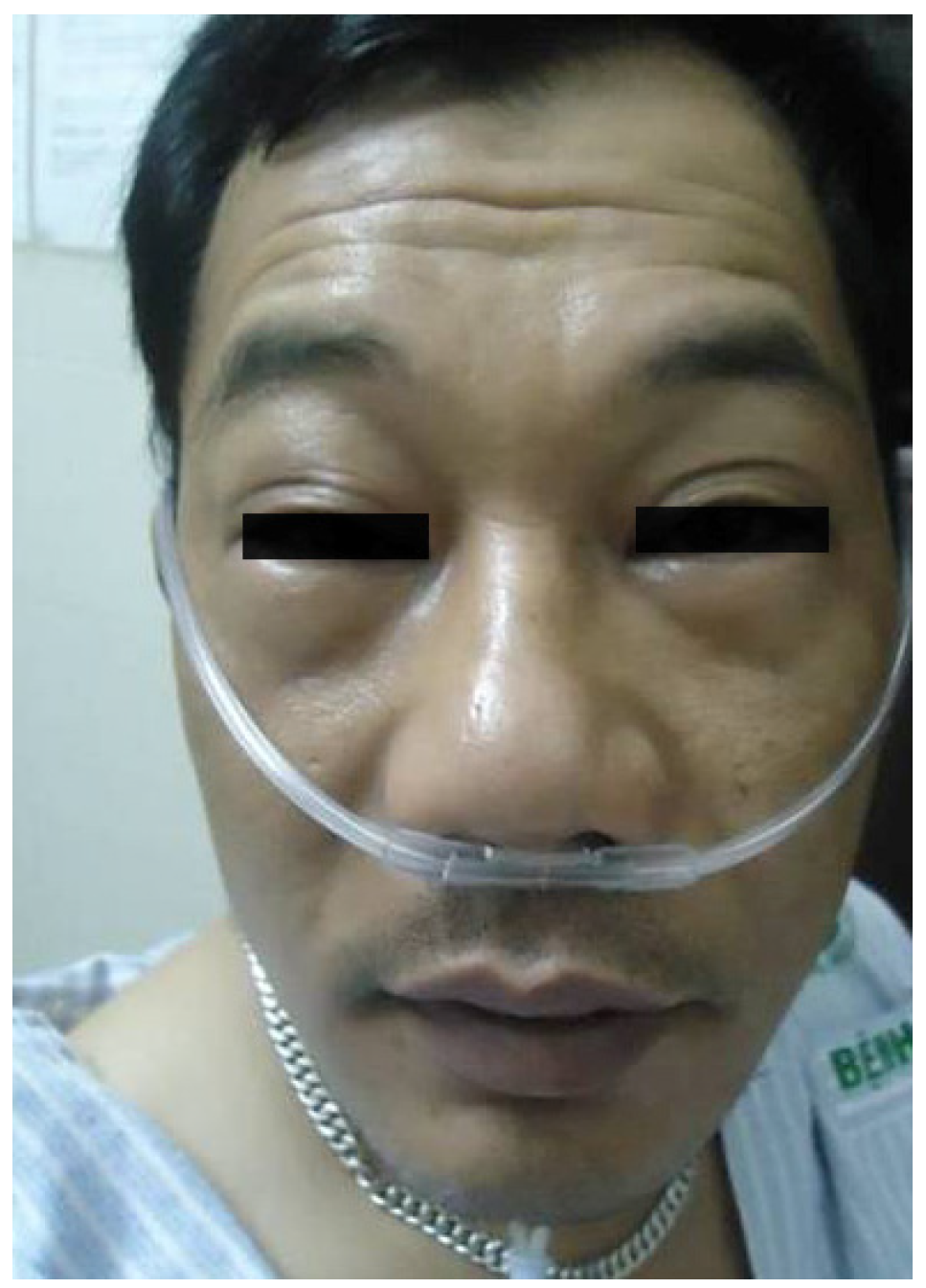
Figure 3.
Angioedema of the face. Although generally non-pruritic, the swelling lasts longer than in urticaria due to fluid accumulation in the tissues. Photograph, courtesy of Dr Sheryl Van Nunen and reproduced with permission of the patient.
Figure 3.
Angioedema of the face. Although generally non-pruritic, the swelling lasts longer than in urticaria due to fluid accumulation in the tissues. Photograph, courtesy of Dr Sheryl Van Nunen and reproduced with permission of the patient.
3. Recognition of β-Lactam Antibiotics by IgE Antibodies
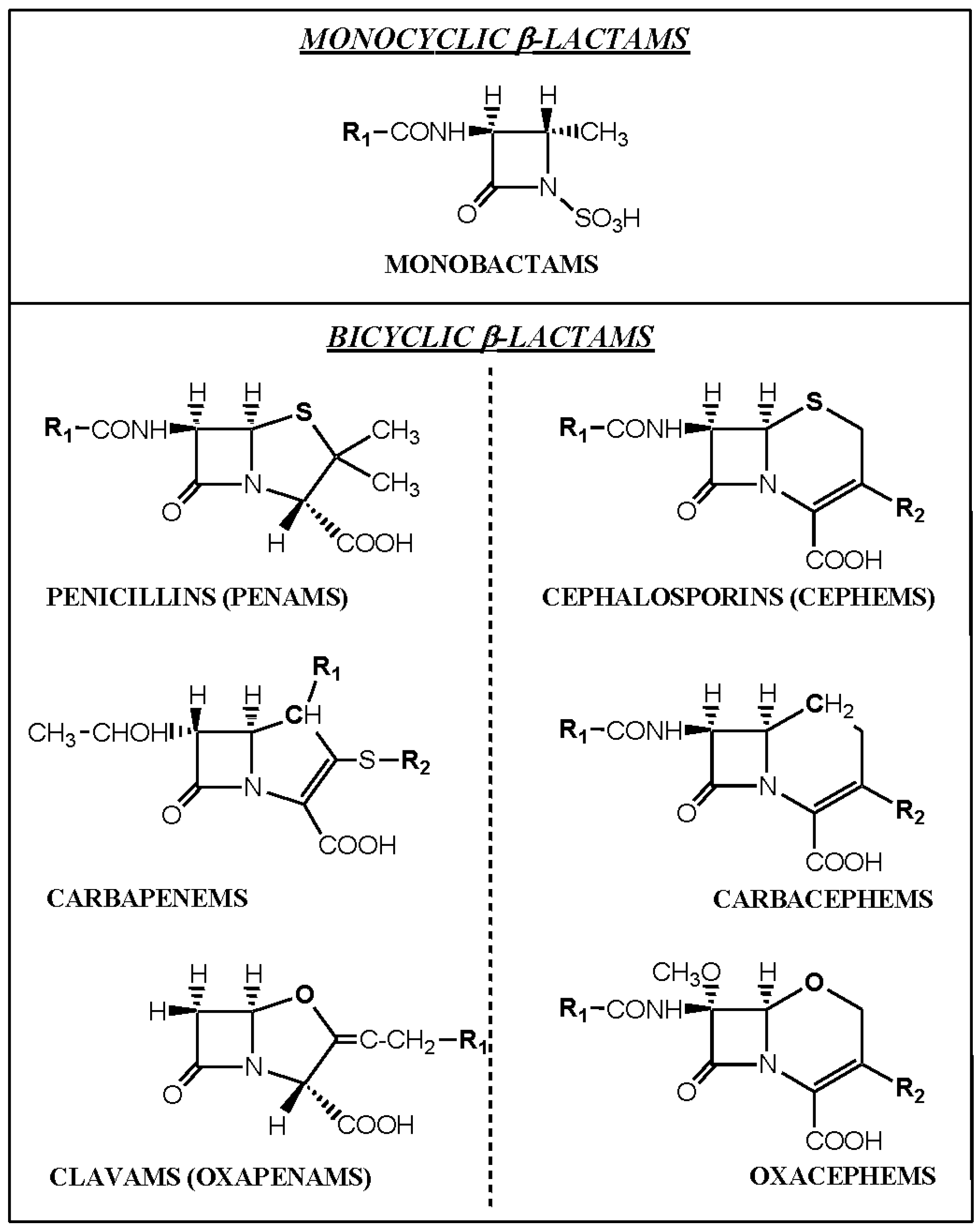
The β-lactam antibiotics include the monobactams (e.g., aztreonam), carbapenems (imipenem, meropenem), clavams (clavulanic acid), carbacephems (loracarbef), and the most important and well known members of the family, the penams (penicillins) and cephems (cephalosporins) (Figure 4). The latter two groups comprise the clinically most important antibiotics used in humans and will henceforth be referred to here as penicillins and cephalosporins. Other less often used antibacterials included in the β-lactam group of drugs are penems which are similar in structure to carbapenems but with a sulfur instead of a carbon at position one of the thiazolidine ring (e.g., faropenem), cephamycins or 7-α-methoxycephalosporins (cefoxitin, cefotetan) and oxacephems or 1-oxacephalosporins (moxalactam) which has an oxygen instead of the sulfur in the cephalosporin nucleus and a 7-α-methoxy group (Figure 4).
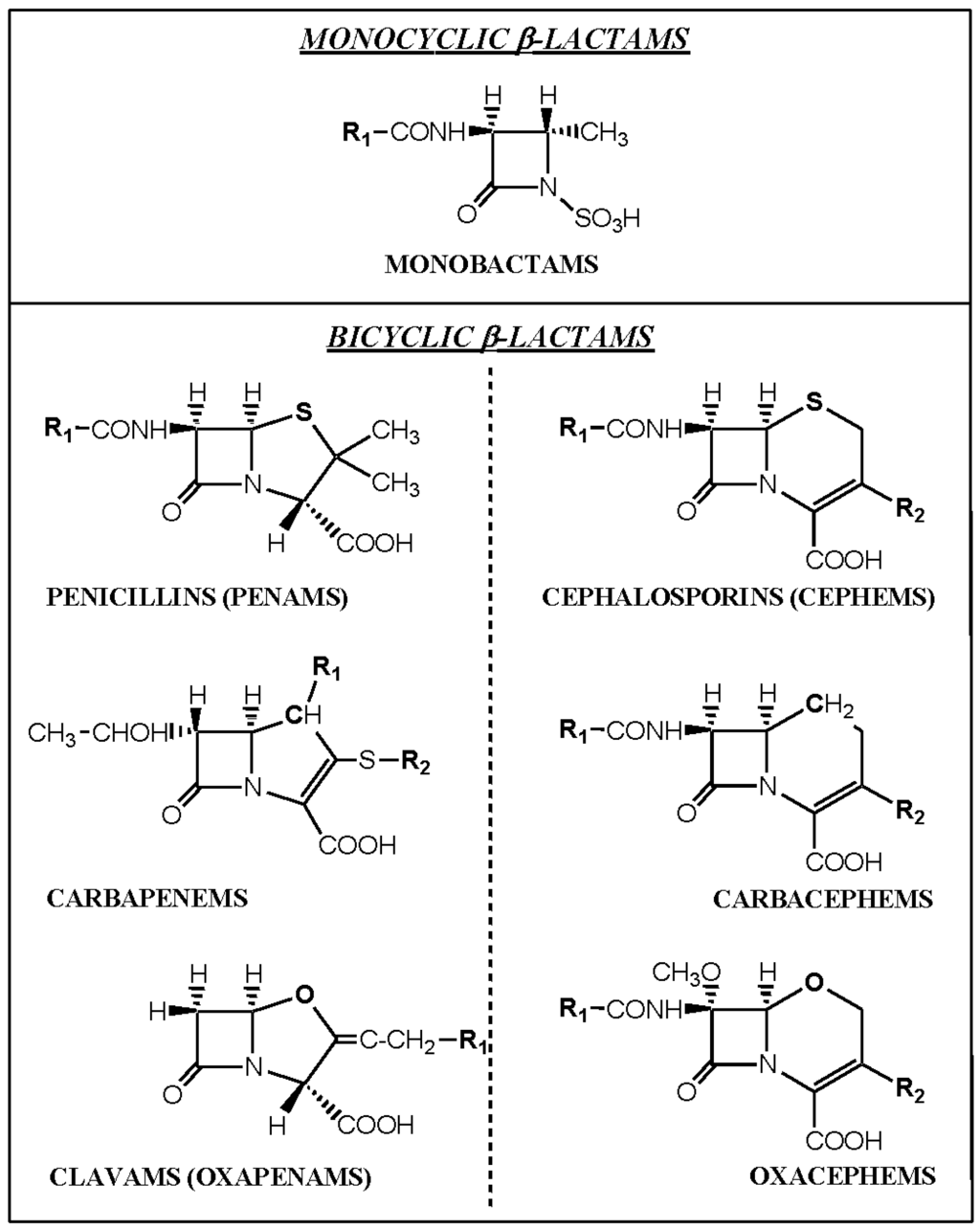
Figure 4.
Structures of the four main groups of β-lactam antibacterials, penams (penicillins), cephems (cephalosporins), monobactams (e.g., aztreonam) and carbapenems (imipenem, meropenem). Other less often used groups of β-lactams are the carbacephems (loracarbef), clavams (clavulanic acid), oxacephems (moxalactam) and (not shown) cephamycins (cefmetazole, cefotetan, cefoxitin).
Figure 4.
Structures of the four main groups of β-lactam antibacterials, penams (penicillins), cephems (cephalosporins), monobactams (e.g., aztreonam) and carbapenems (imipenem, meropenem). Other less often used groups of β-lactams are the carbacephems (loracarbef), clavams (clavulanic acid), oxacephems (moxalactam) and (not shown) cephamycins (cefmetazole, cefotetan, cefoxitin).
3.1. IgE-Mediated Clinical Responses to Penicillins
Penicillins are a well known cause of allergic reactions and the most common cause of drug-induced anaphylaxis, accounting for an estimated 75% of cases of fatal drug-induced anaphylaxis each year in the US. About 10% of patients taking a penicillin report or believe they are allergic to the drug but up to 90% of these patients are not in fact allergic to the medication and the true incidence of hypersensitivity to penicillins is about 1%–2% [5,10,11]. Nevertheless, because of their general lack of toxicity, their efficacy as antibacterials and consequent frequent prescribing, the fact that penicillins can cause all four types of hypersensitivity means that clinicians must remain aware of risk avoidance in their selection of a β-lactam antibiotic. As well as anaphylaxis, immediate type I, or IgE antibody-mediated reactions to penicillins include urticaria (hives) (Figure 2) and angioedema (Figure 3) but types II (cytotoxic), III (immune complex-mediated) and IV (T cell-mediated or delayed) hypersensitivity reactions that are not mediated by IgE also occur. The most commonly seen type II hypersensitivities to penicillins are hemolytic anemia and thrombocytopenia; type III responses include vasculitis and a serum sickness-like reaction; and contact dermatitis, maculopapular exanthema, erythema multiforme and acute generalized erythematous pustulosis are examples of type IV hypersensitivities to the drugs [5].
3.2. Reaction of Penicillins with IgE Antibodies and Identification of Allergenic Determinants
Individual penicillins are distinguished by the composition of the R group side chain (Figure 4). Research over a period of more than 30 years elucidated the pathways for the formation of antigenic and allergenic determinants of benzylpenicillin (penicillin G) including the identification of penicilloyl, penicillenate, penicilloic acid, penamaldate, penicillamine and other determinants of benzylpenicillin [5,12,13,14]. The protein-binding properties of penicillins have been extensively studied. Early findings revealed that most (~95%) of the penicillin molecules that covalently bind protein under physiological conditions form the penicilloyl or so-called ‘major’ determinant. In addition, the main population of antibodies in sera from animals immunized with benzylpenicillin, and from patients following penicillin therapy, has specificity for this determinant. A number of other so-called ‘minor’ determinants [5,13,15] that make up about 5% of administered penicillin have also been implicated as important IgE-binding structures involved in penicillin-induced immediate allergic reactions in some patients.
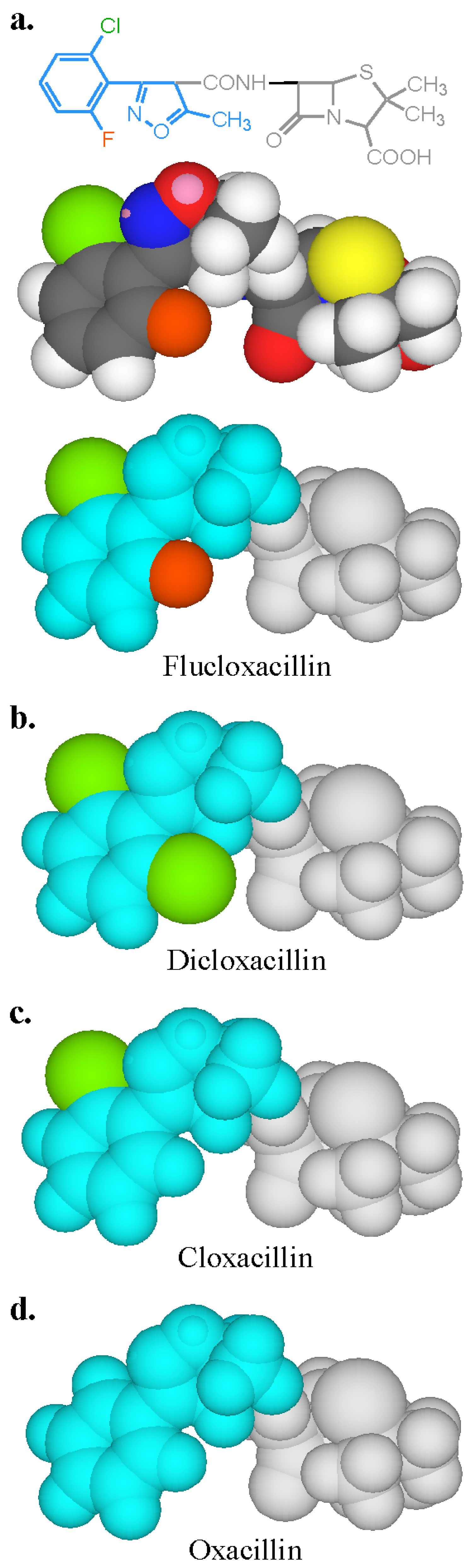
From the perspective of the relevant clinical importance of the penicillin determinants and the corresponding IgE antibody recognition of important penicillin allergenic determinants, the in vitro application of quantitative hapten inhibition methods demonstrated the correlations between fine structural features of determinants and allergic sensitivities in different patients. Examination of the specificities of penicillin-reactive IgE antibodies in the serum from a number of allergic patients revealed a heterogeneous group of allergenic determinants consisting exclusively, or in part, of the side chain groups of penicillins, the β-lactam ring and the thiazoline ring [16,17,18,19,20]. In some patients the entire penicillin molecule comprises the IgE-binding determinant structure. The quantitative immunochemical studies also showed the allergenic importance and ready recognition in vitro of the penicilloyl hapten (Figure 5a) [19], some other structures including the penicillanyl group and the importance of side chain (R) groups not only as allergenic determinant structures but also as the source of cross-reaction between other penicillins (Figure 6) and some cephalosporins (Figure 5b) [19]. Over the years, clinical findings, early experiments on rabbit antibodies to benzylpenicillin and cephalothin and direct binding studies with IgE antibodies in sera of patients allergic to one or other of these drugs, suggested cross-reactivity between the two β-lactams. Employment of benzylpenicilloyl (BPO)- and the so-called cephalosporoyl (CephO)—solid phases in hapten inhibition tests with the two parent drugs, together with molecules representing their side chains viz., phenylacetic acid, benzylamine and 2-thiophene acetic acid; the β-lactam structure without side chain (6-aminopenicillanic acid); and the β-lactam ring structure (azetidinone) indicated that the methylene group in the side chains of each of the two drugs is a dominant feature of the cross-reactive IgE-binding determinant (Figure 5a,b) [19]. The results were similar to earlier quantitative inhibition findings employing an immunoassay for cephalosporin-reactive IgE in the sera of patients allergic to cephalothin [20] and further suggested that as well as the methylene structure, the thiophene ring acts as a bioisostere of the benzene ring in the side chain of benzylpenicillin [19]. Another good example of the preferential recognition of the penicillin R side chain was provided by the demonstration of IgE antibodies complementary to phenylisoxazolyl R substituents in the sera of patients who experienced anaphylaxis following the administration of flucloxacillin [17]. In addition to recognition of flucloxacillin, pronounced cross-reactivity was seen with three structurally related penicillins containing a phenylisoxazolyl side chain, dicloxacillin, cloxacillin and oxacillin. Quantitative inhibition results demonstrated recognition of the 3-(2-chloro-6-fluorophenyl)-5-methyl-4-isoxazolyl group of flucloxacillin and this group, with or without, halogen atoms, was responsible for the clear cross-reactivity between the four different penicillins (Figure 6).
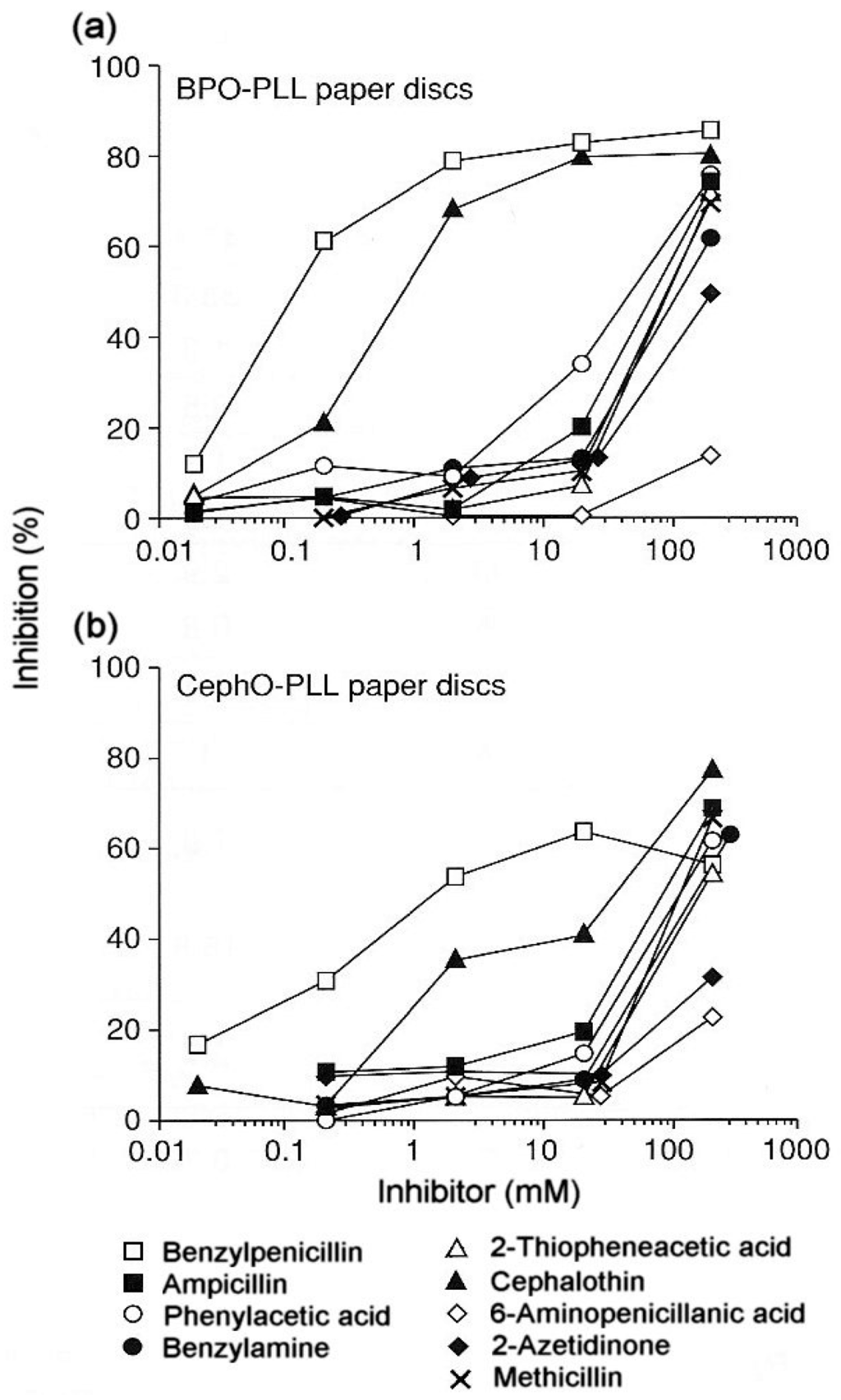
Figure 5.
(a). Demonstration of the reactivity of IgE antibodies in the serum of a patient allergic to penicillins with the benzylpenicilloyl (BPO) determinant and indication of cross-reactivity of the penicillin with cephalothin by the inhibition observed with both drugs. (b). Demonstration that the drug–solid phase conjugate of cephalothin prepared by alkali treatment (as for the preparation of the BPO determinant) retains the capacity to react with the BPO-reactive IgE antibodies (see Section 3.3). As in (a) above, clear inhibition was observed with both benzylpenicillin and cephalothin. Results with some structural analogs, in particular, ampicillin, phenylacetic acid, 2-thiopheneacetic acid and benzylamine indicated that the benzyl side chain of benzylpenicillin and the (2-thienyl)methyl side chain of cephalothin are the cross-reactive determinants with focus on the methylene group in both structures. From Zhao Z et al. [19]. Reprinted with permission from John Wiley and Sons.
Figure 5.
(a). Demonstration of the reactivity of IgE antibodies in the serum of a patient allergic to penicillins with the benzylpenicilloyl (BPO) determinant and indication of cross-reactivity of the penicillin with cephalothin by the inhibition observed with both drugs. (b). Demonstration that the drug–solid phase conjugate of cephalothin prepared by alkali treatment (as for the preparation of the BPO determinant) retains the capacity to react with the BPO-reactive IgE antibodies (see Section 3.3). As in (a) above, clear inhibition was observed with both benzylpenicillin and cephalothin. Results with some structural analogs, in particular, ampicillin, phenylacetic acid, 2-thiopheneacetic acid and benzylamine indicated that the benzyl side chain of benzylpenicillin and the (2-thienyl)methyl side chain of cephalothin are the cross-reactive determinants with focus on the methylene group in both structures. From Zhao Z et al. [19]. Reprinted with permission from John Wiley and Sons.
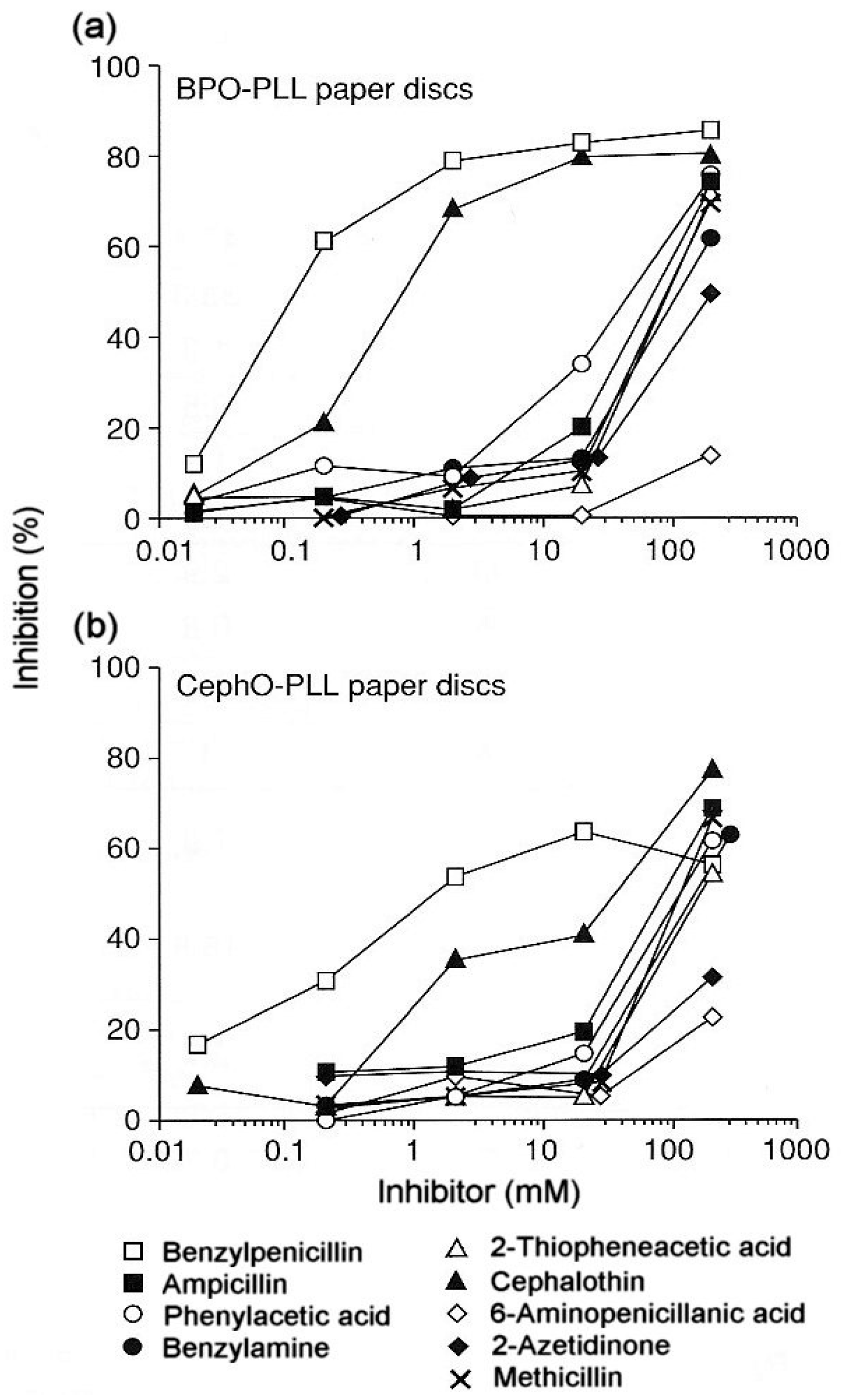
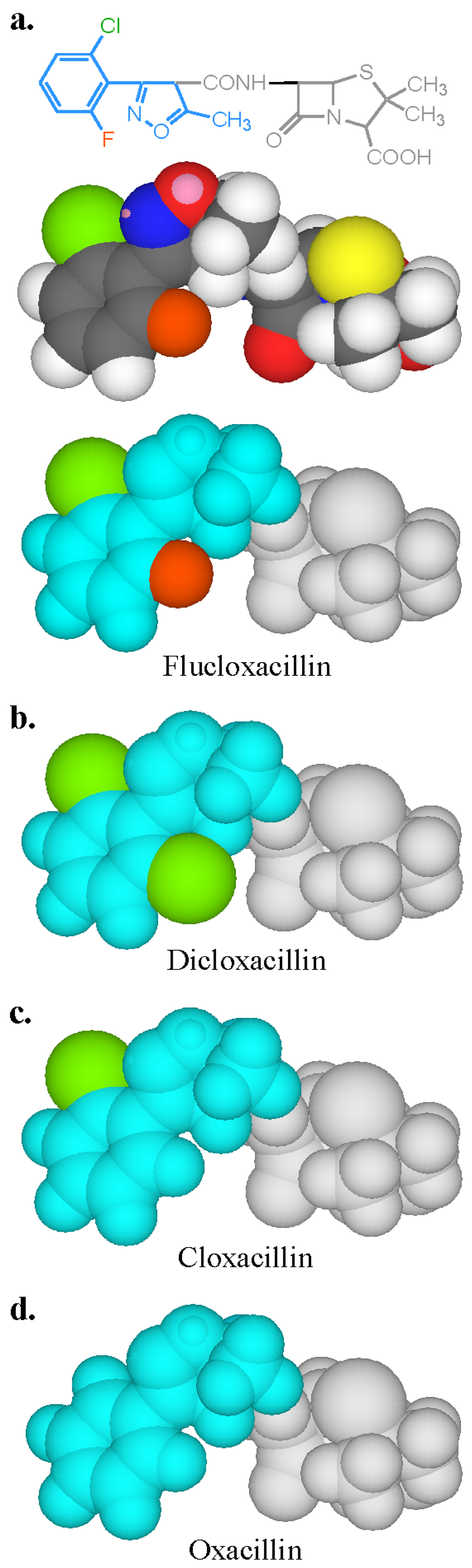
Figure 6.
Two- and three-dimensional structure and models of flucloxacillin and three-dimensional models of dicloxacillin, cloxacillin and oxacillin showing the IgE antibody-binding regions (colored blue, green and orange) on the isoxazolyl penicillins. Chlorine atom is green, fluorine, orange.
Figure 6.
Two- and three-dimensional structure and models of flucloxacillin and three-dimensional models of dicloxacillin, cloxacillin and oxacillin showing the IgE antibody-binding regions (colored blue, green and orange) on the isoxazolyl penicillins. Chlorine atom is green, fluorine, orange.
3.3. Recognition of Cephalosporin R1 and R2 Side Chains by IgE Antibodies
Cephalosporins, which have a six-membered dihydrothiazine ring instead of the five-membered thiazolidine ring of the penicillins, have two side chains—one (R1) attached to the β-lactam ring at position 7 as in the penicillins, and the other (R2), attached at position 3 of the dihydrothiazine ring (Figure 2). Despite the similarity in structure between these two major groups of antibacterials, important differences in chemical reactivity and stability exist. In particular, unlike penicillins, alkali treatment of cephalosporins leads to aminolysis via unstable intermediates that decompose to penaldate and ultimately penamaldate structures [12,21,22], that is, structures comprising only the R1 side chain, the attached amide and parts of the original β-lactam ring remain. By employing a solid phase prepared by alkali treatment of some frequently used cephalosporins, IgE antibodies specific for R1 side chains of some cephalosporins were demonstrated in the sera of patients who experienced immediate allergic reactions, including anaphylaxis, following administration of a cephalosporin (Figure 5b). As with the penicillins, inhibition studies revealed cross-recognition of cephalosporins with the same or structurally related R1 groups. This is demonstrated in Figure 5b where the known cross-reacting benzylpenicillin and cephalothin show clear inhibition of IgE binding to the solid phase prepared by alkali treatment of cephalothin. Because of the lability of the dihydrothiazine ring of cephalosporins and failure to employ and test conjugates that retain an intact R2 side chain, many believe that the R1 side chain group remains the only allergenic structure but employment of drug–solid phase conjugates prepared by linking to the carboxyl group at position 4 using a carbodiimide, revealed clear recognition of intact R2 side chains as well as the R1 group in the sera of some patients with type I hypersensitivities to these drugs [23]. For example, with the cefaclor solid phase linked via position 4 and serum from a cefaclor-allergic patient, inhibition of IgE antibody binding studies showed that the aminobenzyl group at R1 and Cl atom at R2 were required for interaction with the cefaclor-reactive antibodies (group A, Figure 7). Inhibition by cephalothin and cephaloglycin also indicated recognition of the ester group at R2 by a second population of IgE antibodies (group B, Figure 7). Other sera from patients allergic to cephalosporins identified antibodies directed to both side chains but only if the R2 group was ‘small’, for example, a Cl atom or methyl group (group C, Figure 7).
3.4. Summary of IgE Antibody Recognition of the Bicyclic β-Lactams, Penicillins and Cephalosporins
With the aid of molecular models, Figure 8 summarizes similarities and differences in recognition of IgE antibody-binding structures that make up the side chains of some important bicyclic β-lactams. The side chain of penicillins, particularly benzylpenicillin, phenoxymethylpenicillin, ticarcillin, the aminopenicillins ampicillin and amoxicillin and the isoxazolyl penicillins are frequently implicated as allergenic determinant structures by IgE antibodies in the sera of patients allergic to the drugs [5,17]. Note however, in addition to side chain recognition, IgE antibody responses recognize a heterogeneous group of determinants that sometimes include the β-lactam and thiazolidine rings of the penicillin nucleus [16]. The R1 side chains of cephalosporins are the most frequently identified cephalosporin determinants to the extent that some investigators believe that the R2 side chains play no allergenic role. There is, however, evidence both at the laboratory and clinical levels that R2 side chains are in fact allergenic determinants in some allergic patients and these structures interact with complementary IgE antibodies [23]. Interaction of IgE antibodies with the carbacephem, loracarbef, shows a similar pattern of recognition seen with cefaclor where both R1 and R2 structures are identified as allergenic determinants [23].
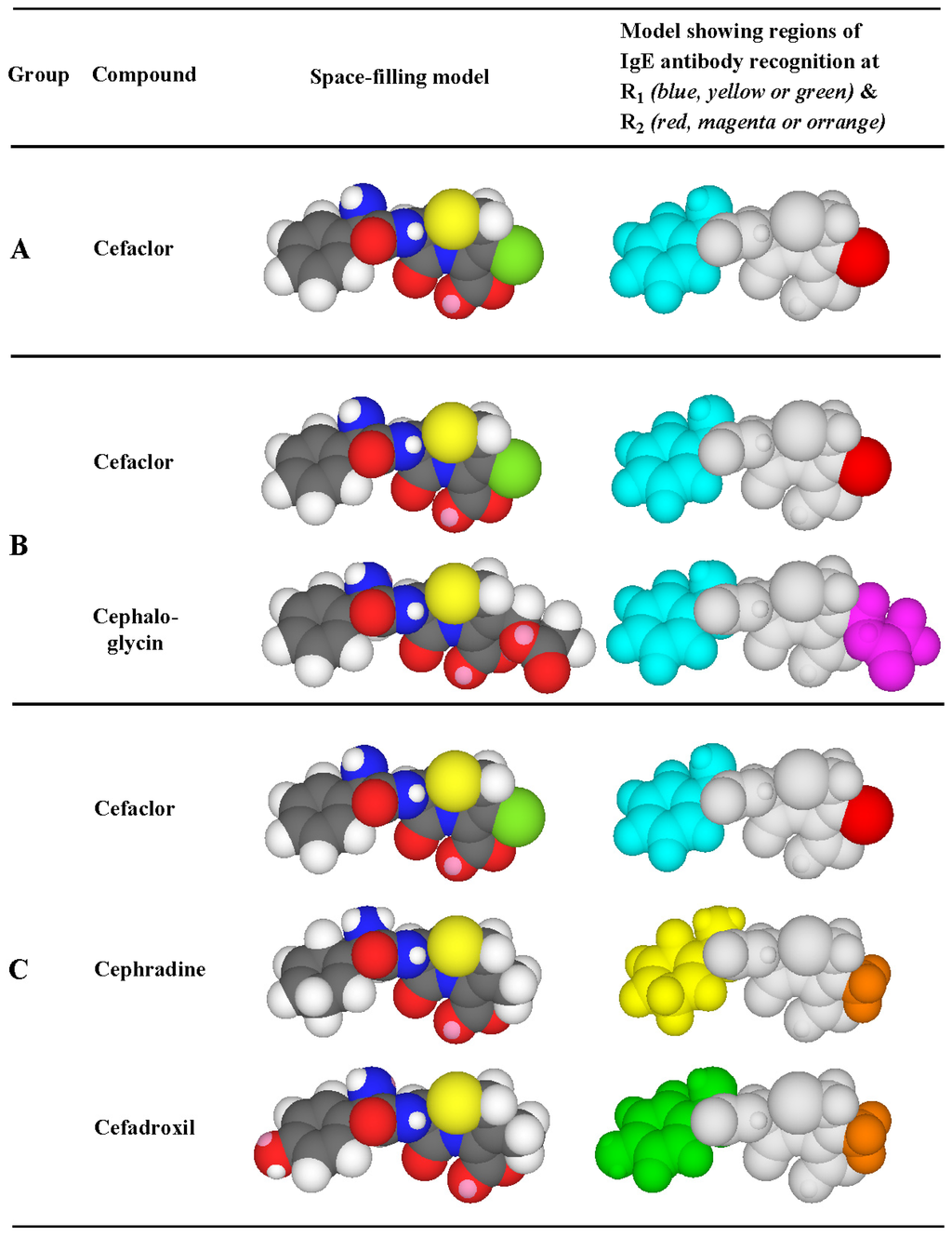
Figure 7.
Three-dimensional Corey, Pauling, Koltun (CPK) space-filling models of cephalosporins highlighting the important side chain structural features recognized by IgE antibodies in the sera of patients allergic to cephalosporins. R1 side chains recognized are shown in blue (aminobenzyl group), yellow (aminocyclohexadienyl group) or green (aminohydroxybenzyl group). R2 side chains recognized are shown in red (Cl atom), magenta (ester group) or orange (methyl group). From Pham NH, Baldo BA [23]. Reprinted with permission from John Wiley and Sons.
Figure 7.
Three-dimensional Corey, Pauling, Koltun (CPK) space-filling models of cephalosporins highlighting the important side chain structural features recognized by IgE antibodies in the sera of patients allergic to cephalosporins. R1 side chains recognized are shown in blue (aminobenzyl group), yellow (aminocyclohexadienyl group) or green (aminohydroxybenzyl group). R2 side chains recognized are shown in red (Cl atom), magenta (ester group) or orange (methyl group). From Pham NH, Baldo BA [23]. Reprinted with permission from John Wiley and Sons.
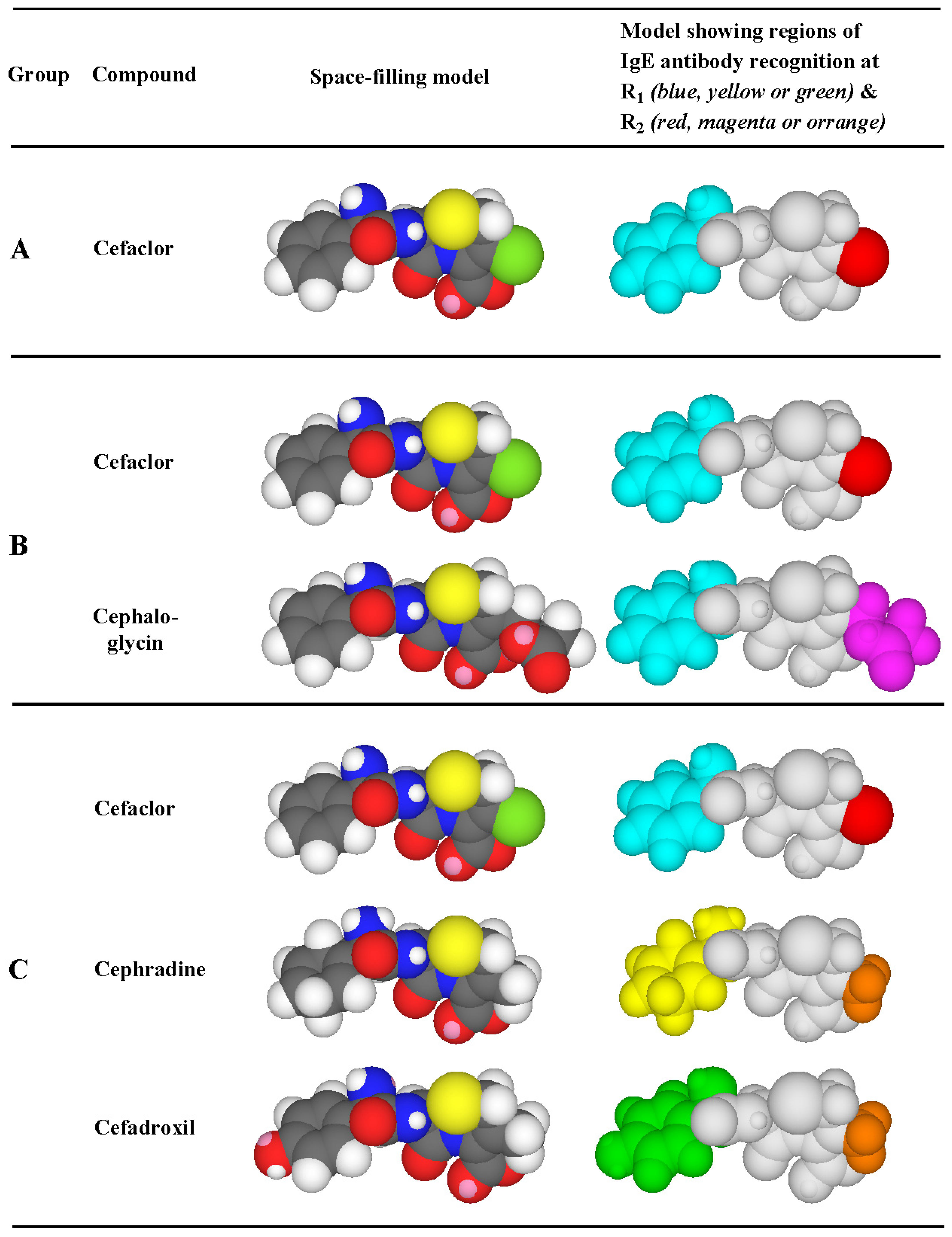
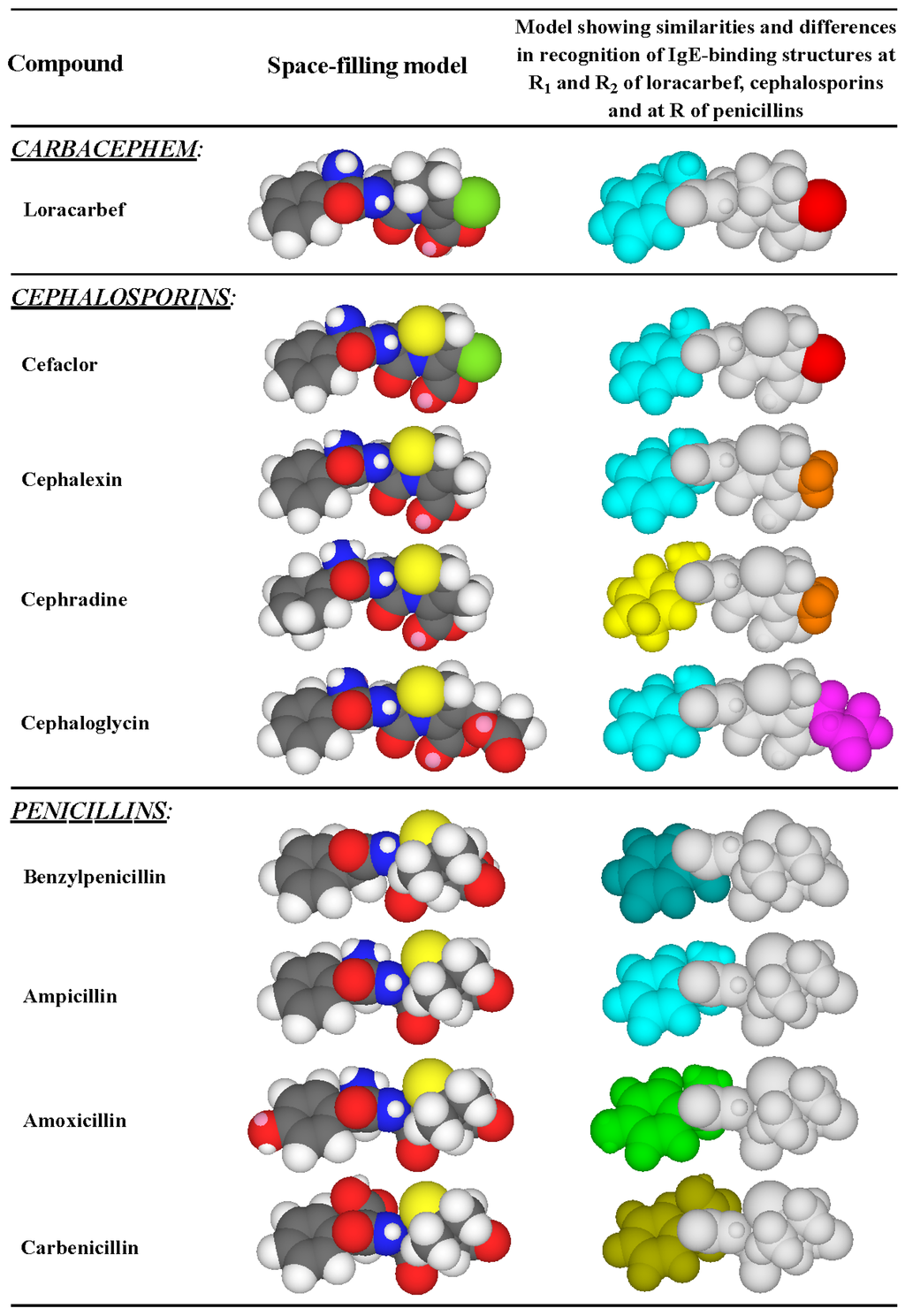
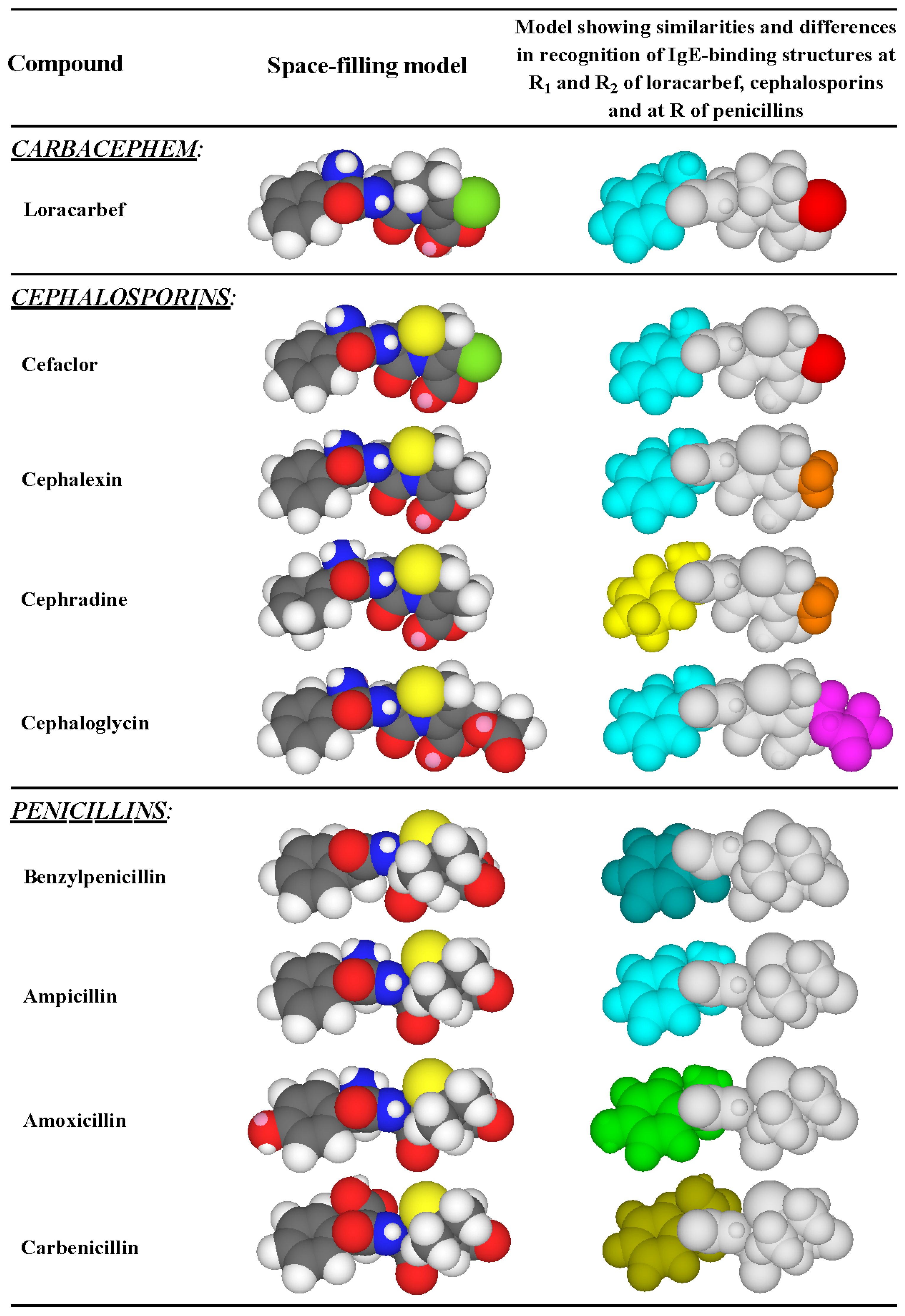
Figure 8.
Side chain reactivity (highlighted in color) of some penicillins, cephalosporins and the carbacephem, loracarbef, with IgE antibodies in the sera of patients allergic to the dicyclic β-lactams. R1 side chains: aminobenzyl group (light blue); aminocyclohexadienyl group (yellow); benzyl group (dark blue); hydroxyaminobenzyl group (green); carboxybenzyl group (yellow-green). R2 side chains: chlorine atom (red); methyl group (orange); ester group -CH2OCOCH3 (magenta).
Figure 8.
Side chain reactivity (highlighted in color) of some penicillins, cephalosporins and the carbacephem, loracarbef, with IgE antibodies in the sera of patients allergic to the dicyclic β-lactams. R1 side chains: aminobenzyl group (light blue); aminocyclohexadienyl group (yellow); benzyl group (dark blue); hydroxyaminobenzyl group (green); carboxybenzyl group (yellow-green). R2 side chains: chlorine atom (red); methyl group (orange); ester group -CH2OCOCH3 (magenta).
4. Immediate Allergic Reactions to Sulfamethoxazole
Sulfonamides used to treat infections, often called ‘sulfa drugs’, are derivatives of p-aminobenzenesulfonamide or sulfanilamide, a structural analog of p-aminobenzoic acid. Structure-activity studies employing a wide range of different sulfonamides together with sera from patients who experienced a type I hypersensitivity reaction to sulfamethoxazole [24] showed that the best recognized compounds by serum IgE antibodies were sulfamethoxazole followed by sulfamerazine and then sulfamethazine. Compounds without a heterocyclic ring at the N1 position, for example, sulfanilamide, were inactive inhibitors of IgE binding to a sulfamethoxazole solid phase (Figure 9) [5,24]. Analysis of the quantitative data led to the conclusion that the fine structural features of the allergenic determinant on sulfamethoxazole is the 5-methyl-3-isoxazolyl group and the methyl substituent is the dominant feature recognized by the IgE antibodies involved in mediating the allergic reaction (Figure 10).
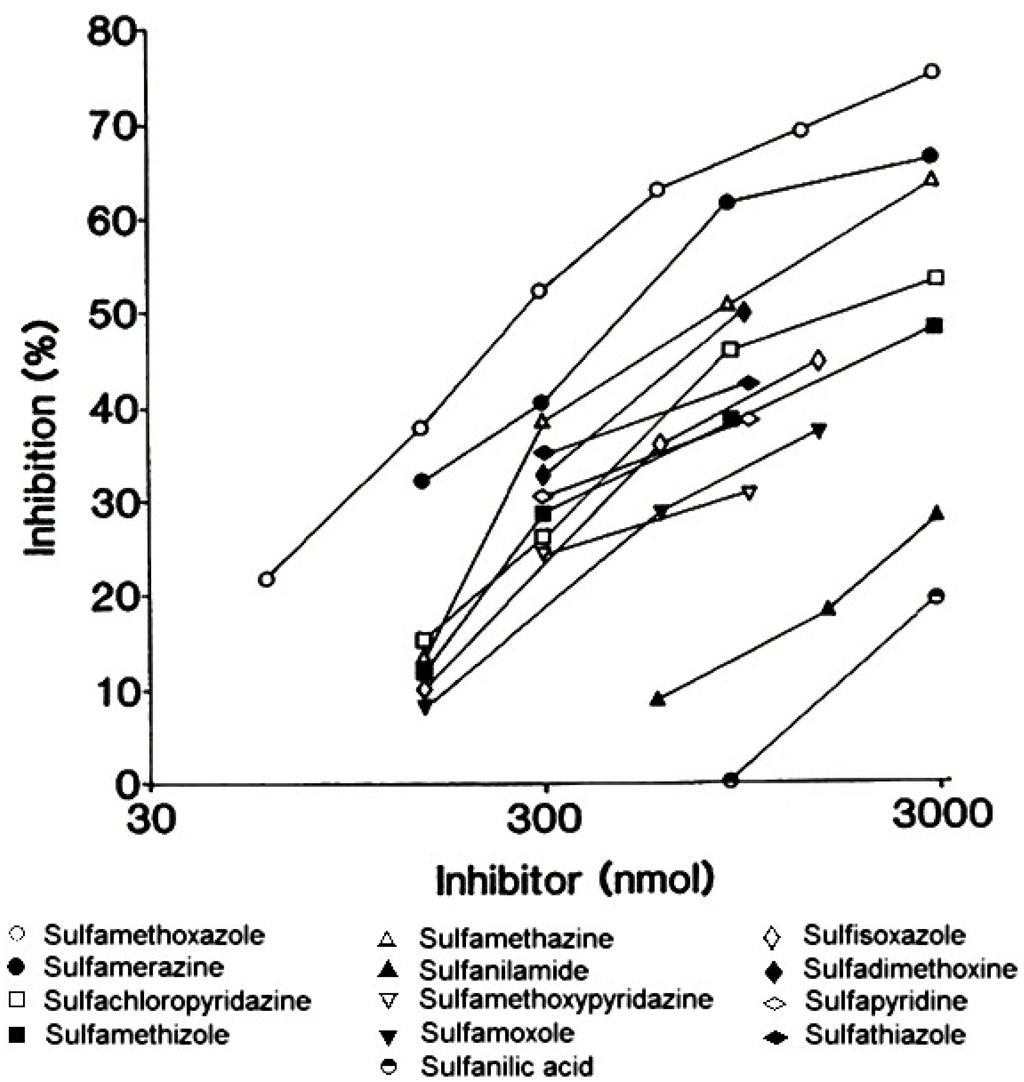
Figure 9.
Quantitative structure-activity comparisons of sulfonamide inhibitors of the binding of IgE antibodies to a sulfamethoxazole solid phase. The most potent inhibitors, that is the structures providing the ‘best fit’ to the antibody combining sites were, in order, sulfamethoxazole, sulfamerazine and sulfamethazine. From Harle DG et al. [24]. Reprinted with permission from Elsevier BV.
Figure 9.
Quantitative structure-activity comparisons of sulfonamide inhibitors of the binding of IgE antibodies to a sulfamethoxazole solid phase. The most potent inhibitors, that is the structures providing the ‘best fit’ to the antibody combining sites were, in order, sulfamethoxazole, sulfamerazine and sulfamethazine. From Harle DG et al. [24]. Reprinted with permission from Elsevier BV.
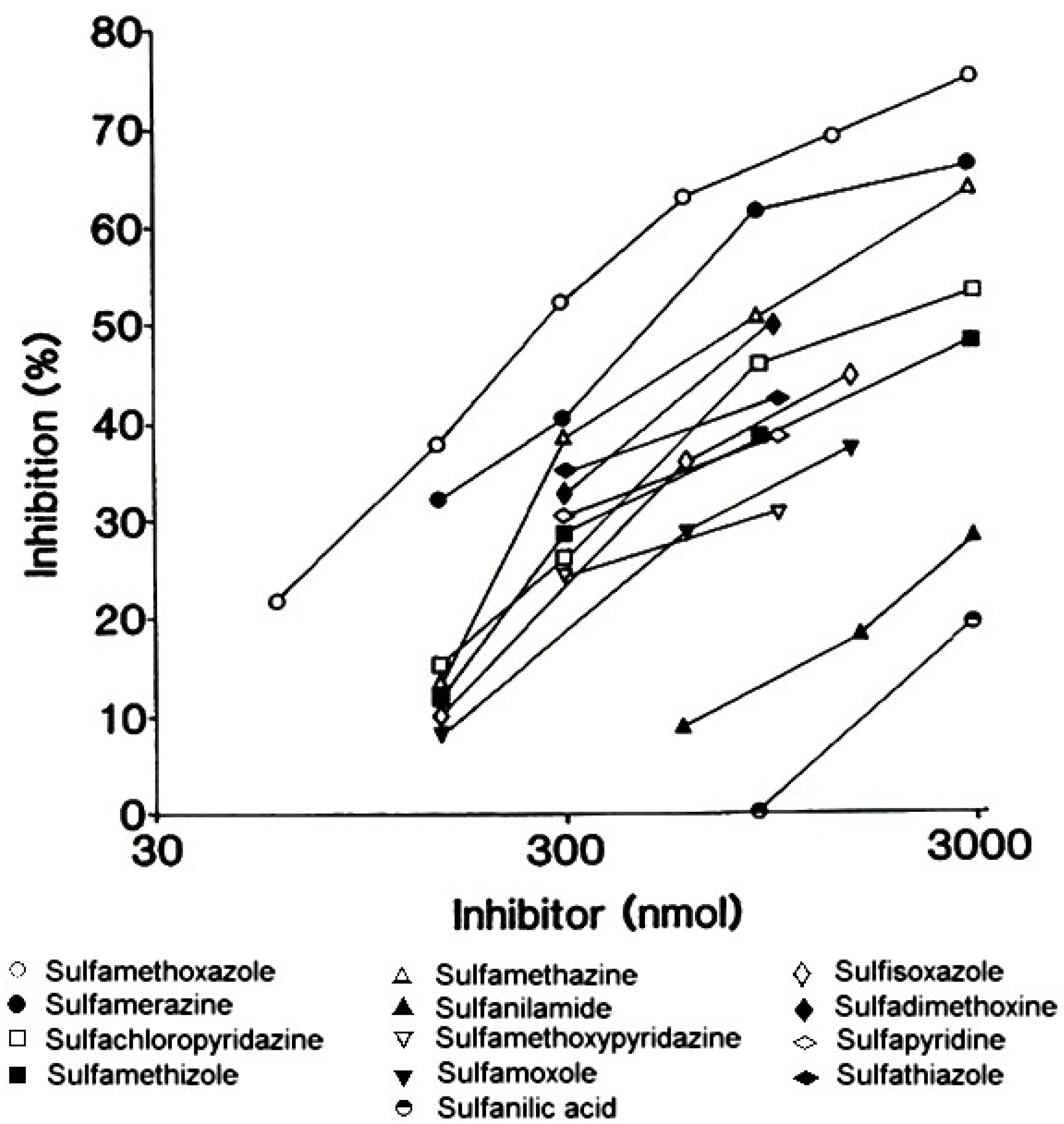
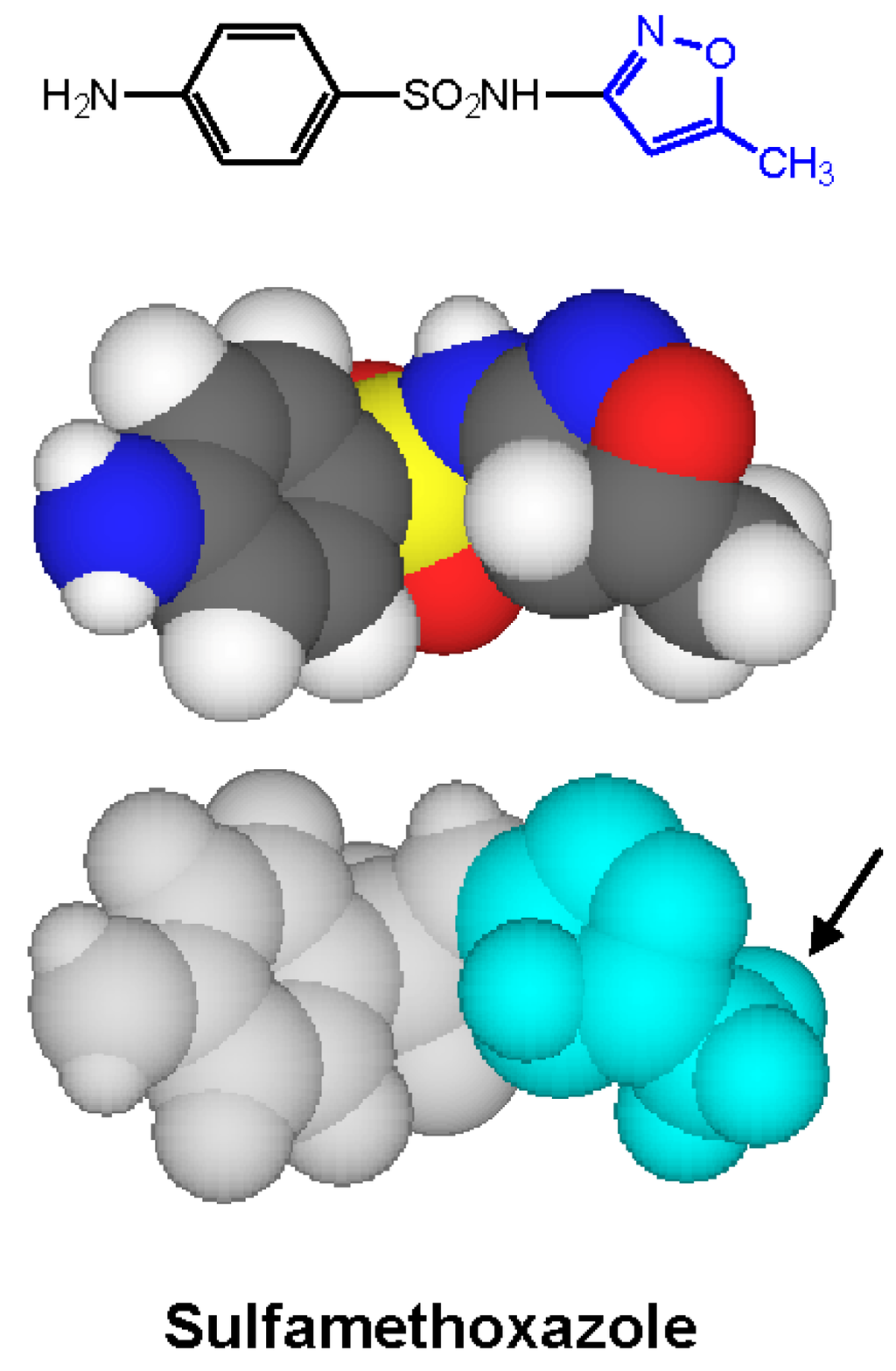
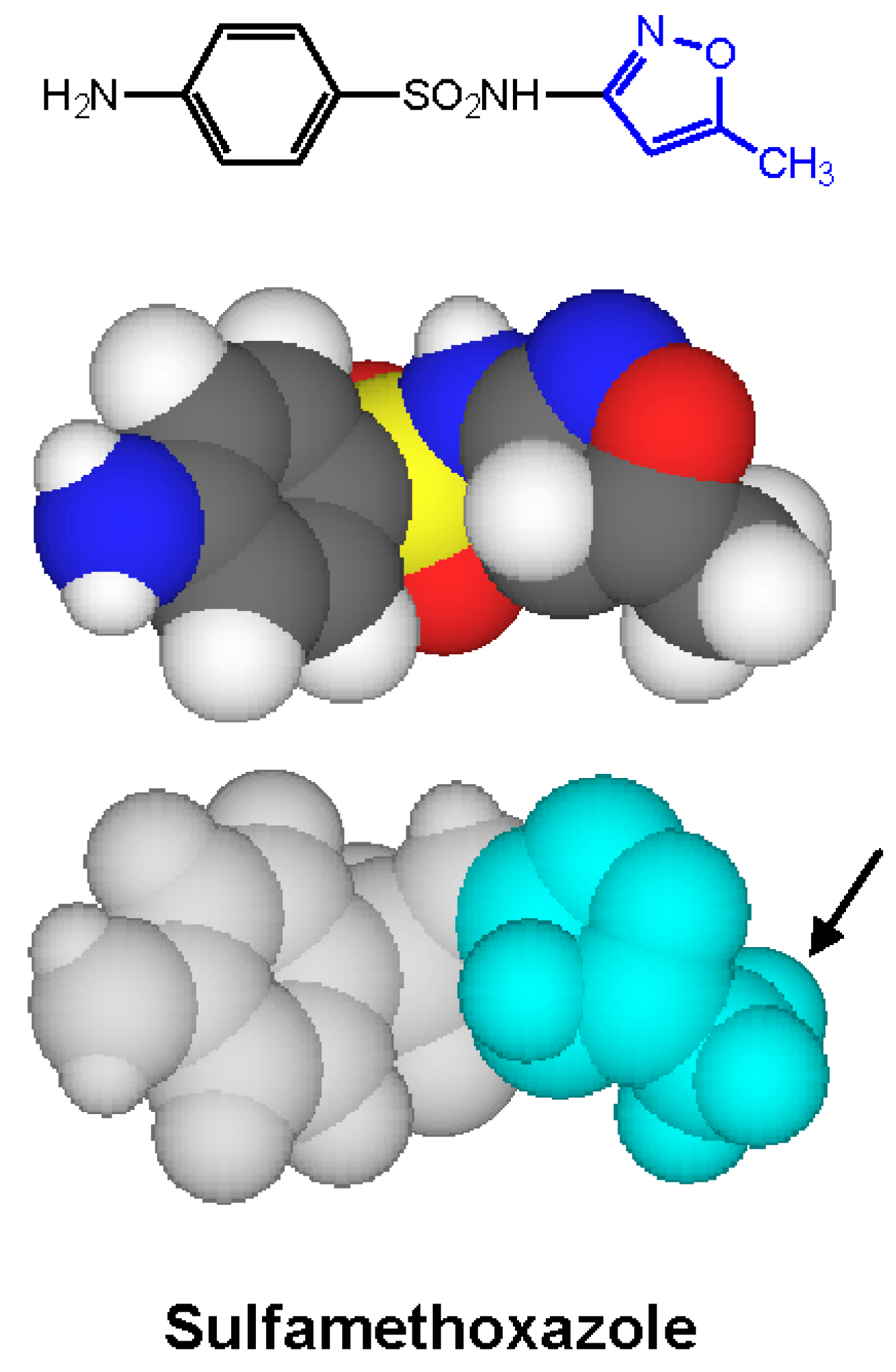
Figure 10.
Two- and three-dimensional structures of sulfamethoxazole with the 5-methyl-3-isoxazolyl IgE-antibody-binding group highlighted (in blue). An important structural feature for recognition by antibody is the methyl substituent (arrowed) β to the point of attachment of the isoxazolyl ring to the N1 nitrogen [24].
Figure 10.
Two- and three-dimensional structures of sulfamethoxazole with the 5-methyl-3-isoxazolyl IgE-antibody-binding group highlighted (in blue). An important structural feature for recognition by antibody is the methyl substituent (arrowed) β to the point of attachment of the isoxazolyl ring to the N1 nitrogen [24].
5. IgE antibody-Mediated Reactions to Trimethoprim
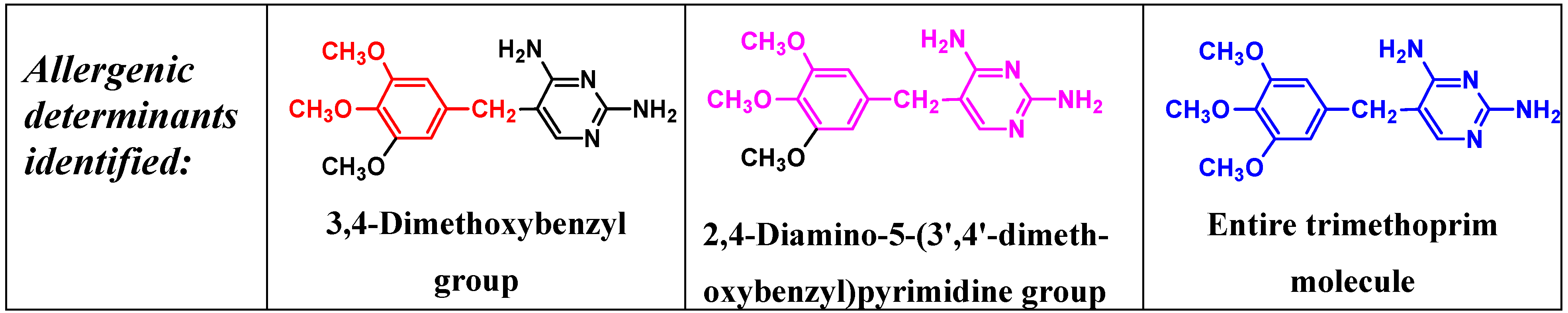
IgE antibodies to the antibacterial trimethoprim (5-(3,4,5-trimethoxybenzyl)pyrimidine-2,4-diamine) were identified over 25 years ago in sera from patients who experienced anaphylaxis to the drug. Antibodies were detected with a drug-solid phase complex prepared by covalently coupling trimethoprim via a spacer arm to Sepharose [25]. The assay has proven to be a useful test to aid the diagnosis of immediate allergic reactions to trimethoprim. The known heterogeneity of the humoral immune response to drugs [5] is well illustrated by studies so far undertaken on identification of the allergenic determinants of trimethoprim where fine structural differences are recognized by the IgE antibodies in sera from different patients [25,26,27]. Even with limited numbers of sera from different allergic patients, three different IgE-binding determinants have so far been identified. Sera were classified into these groups by employing a series of carefully selected structural analogs of trimethoprim side-by-side in quantitative inhibition experiments. Strong inhibition by diaveridine, which differs in structure from trimethoprim by absence of a single methoxy group, 3,4,5-trimethoxycinnamic acid and 3,4-dimethoxyphenylethylamine along with trimethoprim, was observed with some sera (Figure 11) indicating that the structure most complementary to the trimethoprim-reactive IgE antibodies was the 3,4-dimethoxybenzyl group. This structure represents almost one half of the trimethoprim molecule (Figure 12). Reinforcing this conclusion was the almost complete absence of inhibition seen with structures representing the other end of the trimethoprim molecule. For the two other identified groups of sera, potent inhibition only with trimethoprim and diaveridine showed that the determinant recognized by IgE of one of the groups was the 2,4-diamino-5-(3',4'-dimethoxybenzyl)pyrimidine structure while inhibition by trimethoprim alone indicated that these sera had IgE antibodies with combining sites complementary to the entire trimethoprim molecule (Figure 12) [26,27]. It will be interesting to see if examinations of further sera from trimethoprim-allergic patients reveal additional antibody combining site heterogeneity or if the three determinant structures so far identified remain the full extent of the trimethoprim type I hypersensitivity recognition pattern.
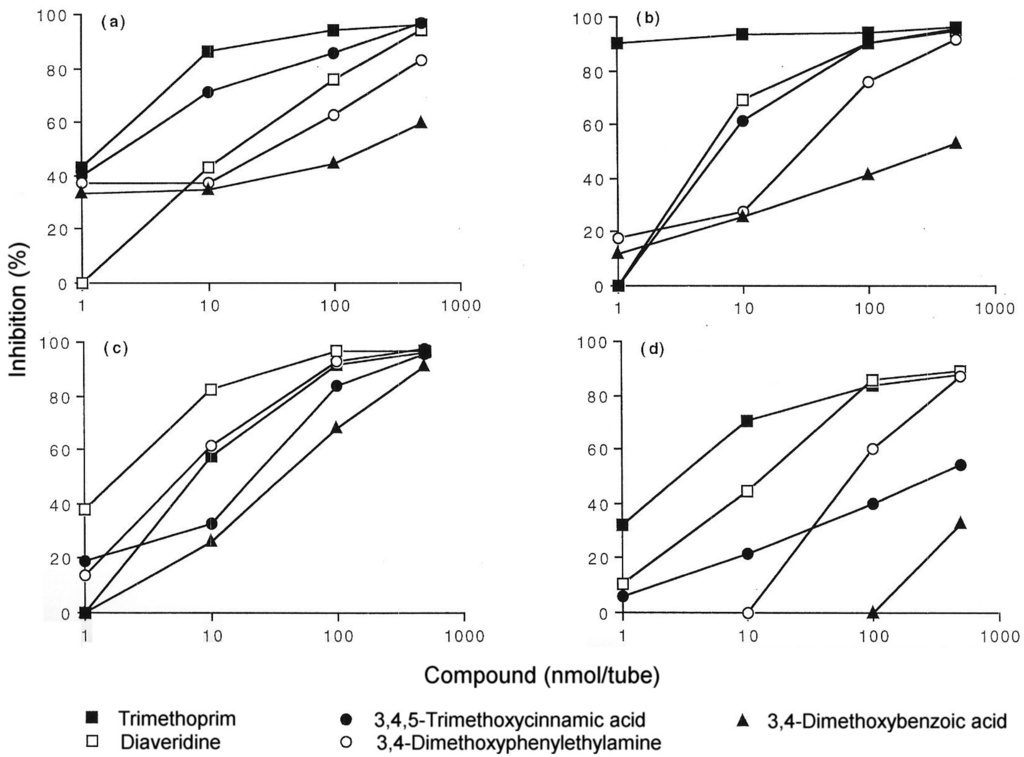
Figure 11.
Concentration-dependent inhibition of IgE binding to trimethoprim-Sepharose complex by trimethoprim and some selected structural analogs. Results shown were obtained with four different sera (a, b, c and d) from patients demonstrating immediate hypersensitivity reactions to the drug. From Pham NH et al. [27]. Reproduced with permission from John Wiley and Sons.
Figure 11.
Concentration-dependent inhibition of IgE binding to trimethoprim-Sepharose complex by trimethoprim and some selected structural analogs. Results shown were obtained with four different sera (a, b, c and d) from patients demonstrating immediate hypersensitivity reactions to the drug. From Pham NH et al. [27]. Reproduced with permission from John Wiley and Sons.
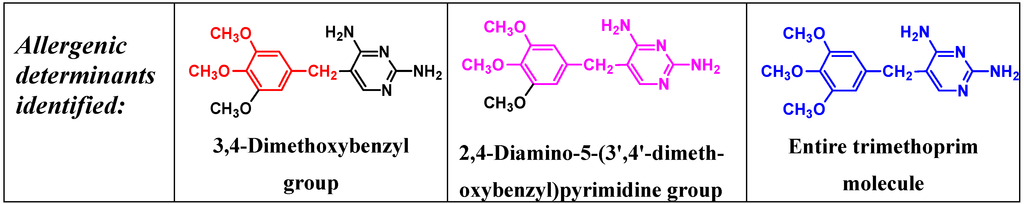
Figure 12.
Three allergenic (IgE antibody-binding) determinants of trimethoprim identified so far in sera from patients showing immediate type I reactions to the drug.
Figure 12.
Three allergenic (IgE antibody-binding) determinants of trimethoprim identified so far in sera from patients showing immediate type I reactions to the drug.
6. Anaphylaxis to Neuromuscular Blocking Drugs
Anaphylaxis to neuromuscular blocking drugs (NMBDs) is the most common cause of anaphylaxis during anesthesia [5,28,29]. Previous exposure to a NMBD is not a prerequisite for a reaction to occur since only about 15%–50% of patients have received the drug beforehand. Atopy and previous anesthesia are not risks but female gender is since up to four times as many females as males experience the reactions [5,28,30].
6.1. IgE Antibodies, NMBDs and Substituted Ammonium Groups
The molecular basis of anaphylaxis provoked by NMBDs is recognition by IgE antibodies of tertiary or quaternary ammonium ions present on all NMBDs [31,32,33,34], and this accounts for the allergenic cross-reactivity seen between all NMBDs. The antibodies do not recognize primary and secondary amino groups. Figure 13 shows the structure of the commonly used NMBD atracurium, a non-depolarizing agent belonging to the tetrahydroisoquinolinium class of NMBDs. All other NMBDs contain at least two tertiary and/or quaternary ammonium ions separated by a sufficient distance, for example, the optimum distance for blockade at the neuromuscular junction, whether depolarizing or not, is 2–2.1 nm seen in the ten-carbon 2 nm length of decamethonium which shows potent activity. The distances between the ammonium ions of NMBDs is sufficient to bridge adjacent mast cell-bound IgE molecules (Figure 1) thus inducing mediator release without the drug needing to be bound to a carrier protein in vivo [5,32,33,34]. Some experimental support for this suggestion has been forthcoming [35].
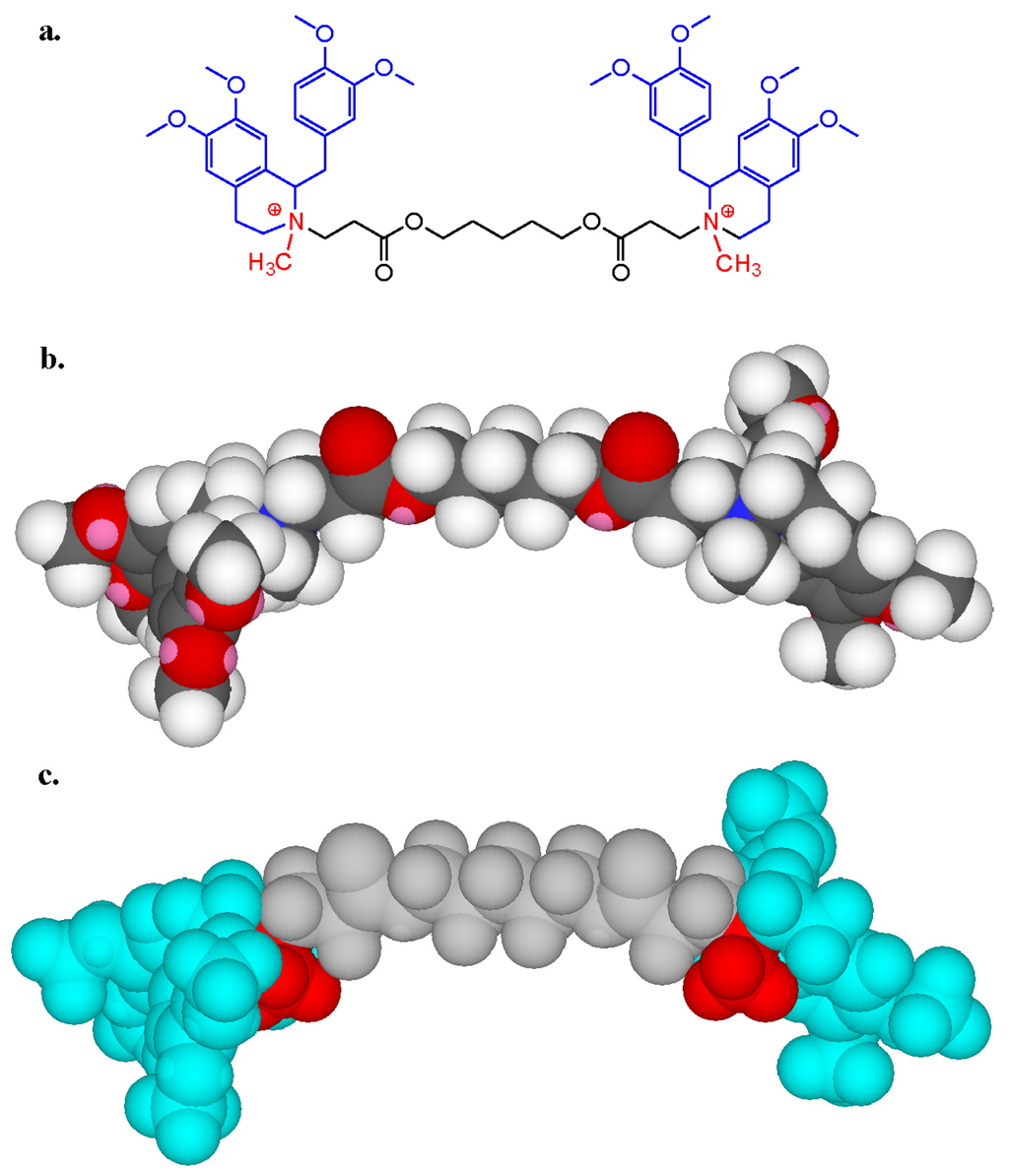
Figure 13.
Two- (a) and three-dimensional (b and c) structures of the non-depolarizing neuromuscular blocking drug (NMBD) atracurium which has two quaternary methylammonium groups (in red in a and c), each forming part of a tetrahydroisoquinoline ring (blue in a and c) and separated by a 13 atom chain (grey in a and c). A 3,4-dimethoxybenzyl substituent (blue in a and c) is attached to each of the tetrahydroisoquinoline rings.
Figure 13.
Two- (a) and three-dimensional (b and c) structures of the non-depolarizing neuromuscular blocking drug (NMBD) atracurium which has two quaternary methylammonium groups (in red in a and c), each forming part of a tetrahydroisoquinoline ring (blue in a and c) and separated by a 13 atom chain (grey in a and c). A 3,4-dimethoxybenzyl substituent (blue in a and c) is attached to each of the tetrahydroisoquinoline rings.
Because tertiary and quaternary ammonium ions occur widely on many drugs and other chemicals, IgE antibodies that react with NMBDs also show a wide spectrum of recognition in vitro of drugs and other agents such as disinfectants, cosmetics and industrial materials commonly encountered in the domestic and workplace environments [32,36]. Drugs that may react with sera from patients allergic to a NMBD are opioids including morphine, codeine and pholcodine; phenothiazine antihistamines such as promethazine and the antipsychotic chlorpromazine; the acetylcholinesterase inhibitor neostigmine; acetylcholine receptor antagonists trimethaphan campsylate and pentolinium; the tetracycline antibiotics; some quinolone antibacterials; a number of different alkaloids including caffeine, cocaine, nicotine and atropine; local anesthetics such as procaine; and many other commonly used pharmacologic agents (Table 1) [5,32,34,36].

Table 1.
Examples of some compounds containing substituted ammonium groups that react with NMBD-reactive IgE antibodies in the sera of patients who experienced an anaphylactic reaction following the administration of a NMBD during anesthesia. Ammonium groups are highlighted. R = alkyl.
| Trialkylamines | NR3 |
| Tetraalkylammonium salts | N+R4 , RN+(Rי)3 |
| Choline | (CH3) 3N+CH2CH2OH |
| Acetylcholine | (CH3) 3N+CH2CH2OCOCH3 |
| Promethazine | 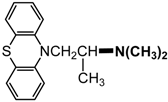 |
| Neostigmine | 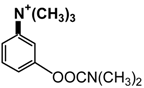 |
| Morphine | 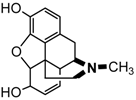 |
6.2. In Vitro Tests to Help Confirm Diagnosis of Anaphylaxis to NMBDs and Cross-Reactivity of NMBDs
Although the NMBDs alcuronium and d-tubocurarine coupled directly to solid supports were used in the initial radioimmunoassay demonstrations of the presence of IgE antibodies to these drugs [31,32,37], other NMBDs such as succinylcholine, decamethonium and gallamine contain no suitable functional groups that can be used to effect direct coupling of drug to the solid phase. To overcome this restriction, analogs containing identical terminal structures as the NMBDs but also groups amenable for conjugation to a suitable insoluble phase were utilized. Thus, choline can be employed for preparation of a drug–solid phase to detect IgE antibodies to succinylcholine and decamethonium [38] since both NMBDs contain the same terminal four (succinylcholine) or three (decamethonium) groups, including the quaternary nitrogen, that occur in choline. The same strategy of using the identical terminal group of triethylcholine to mimic the terminal quaternary triethylammonium groups of gallamine was also employed to develop a diagnostic test for the detection of gallamine-reactive IgE antibodies in the sera of patients allergic to the NMBD [38]. Figure 14 and Figure 15 show typical quantitative inhibition results obtained when six different NMBDs were assessed for their reactivities and inhibition potencies in immunoassay studies when the choline solid phase was employed with serum from a patient allergic to succinylcholine and when the triethylcholine support was used with serum from a gallamine-allergic patient, respectively. Due to the presence of substituted ammonium ions in each of the NMBDs, all the NMBDs cross-reacted, shown by inhibition to greater or lesser extent of the binding of IgE antibodies. This is demonstrated in the comparative quantitative inhibitory results set out in Table 2 and Table 3 where, as expected, the di- and trimethylammonium groups of d-tubocurarine and succinylcholine and decamethonium, respectively, were good inhibitors of IgE from patients who experienced an anaphylactic reaction to succinylcholine (Figure 14, Table 2), and gallamine was the most potent inhibitor of the IgE antibodies from a gallamine-allergic patient (Figure 15, Table 3). In both of these studies, alcuronium proved the poorest inhibitor and this is what would be expected given the structural difference of the ammonium groups (allylammonium) on this compound.
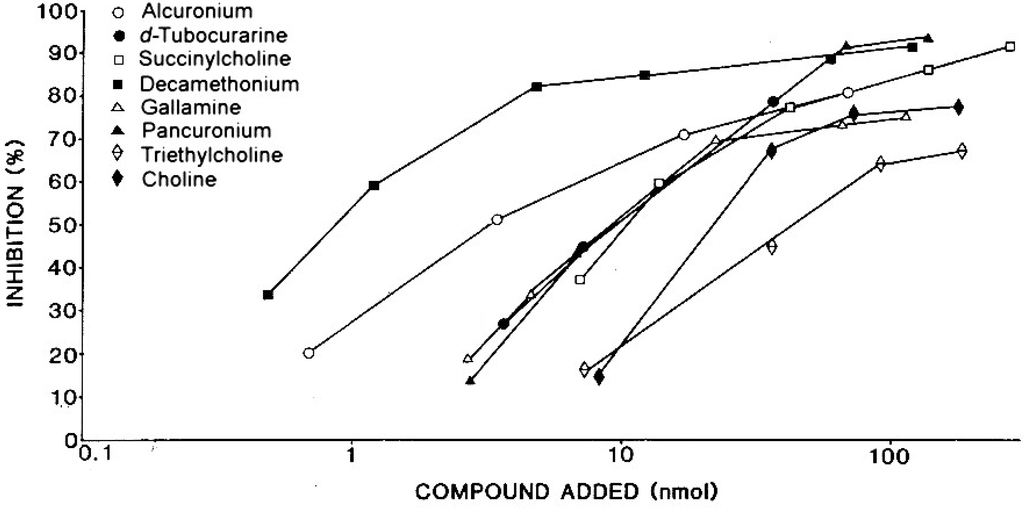
Figure 14.
Demonstration of recognition of NMBDs by IgE antibodies in serum from a patient allergic to succinylcholine and dose-dependent inhibition results showing cross-reactions between the drugs using a choline-Sepharose covalent complex as solid phase in the immunoassay. See also Table 2. From Harle DG et al. [38]. Reproduced with permission from Elsevier Limited.
Figure 14.
Demonstration of recognition of NMBDs by IgE antibodies in serum from a patient allergic to succinylcholine and dose-dependent inhibition results showing cross-reactions between the drugs using a choline-Sepharose covalent complex as solid phase in the immunoassay. See also Table 2. From Harle DG et al. [38]. Reproduced with permission from Elsevier Limited.
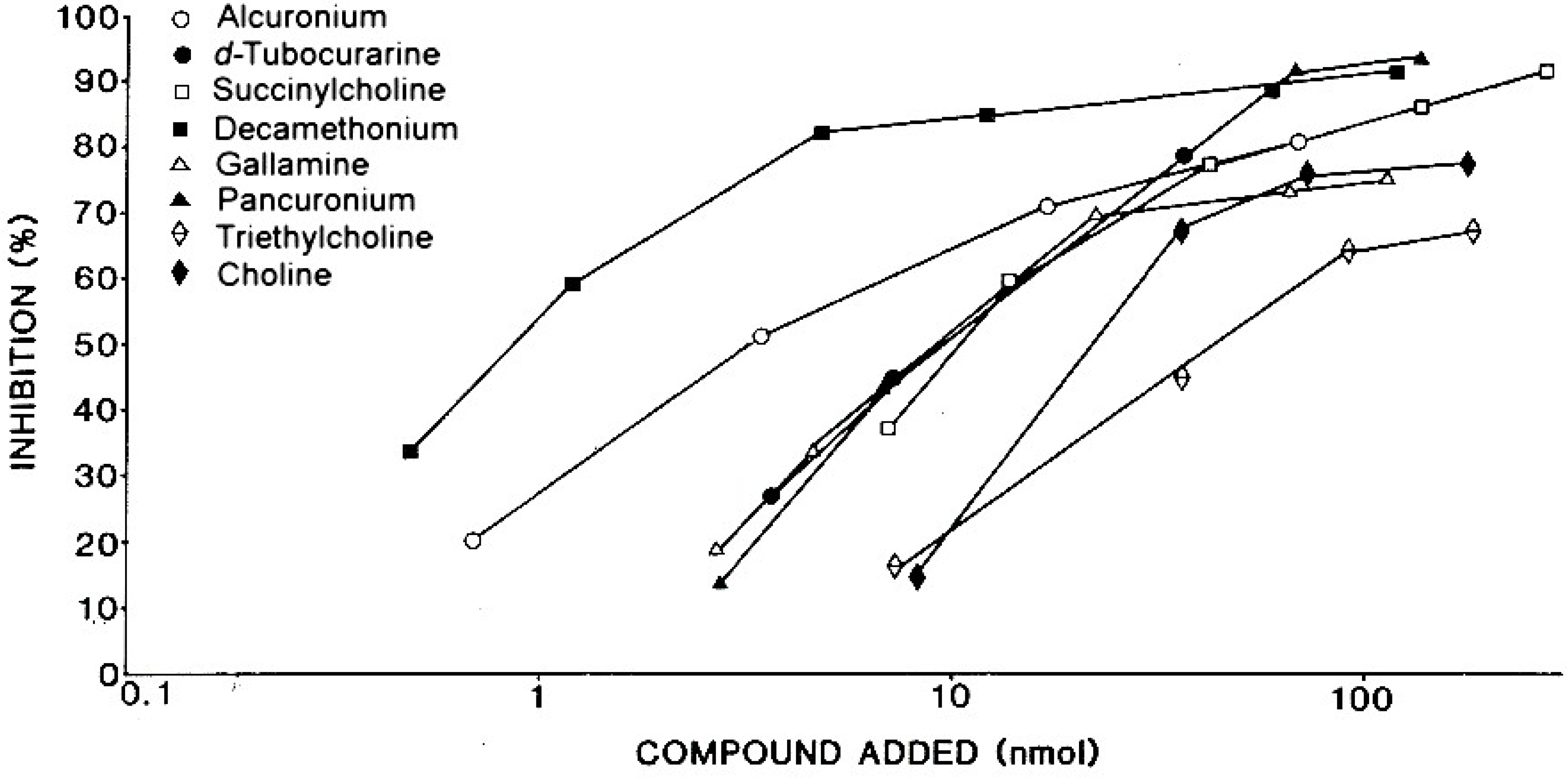

Table 2.
Cross-reactivity between neuromuscular blocking drugs (NMBDs). Inhibition by different NMBDs of IgE antibodies to succinylcholine.
| Patient’s Serum 3 | Amount of drug 1 for 50% inhibition 2 | |||||
|---|---|---|---|---|---|---|
| Succinylcholine | Decamethonium | d-Tubocurarine | Pancuronium | Gallamine | Alcuronium | |
| 1 | 4.7 | 0.5 | 1.3 | 17.0 | 1.6 | 88.0 |
| 2 | 6.0 | 0.9 | 1.8 | 4.7 | 6.8 | 37.0 |
| 3 | 9.8 | 1.9 | 0.7 | 7.7 | 8.4 | 58.0 |
| 4 | 7.3 | 1.3 | 10.0 | 14.0 | 6.9 | 41.0 |
| 5 | 11.0 | 0.8 | 9.7 | 9.8 | 9.6 | 3.3 |
1 nmol per tube. 2 Inhibition of binding of IgE antibodies to a choline solid phase complex. 3 Sera from patients who experienced an anaphylactic reaction to a NMBD during anesthesia. Refer to Figure 14.
Figure 15.
Demonstration of recognition of NMBDs by IgE antibodies in serum from a patient allergic to gallamine and dose-dependent inhibition results showing strong inhibition by gallamine and cross-reactions between the drugs using a triethylcholine-Sepharose covalent complex as solid phase in the immunoassay. Refer also to Table 3.
Figure 15.
Demonstration of recognition of NMBDs by IgE antibodies in serum from a patient allergic to gallamine and dose-dependent inhibition results showing strong inhibition by gallamine and cross-reactions between the drugs using a triethylcholine-Sepharose covalent complex as solid phase in the immunoassay. Refer also to Table 3.
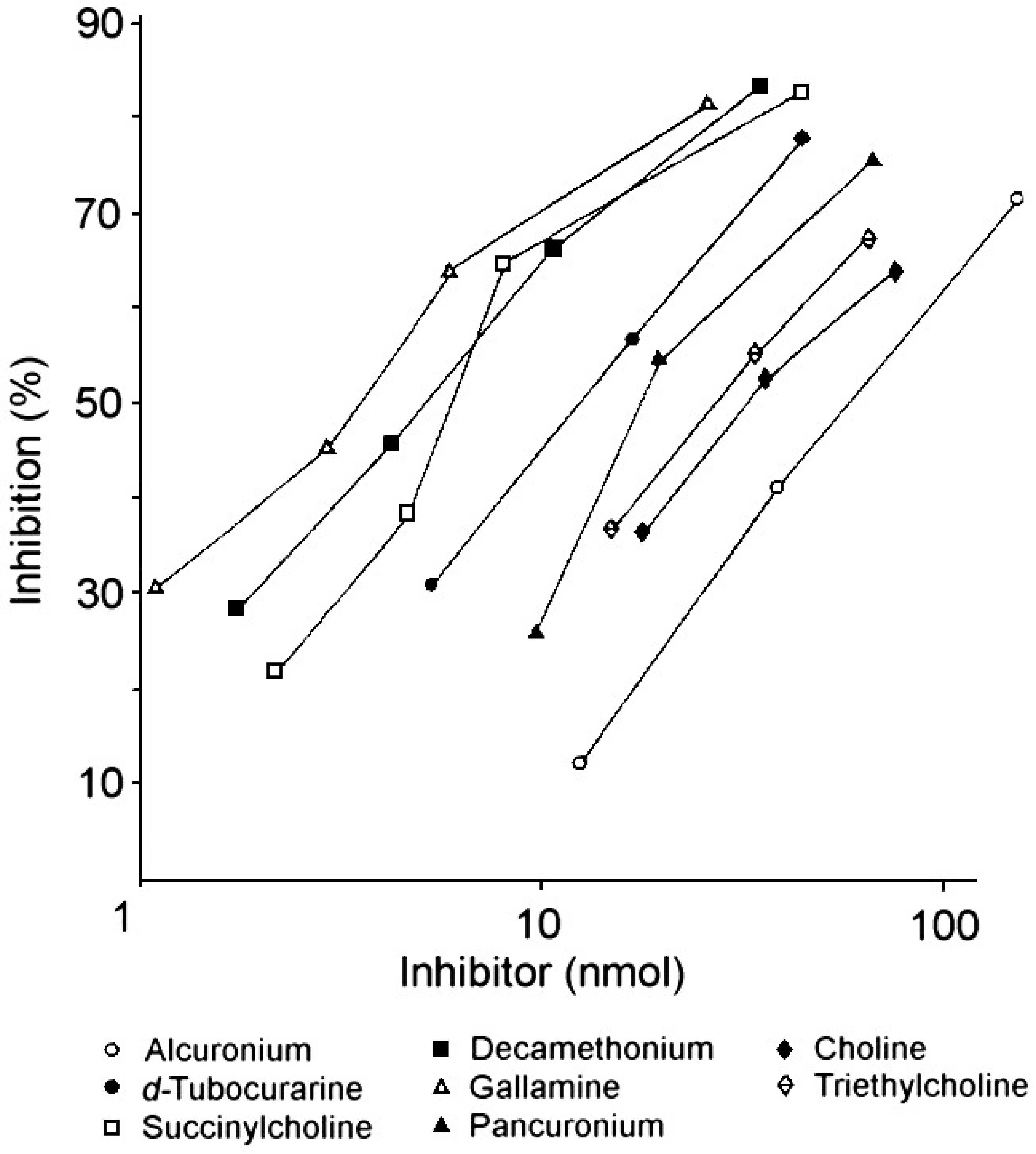

Table 3.
Demonstration of specificity and allergenic cross-reactivity between NMBDs demonstrated by IgE antibodies to gallamine.
| Drug or analog | Amount (nmol) for 50% inhibition 1 of IgE 2 binding |
|---|---|
| Gallamine | 4.2 |
| Succinylcholine | 8.1 |
| Decamethonium | 7.3 |
| d-Tubocurarine | 12.0 |
| Pancuronium | 14.0 |
| Alcuronium | 74.0 |
| Choline | 27.0 |
| Triethylcholine | 22.0 |
1 Inhibition of IgE antibody binding to a triethylcholine solid phase complex. 2 NMBD-reactive IgE antibodies in the serum of a subject allergic to gallamine. Refer to Figure 15.
Although succinylcholine is still widely used in anesthesia, newer NMBDs, in particular, the aminosteroids pancuronium, vecuronium and rocuronium (Figure 16) and tetrahydroisoquinolines such as atracurium (Figure 13), have now largely taken over the market. Following a number of reports of anaphylaxis upon administration of rocuronium [39], an immunoassay to detect IgE antibodies to this NMBD has been introduced [40]. Some investigators believe that rocuronium presents an increased risk of anaphylaxis [41,42,43] but others dispute this, claiming the apparent increase in allergic reactions is due to the drug's greater market share [44,45]. An immunoassay for the detection of IgE antibodies to atracurium has recently been examined using sera from a few patients but more studies with the test are needed before its value and reliability can begin to be properly assessed [46].
Because of morphine's capacity to cross-react with NMBDs via its tertiary ammonium group, it has found wide application as a test for the detection of NMBD-reactive IgE antibodies [5,32,34]. A commercially available assay has been introduced for this purpose.
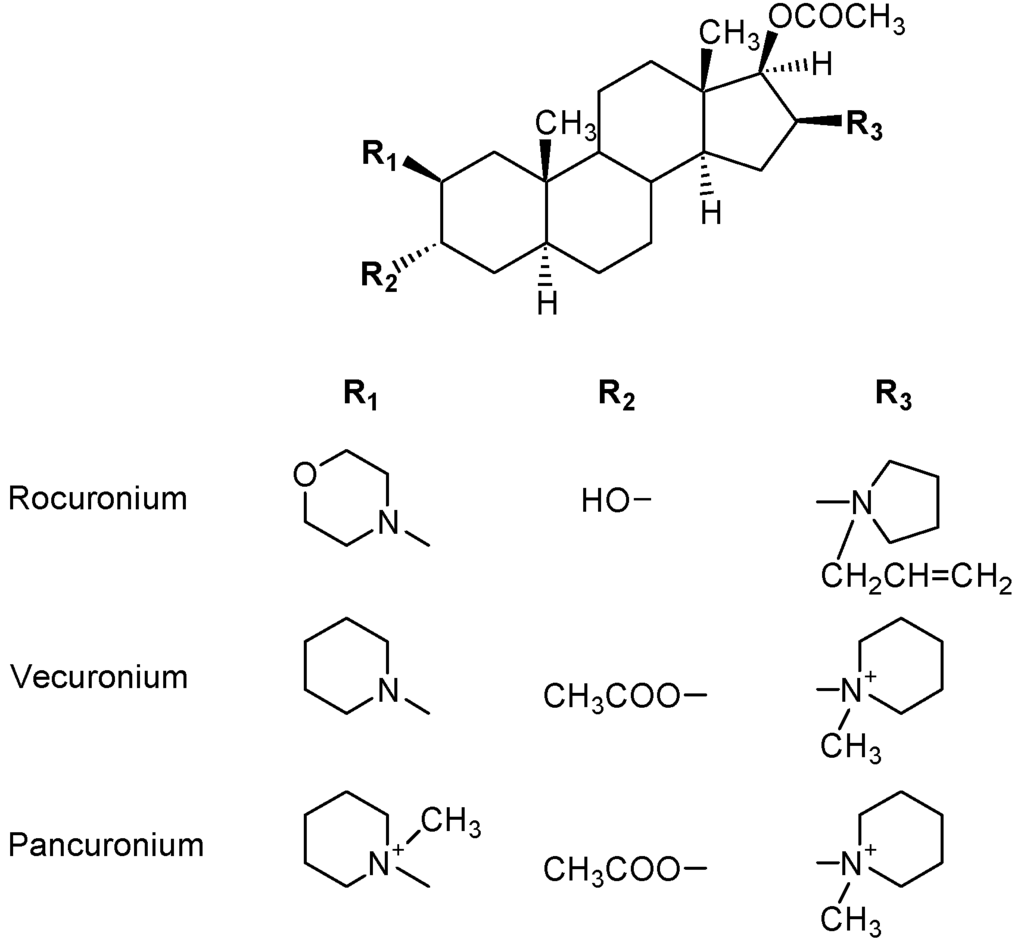
Figure 16.
Structures of the non-depolarizing and competitive aminosteroid NMBDs rocuronium, vecuronium and pancuronium. From Baldo B.A. et al. [39]. Reproduced with permission from Bentham Science Publishers.
Figure 16.
Structures of the non-depolarizing and competitive aminosteroid NMBDs rocuronium, vecuronium and pancuronium. From Baldo B.A. et al. [39]. Reproduced with permission from Bentham Science Publishers.
7. Recognition of Polyamines and PrimaryAmines by IgE Antibodies
The discovery of IgE antibodies with apparent specificity for poly-L-lysine (PLL) in some sera screened for suspected drug allergies [47] indicated that primary, as well as tertiary and quaternary amines, can be recognized by IgE immunoglobulin known for its central role in immediate, type I allergic reactions. The origin of these antibodies is not known and the clinical details of the patients with the positive sera do not provide any obvious clues. Of 9 sera found to react with PLL and other polyamines, allergies were suspected to penicillins (2 patients); cephalosporins (2 patients, 1 also with suspected trimethoprim allergy); ciprofloxacin (1 patient); grass pollens and house dust mite (1 patient); and ‘antibiotics’ (3 patients). Radioimmunoassay inhibition studies showed that the two amino groups, but not the carboxyl group, in lysine contributed to antibody binding and that compounds containing three primary amino groups (e.g., 4-aminomethyl-1,8-octanediamine) were better inhibitors than compounds containing two primary amino groups. Even a single amino group was found to be recognized (e.g., ethylamine) although more weakly. Table 4 lists side-by-side the inhibition results obtained with a range of different primary amines, polyamines and some compounds containing substituted ammonium groups when tested with two of the positive sera. Overall, sera were separated into two groups – those containing antibodies clearly inhibited by L-lysine, d-lysine and N-methyl-L-lysine and those poorly inhibited by these compounds. Serum 1 and serum 2 in Table 4 are examples of the first and second groups, respectively. Secondary and tertiary amines, for example, N-methylethanolamine, N,N-dimethylethanolamine and N,N,N',N'-tetramethylethylenediamine, were weakly to moderately inhibitory with some of the sera (Table 4) but not all. Considering the inhibitory profiles of all 9 sera, the overall inhibitory potencies of amines was primary > seconday > tertiary > quaternary [47].
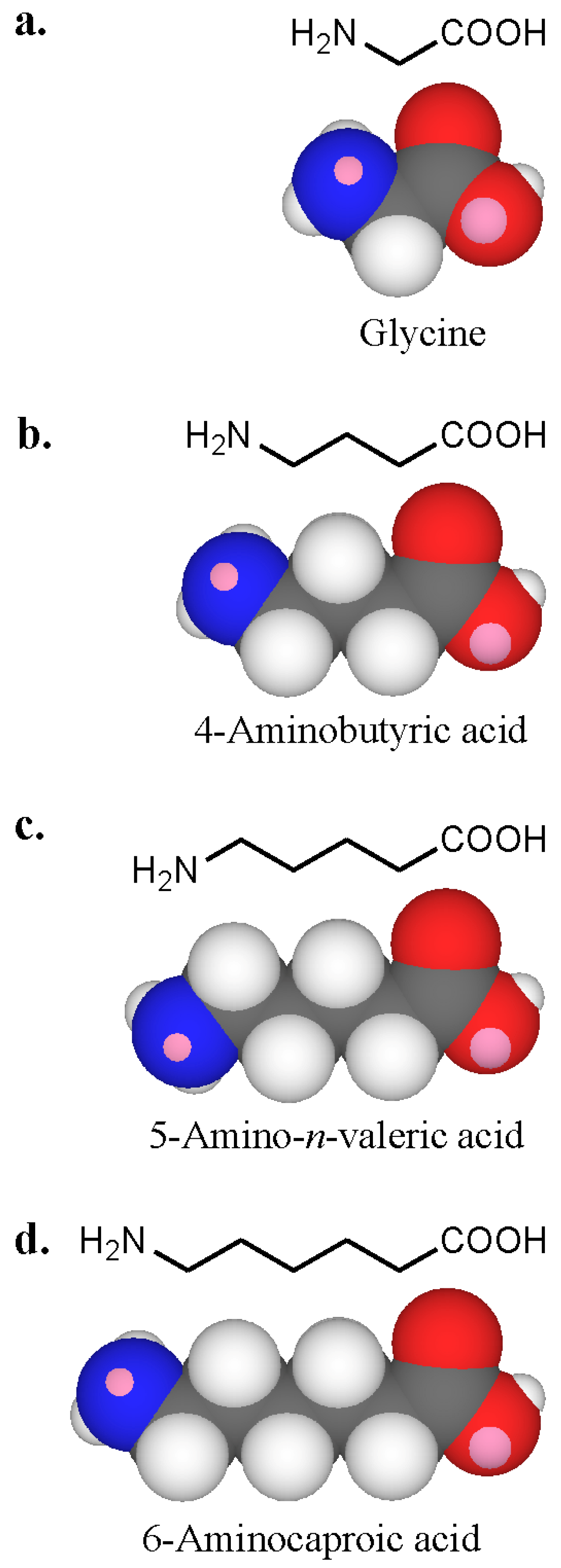
To see whether neighbouring groups affected antibody recognition, amines containing one or more carboxyl groups were examined. Amino acids with increasing chain length between the amino and carboxyl groups were selected ranging from a separation by one carbon (glycine), to three carbons (4-aminobutyric acid), four carbons (5-aminovaleric acid) and five carbons (6-aminocaproic acid) (Figure 17). With serum 1 containing IgE antibodies strongly inhibited by l- and d-lysines, all four amino acids showed clear inhibition with an order of potency 6-aminocaproic acid > 5-aminovaleric acid > 4-aminobutyric acid and glycine [47]. No inhibition was seen with some of the other sera. Inhibition with the amino acids and the diaminoacid 2,6-diaminopimelic acid suggested that charge distribution is involved in antibody recognition. Amino acids are amphoteric and exist as zwitterions or hybrid ions with the carboxyl group being the proton donor and the amino the proton acceptor. Reactivity of IgE in serum 1 with primary amino groups appeared to be influenced by near neighbour effects of carboxyl groups – antibody recognition correlated with increased distance between the carboxyl and amino groups.

Table 4.
Recognition of primary and substituted amines by IgE antibodies.1
| Compounds | Structure | IC30 (mM) 1 | IC50 (mM) 1 | Inhibition (%) at 40 mM 1 | |||
|---|---|---|---|---|---|---|---|
| Serum 1 | Serum 2 | Serum 1 | Serum 2 | Serum 1 | Serum 2 | ||
| Poly-L-lysine49 | (L-Lysine)49 | 2 | 3.2 | 93 | |||
| Poly-L-lysine18 | (L-Lysine)18 | 6.1 | 0.18 | 19 | 0.39 | 66 | 94 |
| L-Lysine | NH2(CH2)4CH(NH2)COOH | 3.1 | 90 | 13 | 130 | 76 | 0 |
| D-Lysine | NH2(CH2)4CH(NH2)COOH | 3.1 | 90 | 12 | 140 | 68 | 0 |
| N-Methyl-L-lysine | CH3NH(CH2)4CH(NH2)COOH | 50 | 105 | > 200 | > 200 | 26 | 9 |
| Ethylamine | CH3CH2NH2 | 4.2 | 81 | 19 | 130 | 54 | 5 |
| 1,3-Diaminopropane | NH2(CH2)3NH2 | 7.1 | 18 | 29 | 30 | 56 | 57 |
| 1,5-Diaminopentane | NH2(CH2)5NH2 | 2.6 | 20 | 5.2 | 36 | 62 | 54 |
| 4-Aminomethyl-1,8-octanediamine | NH2(CH2)4CH(CH2NH2)(CH2)3NH2 | 6 | 8.1 | 18 | 13 | 65 | 84 |
| Spermine | NH2(CH2)3NH(CH2)4NH(CH2)3NH2 | 100 | 16 | > 200 | 21 | 19 | 77 |
| Ethanolamine | HO(CH2)2NH2 | 45 | 75 | > 200 | 130 | 28 | 14 |
| N-Methylethanolamine | HO(CH2)2NHCH3 | 90 | 75 | > 200 | 135 | 20 | 13 |
| N,N-Dimethylethanolamine | HO(CH2)2N(CH3)2 | 19 | 80 | 39 | > 200 | 53 | 17 |
| N,N,N',N'-Tetramethylethylenediamine | (CH3)2NCH2CH2N(CH3)2 | 100 | 90 | > 200 | 190 | 15 | 8 |
| Choline | (CH3)3N(OH)(CH2)2OH | 110 | 190 | > 200 | > 200 | 12 | 17 |
| Glycine | NH2CH2COOH | 115 | > 200 | > 200 | > 200 | 8 | 0 |
| 4-Aminobutyric acid | NH2(CH2)3COOH | 61 | > 200 | 120 | > 200 | 18 | 0 |
| 5-Amino-n-valeric acid | NH2(CH2)4COOH | 10 | > 200 | 35 | > 200 | 52 | 0 |
| 6-Aminocaproic acid | NH2(CH2)5COOH | 21 | > 200 | > 200 | > 200 | 37 | 0 |
| 2,6-Diaminopimelic acid | HOOCCH(NH2)(CH2)3CH(NH2)COOH | 85 | > 200 | 12 | |||
| L-Lysine methyl ester | NH2(CH2)4CH(NH2)COOCH3 | 18 | 45 | 46 | |||
1 Inhibition results from immunoassays using sera and poly-L-lysine49–solid phase complex. Data from [47].
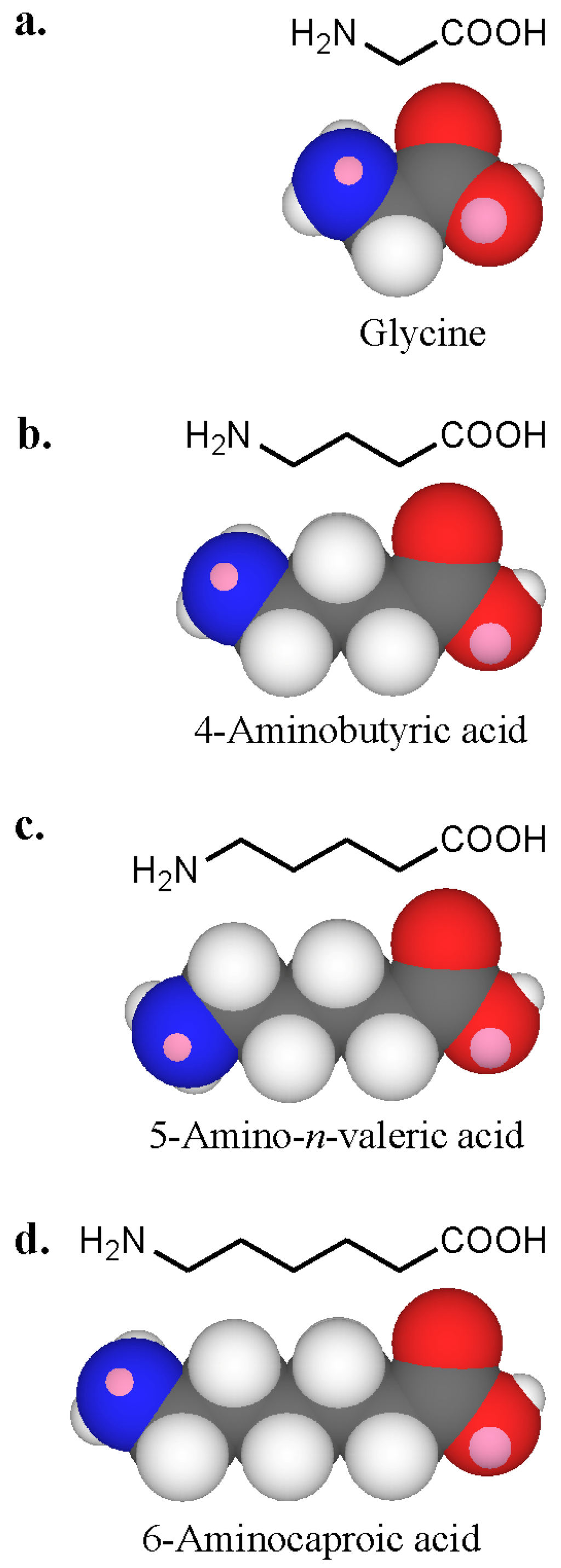
Figure 17.
Three-dimensional CPK models of amino acids of increasing carbon chain length (a to d, glycine to 6-aminocaproic acid) showing the proximity of the carboxyl group to the amino group in each compound.
Figure 17.
Three-dimensional CPK models of amino acids of increasing carbon chain length (a to d, glycine to 6-aminocaproic acid) showing the proximity of the carboxyl group to the amino group in each compound.
Although detected in sera from allergic patients, particularly patients with suspected drug allergies, there is no evidence, for the time being at least, that these antibodies underlie allergic or other adverse reactions or are complementary to some amino acids in some peptides/proteins. These results do, however, have bearing on the use of PLLs as carriers for ‘small’ molecules or haptens in drug conjugates (e.g., as in PLO–polylysine conjugates) employed for immunoassay studies where so-called “false-positives” may result due to antibody recognition of PLLs.
Polyamines occur in all living organisms, where they stabilize membranes, alter intracellular Ca2+ levels and are necessary for optimal growth and replication of cells. It remains unclear what biologic role(s) if any polyamine-reactive antibodies of the IgE class might have but recognition of such compounds by IgE already known to be involved in immediate allergic responses and suggested to have roles in parasite control and rejection and possibly even cancer, is intriguing and warrants further study.
8. Anaphylaxis to the Induction Agent Thiopentone and Identification of Allergenic Determinants
Thiopentone is an ultrashort acting barbiturate used for induction of anesthesia. Although now largely replaced by propofol, it is still seen as the classic drug used in rapid sequence inductions and it is still occasionally used in electroconvulsive therapy. Allergic reactions to the drug are rare and when they do occur it is usually after multiple previous exposures [48]. IgE antibodies to thiopentone were first identified in the mid 1980s in the sera of patients who experienced a life-threatening anaphylactic reaction following induction of anesthesia with the drug [49]. Extensive quantitative inhibition investigations employing a range of carefully selected analogs, in particular, the barbiturates pentobarbitone, barbitone, methohexital, thiobarbituric acid and barbituric acid, identified the structures of two allergenic determinants on opposite sides of the thiopentone molecule—position 1 of the pyrimidine ring with its attached sulfur atom (Figure 18a) and the ethyl and secondary pentyl groups at position 5 (Figure 18b). Pentobarbitone, which is identical in structure to thiopentone except for an oxygen instead of a sulfur at position 2, was a key inhibitor in the identification of the determinant at position 5 while 2-mercaptopyrimidine (Figure 18c) was important in identifying the thio region as a second allergenic structure [50,51].
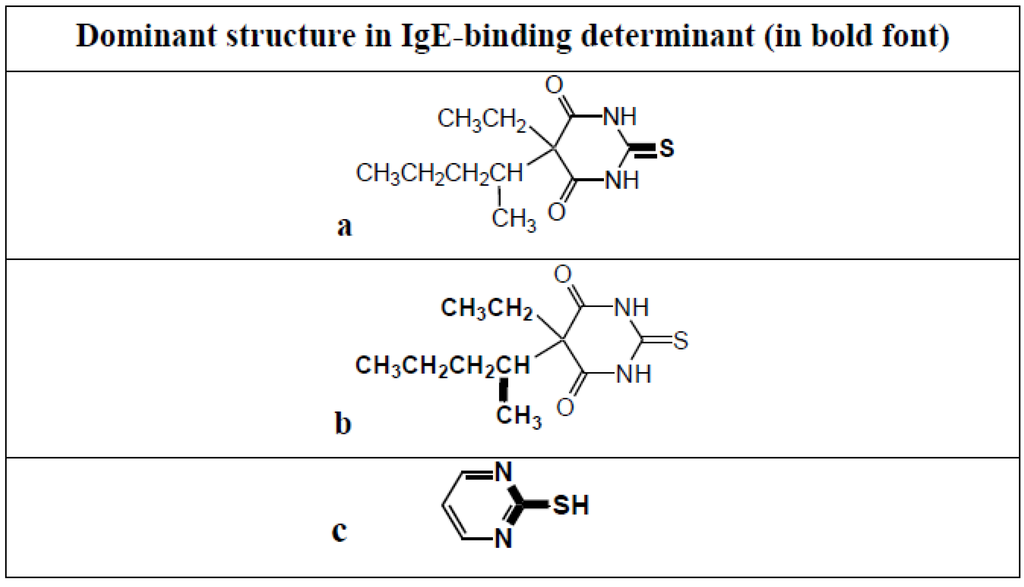
Figure 18.
Thiopentone allergenic (IgE antibody-binding) determinants identified using quantitative hapten inhibition studies together with sera from patients who experienced anaphylaxis to the induction agent. Determinants (shown in bold) are the regions of the pyrimidine ring encompassing the attached sulfur atom (a) and the ethyl and secondary pentyl groups at position 5 of the ring (b). 2-Mercaptopyrimidine (c), unlike thiopentone, inhibits binding of IgE in sera from patients allergic to NMBDs to the ‘thiopentone’ solid phase.
Figure 18.
Thiopentone allergenic (IgE antibody-binding) determinants identified using quantitative hapten inhibition studies together with sera from patients who experienced anaphylaxis to the induction agent. Determinants (shown in bold) are the regions of the pyrimidine ring encompassing the attached sulfur atom (a) and the ethyl and secondary pentyl groups at position 5 of the ring (b). 2-Mercaptopyrimidine (c), unlike thiopentone, inhibits binding of IgE in sera from patients allergic to NMBDs to the ‘thiopentone’ solid phase.
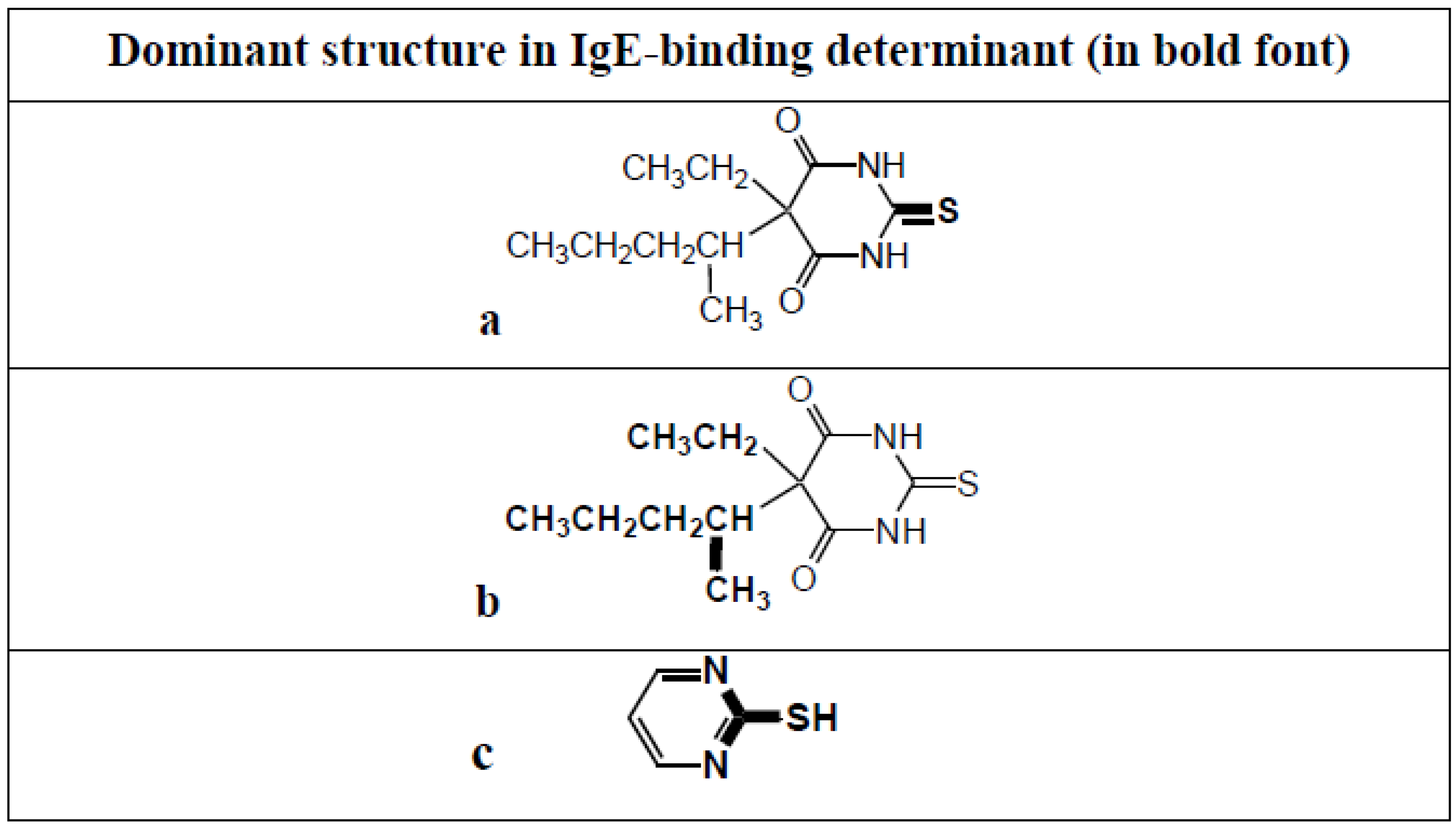
Although the thiopentone immunoassay is a valuable test in helping to confirm immediate allergic reactions to the hypnotic [52,53,54], the test sometimes detects false-positive reactions due to reactivity of the thiopentone–solid phase with IgE antibodies in the sera of subjects allergic to NMBDs. Inhibition of IgE binding to the drug solid phase by 2-mercaptopyrimidine but not thiopentone or thiobarbituric acid indicated that the NMBD-reactive IgE antibodies were recognizing the ring nitrogens of the pyrimidine nucleus (Figure 18c). The ring nitrogens become accessible to antibody binding on the solid phase following alkali treatment during the coupling procedure but are sterically hindered on free thiopentone [48,50,51].
9. Less Well Studied IgE Antibody Responses to Other Drugs
9.1. Quinolones
Quinolones are a family of synthetic antibacterials based on the 4-quinolone and 1,8-naphthyridine nuclei. Drug examples of each of these structures, respectively, are norfloxacin and nalidixic acid. A third category of quinolones represented by, for example, cinoxacin, is based on cinnoline (Figure 19). Immediate hypersensitivity reactions to quinolones occur with a frequency of from 0.4 to 2%. There are a number of reports of anaphylaxis to quinolones, especially to ciprofloxacin, and reactions following first exposure to the drugs are well known. In an attempt to improve bioavailability and broaden the antibacterial spectrum, the range of quinolones has been continually expanded. Four generations of the drugs have now been synthesized represented, for example, by pipemidic acid, ciprofloxacin, levofloxacin and moxifloxacin, respectively.
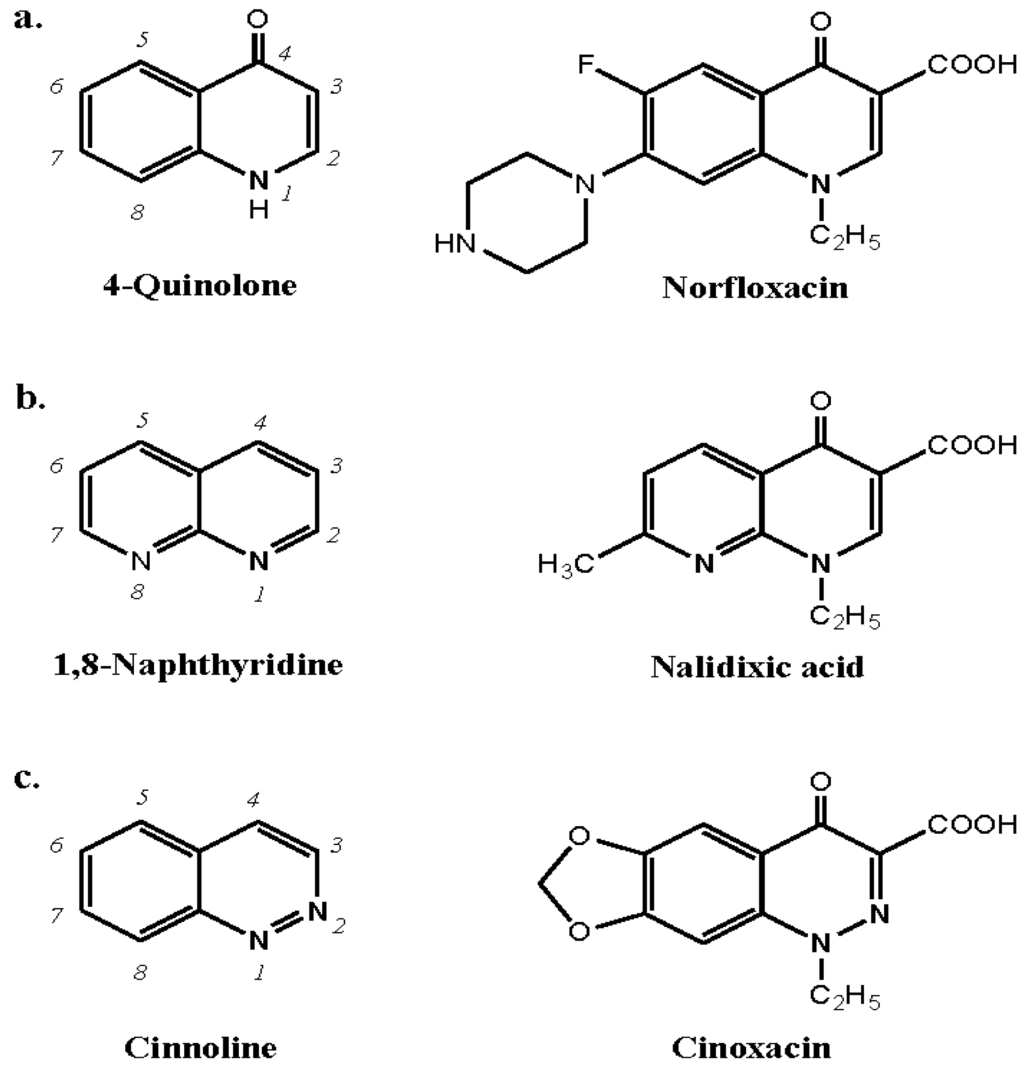
Figure 19.
Structures of the broad family of quinolone antibacterial drugs based on the 4-quinolone, 1,8-naphthyridine and cinnoline nuclei together with examples of a drug from each class, viz., norfloxacin, nalidixic acid and cinoxacin, respectively.
Figure 19.
Structures of the broad family of quinolone antibacterial drugs based on the 4-quinolone, 1,8-naphthyridine and cinnoline nuclei together with examples of a drug from each class, viz., norfloxacin, nalidixic acid and cinoxacin, respectively.
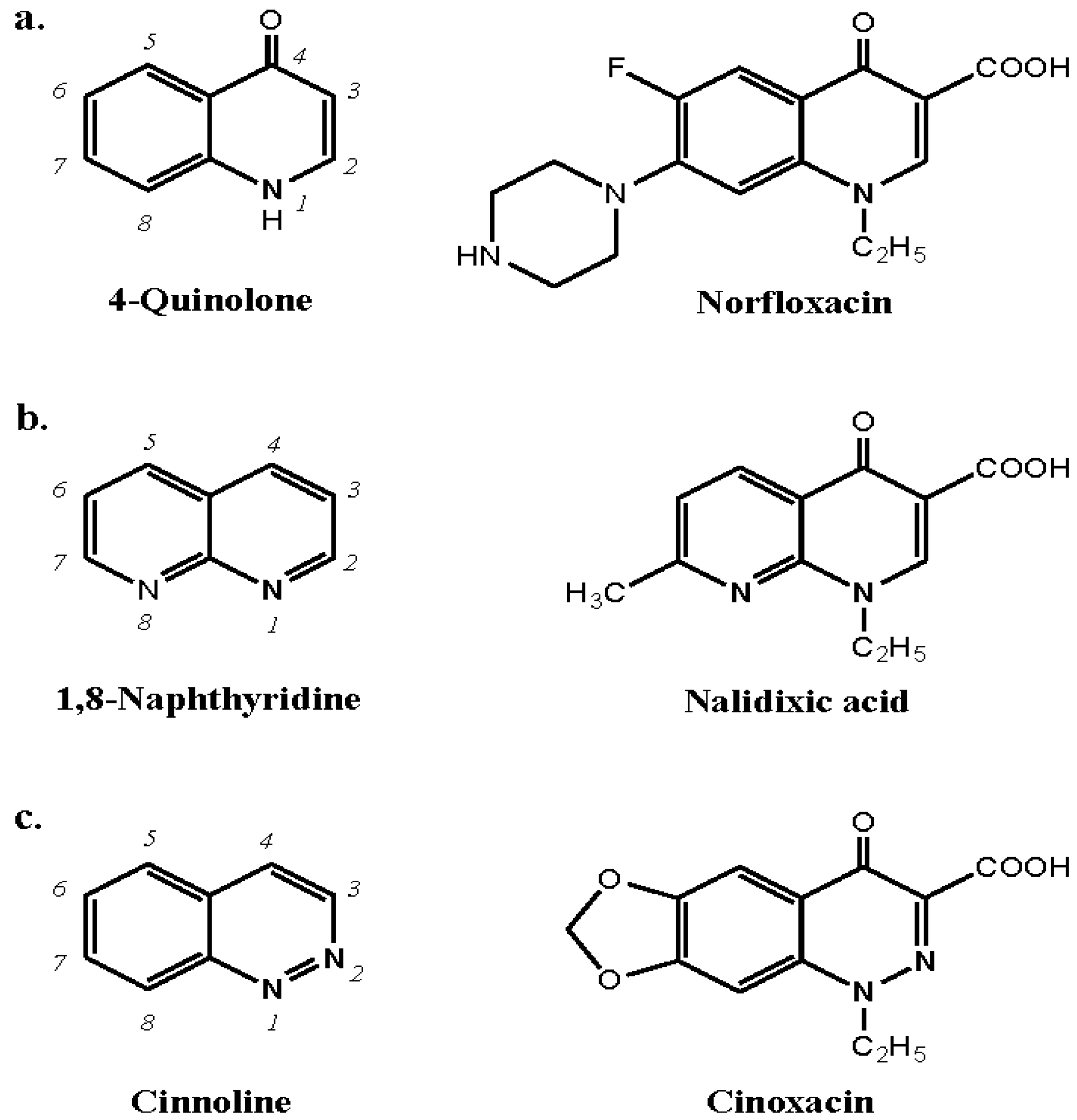
In the first apparently successful attempt to demonstrate IgE antibodies to the quinolones pipemidic acid and norfloxacin, drug–solid phase complexes were prepared in the author’s laboratory in 1995-6 by linking the carboxyl group at position 3 to the support via carbodiimide/human serum albumin and through the piperazine substituent at position 7 to bis-oxirane-activated Sepharose. A comparison of the IgE binding of each of these drug conjugates showed positive reactions only with the conjugate linked via the piperazine group at position 7. Conjugates prepared by coupling through the 3-carboxyl group were clearly unreactive indicating that the allergenic structures are composed of all, or part of, the face of the molecule containing the 3-carboxy group and opposite the side of the molecule with the attached piperazine ring [5,55]. Inhibition studies with the appropriate quinolone and structural analogs appeared to confirm the specificity and diagnostic value of the Sepharose conjugates for the detection of quinolone-reactive IgE antibodies but reaction of the Sepharose conjugates with some apparently ‘normal’ sera (i.e., sera from subjects who were not allergic including to quinolones) cast doubt on the assay's specificity and general applicability [5]. It should also be noted that positive skin tests to quinolone drugs in normal, healthy, non-allergic controls have been observed in a number of careful investigations [5,56,57,58,59]. Nevertheless, in a study from Europe in 2004, the quinolone-Sepharose assay was used to detect positive reactions in 30 of 55 patients with an immediate allergic reaction to a quinolone drug [60] and this was followed by application of the same assay to detect IgE antibodies to ciprofloxacin, levofloxacin and/or moxifloxacin in 12 of 38 patients with severe allergic reactions to the antibacterials [61].
Recently it was claimed that immediate hypersensitivity to quinolones is frequently linked to NMBD sensitization [62] but this is likely to simply be a reflection of cross-reactivity of substituted ammonium groups on both NMBDs and quinolones detected by the ammonium group-reactive IgE antibodies present in the sera of patients allergic to NMBDs [63,64,65]. Clearly, questions remain concerning the involvement, detection and specificity of IgE antibodies to quinolones and the relevance of these antibodies detected in skin tests and immunoassays in both allergic and non-allergic subjects remains to be determined.
9.2. Chlorhexidine
The antiseptic and disinfectant chlorhexidine is widely used, domestically, industrially and in medicine in a myriad of products. Skin and mucous membrane contact with chlorhexidine is therefore not uncommon and due to its capacity to induce serious anaphylactic reactions in the occasional sensitive individual, clinicians are now much more aware of the allergenic potential of this agent sometimes described as a covert allergen source [5]. The first reported cases of anaphylaxis to chlorhexidine were in 1985 and subsequent investigations showed a high incidence of patients sensitized via the urethral route and mucous membrane exposure in general [66,67]. Allergic IgE antibody-mediated sensitivity to the drug can be demonstrated by positive skin tests and tests for specific serum IgE antibodies. Following the development in Japan in 1986 of a specific immunoassay for detecting chlorhexidine-reactive IgE by linking a chlorhexidine-albumin conjugate to paper discs and demonstration of specificity by inhibition with chlorguanide and the parent drug [68], experiments to identify allergenic determinants were subsequently undertaken using chlorhexidine linked to a bis-oxirane-activated solid phase [69]. To identify the fine structural features of the determinant(s) complementary to chlorhexidine-reactive IgE antibodies, a number of analogs were selected for examination. These included chlorguanide [1-(4-chlorophenyl)-2-(N'-propan-2-ylcarbamimidoyl) guanidine] which represents almost half the chlohexidine molecule, alexidine, aminoguanide and arginine to mimic interior structures and some chlorophenyl derivatives to test for recognition of the terminal structures (Figure 20). Only the parent drug, chlorguanide and alexidine produced clear dose-dependent inhibition; compounds selected to represent the terminal structures of chlorhexidine, for example, 4-chloroacetanilide and 4-guanidinobenzoic acid, were without activity. On a molar basis, chlorhexidine was 100–200 times as potent an inhibitor as chlorguanide and alexidine and, overall, the results showed that the parent drug was the 'best fit' for the IgE antibody combining sites [69].
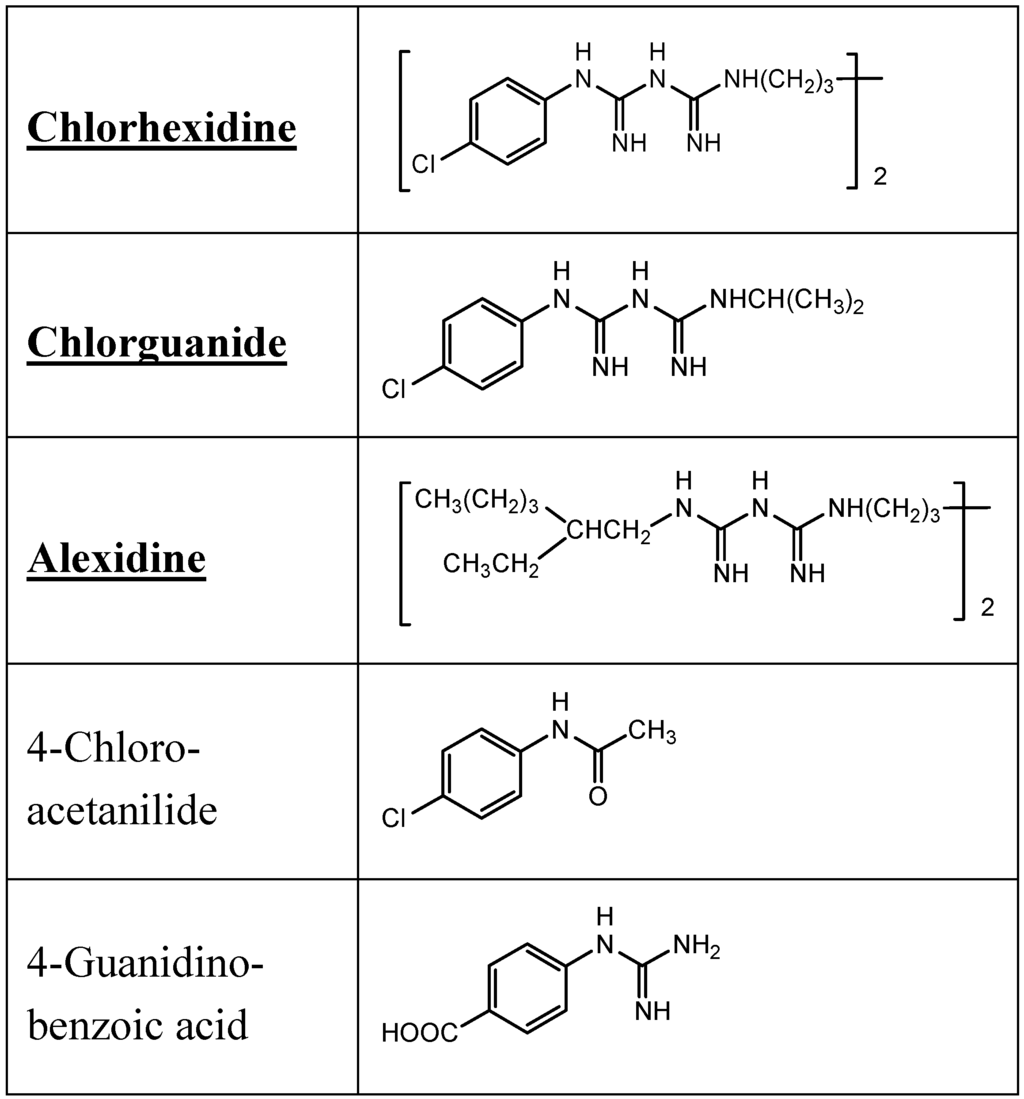
Figure 20.
Structures of chlorhexidine, chlorguanide and alexidine, compounds that strongly inhibit binding to chlorhexidine of IgE antibodies from a patient who experienced an anaphylactic reaction to the antibacterial. By contrast, structures representing the terminal groupings of chlorhexidine, 4-chloroacetanilide and 4-guanidinobenzoic acid, were devoid of inhibitory activity. Overall, these findings led to the conclusion that the entire chlorhexidine molecule was complementary to the antibody combining sites.
Figure 20.
Structures of chlorhexidine, chlorguanide and alexidine, compounds that strongly inhibit binding to chlorhexidine of IgE antibodies from a patient who experienced an anaphylactic reaction to the antibacterial. By contrast, structures representing the terminal groupings of chlorhexidine, 4-chloroacetanilide and 4-guanidinobenzoic acid, were devoid of inhibitory activity. Overall, these findings led to the conclusion that the entire chlorhexidine molecule was complementary to the antibody combining sites.
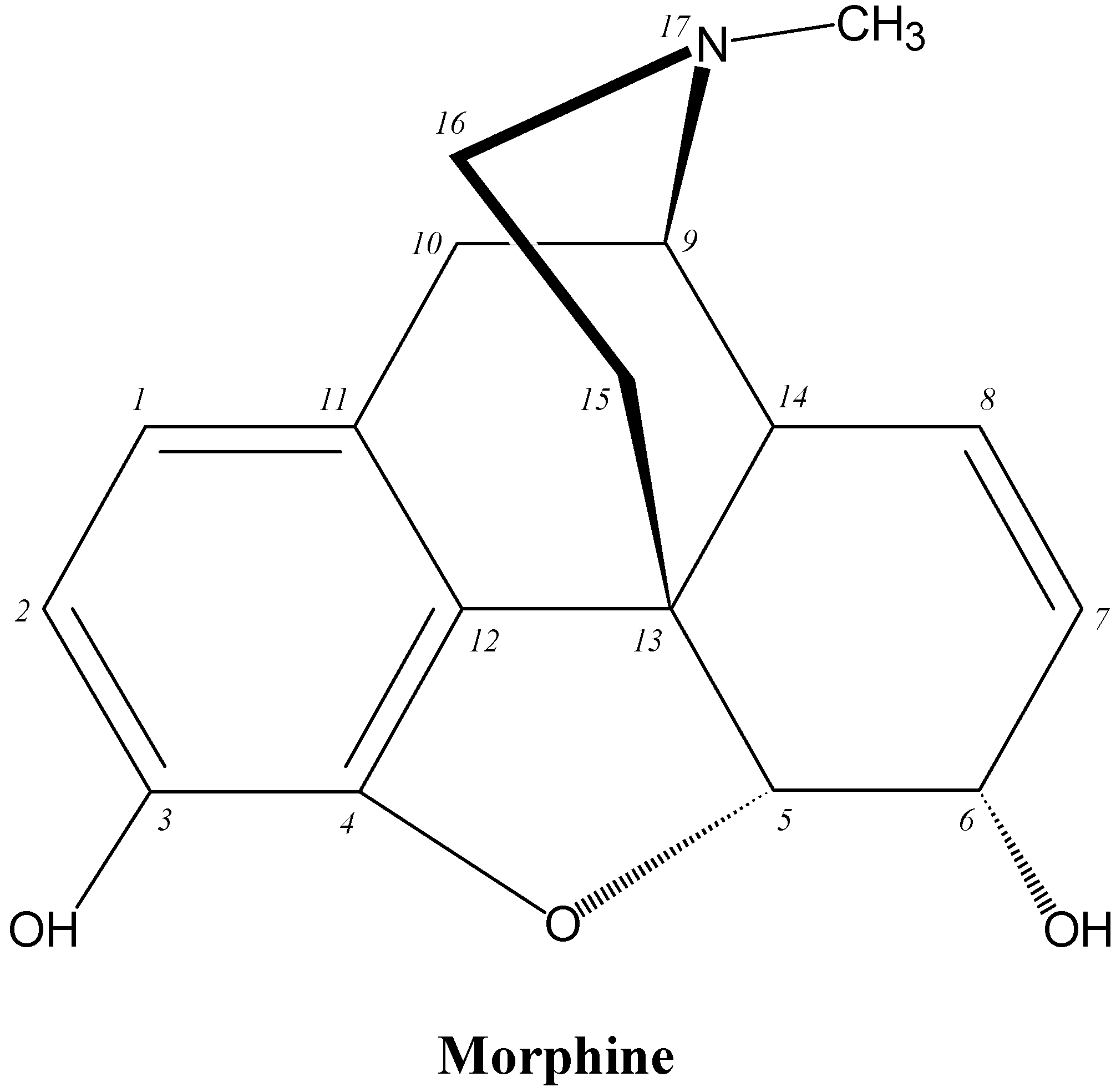
9.3. Opioid Analgesics
Opioid analgesics (OADs) are near the top of any list of the most frequently used drugs in hospitals and although a number of these drugs are potent releasers of histamine (e.g., morphine), producing anaphylactoid-like reactions involving flushing, rash, urticaria, pruritus, mucous production and hypotension, IgE antibody-mediated reactions to OADs are rarely seen [5,70]. Given the widespread prescribing of OADs, their frequent administration, allergy-like symptoms seen in anaphylactoid reactions to the drugs, their heavy use by addicts and resultant deaths due to no clearly demonstrable cause, the apparent lack of allergenicity of OADs seems surprising. It has been suggested that because both the natural and synthetic opioids act on the same receptors in vivo, the synthetic drugs may be seen as ‘self’ components and not foreign antigens [5,70]. Clearly established cases of true type I allergic reactions to the drugs are hard to find and there appears to be only one report describing such a reaction and defining the fine structural features of the drug's allergenic determinant(s). Comparative inhibitory potencies of structural analogs of morphine revealed that a morphine determinant recognized by the complementary IgE antibodies comprised the N-methylpiperidine ring (ring D) attached to the cyclohexenyl ring (ring C), the double bond at position 7-8 of ring C and the hydroxyl group at carbon 6 (Figure 21) [71]. There are a few rare reports of the detection of IgE antibodies to some of the semisynthetic and synthetic OADs, particularly, heroin, meperidine (pethidine), fentanyl and methadone [5] but structural recognition information is lacking.
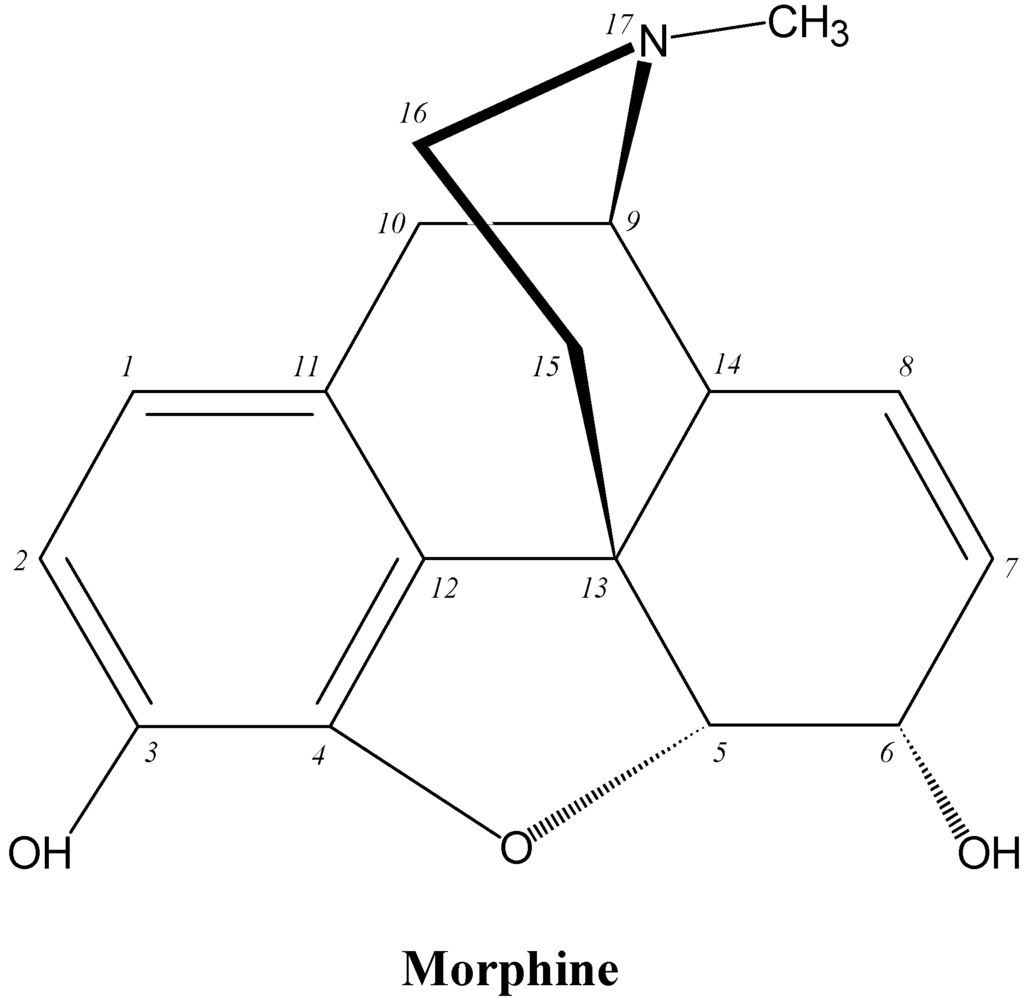
Figure 21.
Structure of the opioid analgesic drug morphine.
Figure 21.
Structure of the opioid analgesic drug morphine.
9.4. Non-Steroidal Antiinflammatory Drugs
Of the large number of structurally different non-steroidal antiinflammatory drugs (NSAIDs) administered throughout the world, IgE antibody-mediated type I allergic reactions to these agents are mainly confined to aspirin (acetylsalicylic acid) and the pyrazolone group [5]. The salicyloyl determinant and O-methylsalicoyl derivative, each in solid phase form were used to demonstrate the presence of complementary IgE antibodies in the sera of patients with a history of aspirin sensitivity that had been confirmed by oral challenge. Inhibition of antibody binding to the salicyloyl determinant by salicylic acid and 2-aminophenol and inhibition of binding to the O-methylsalicoyl solid phase by O-methylsalicylic confirmed the specificities of the reactions [72].
Reports of anaphylaxis to the pyrazolones, phenazone (antipyrine), propyphenazone (4-isopropylpyrine), aminophenazone (aminopyrine, amidopyrine) and metamizole (dipyrone) date back more than 40 years. A solid phase immunoassay for the detection of IgE antibodies to the 1-phenyl-2,3-dimethyl-3-pyrazoline-5-one common core structure of the pyrazolone drugs has been developed [73] and propyphenazone-specific IgE antibodies were detected with a conjugate prepared by coupling N-demethylpropyphenazone to human serum albumin [74]. The latter assay was found to be specific for propyphenazone since that pyrazolone, but no other, inhibited IgE antibody binding to the drug-solid phase.
9.5. Macrolide Antibiotics
The incidence of “allergies” to macrolide antibiotics has been estimated to be up to 3% but it is not always clear how many of the reactions are true type I IgE antibody-mediated responses. Symptoms seen include urticaria, angioedema and there are rare reports of anaphylaxis. The existence of IgE antibodies to macrolide antibiotics, especially spiramycin, roxithromycin, clarithromycin and erythromycin has been inferred from positive prick test results and asthma to spiramycin in an occupational setting was confirmed by skin and/or challenge tests [75]. There is at least one report of the detection of IgE antibodies to erythromycin inhibited by the drug, a case of anaphylaxis detected with the Prausnitz-Kustner (P-K) test and a demonstration of IgE antibodies to erythromycin that were specifically inhibited by, and cross-reactive with, diacetylmidecamycin [76,77]. The rarity of cases of immediate hypersensitivity to the macrolides has no doubt contributed to the paucity of knowledge of the structures that interact with IgE antibodies.
9.6. Rifamycins
Rifamycins, chiefly rifamycin SV and rifampicin, can produce a variety of adverse reactions and true hypersensitivity reactions including a ‘flu’-like syndrome, hemolytic anemia, thrombocytopenia, vasculitis, renal failure, urticaria, contact dermatitis and type IV toxidermias [78]. Anaphylaxis to rifamycin SV and rifampicin, usually after local application, has been investigated by skin testing and/or the P-K, basophil activation, and radioallergosorbent tests. In one application of the latter test, rifamycin SV was conjugated to poly-l-lysine followed by reductive amination to reduce the Schiff base and coupling to cellulose to produce a drug–solid phase for detecting serum IgE antibodies [79].
Comprehensive investigations designed to identify rifamycin structures that react with complementary IgE antibodies have not so far been undertaken although IgE antibodies that recognized the nucleus of rifamycin SV but not the 4-methylpiperazine side chain were demonstrated over 35 years ago [80]. More recently, IgE antibodies that reacted with rifamycin SV and rifampicin were detected and rifapentine, rifabutin and rifaximin were shown to inhibit antibody binding to a rifampicin—Sepharose solid phase [81].
10. Conclusions
Of the many drugs known to have provoked anaphylactic and other type I IgE antibody-mediated reactions in humans, relatively few laboratories have pursued immunochemical investigations designed to detect the reactions with specific tests and identify the structures responsible for the induction of the antibody responses. The largest bodies of such work so far have been undertaken with the penicillin group of β-lactam antibiotics and the NMBDs but even with the former long-studied drugs, immunochemical insights into the relative importance of allergenic structures has waned in recent years following the impressive early contributions delivered over 30-40 years ago [5,12,13,14]. The deficiency of information on β-lactam allergenic structures is readily apparent with the cephalosporins where there is still no agreement on whether or not R2 side chains contribute to the allergenicity of these drugs [5,23,82,83]. In addition, knowledge of allergenic structures of other important β-lactams, particularly monobactams, carbapenems and clavams, has barely begun to accumulate [5,84].
For the sulfonamides, their decreased usage over recent years (except in HIV-associated therapies) and identification over 25 years ago of the IgE-binding structures on sulfamethoxazole [24], has seen a heavy emphasis in attempts to elucidate mechanisms of non-IgE-mediated adverse responses to the drug. This large research investment, aimed at both understanding the pathogenesis of reactions and using sulfamethoxazole as a model for the immune response to drug- and/or metabolite-protein adducts, does not appear to have yielded the hoped-for new insights or progressed the field well beyond the understanding of the role of nitroso and hydroxylamine metabolites obtained in the early to mid 1990s [5,85,86]. For the drug often administered with sulfamethoxazole, viz., trimethoprim, research investigations of its roles in IgE-mediated responses or other hypersensitivity reactions have been surprisingly few. Given the author's experience of small but steady numbers of cases of suspected immediate reactions to the drug, the existing proven and reliable drug-specific immunoassay for the detection of trimethoprim-reactive IgE antibodies introduced 25 years ago, and the identification of the drug’s IgE-binding structures [26,27], one might expect to have seen more attention paid to this drug both in clinical diagnosis and laboratory testing.
The NMBDs provide one of the more fascinating stories that have emerged over the last 30 years both from the clinical aspect of drug allergy and the underlying science [5,28,34]. Identification of substituted (tertiary and quaternary) ammonium ions as the structures recognized on all NMBDs by the IgE antibodies in the sera of patients who experienced an anaphylactic reaction to a NMBD during anesthesia, has led on to intriguing questions and suggested explanations concerning the origin of the antibodies and to the increasing number of reports of IgE to ammonium groups on other drugs [5,32,34,36,62,63,64,65]. Further studies should determine any additional clinical relevance or otherwise of these antibodies. Although the ammonium groups on NMBDs are the basis of antibody recognition and allergenic cross-reactivity between the different NMBDs, IgE combining sites often appear to recognize additional structures and this is reflected in cross-reactions observed in vitro and in vivo [5,34]. Unlike the situation with the immunoassays developed for the detection of specific IgE antibodies to some other drugs, tests for serum IgE antibodies to NMBDs have been widely taken up and applied together with skin testing in the diagnosis of suspected cases of NMBD-induced anaphylaxis [5,34].
In a recent development, the γ-cyclodextrin sugammadex was chemically modified to encapsulate rocuronium and reverse neuromuscular blockade by forming a stable host-guest inclusion complex and removing the drug from the neuromuscular junction [87,88]. Whether encapsulation of rocuronium in this manner can mitigate an existing anaphylactic reaction induced by rocuronium remains to be proven but it has already been suggested that allergenicity may be retained due to incomplete encapsulation of the molecule in the inclusion complex form. Interestingly, however, there are now at least 11 reports of reversal of rocuronium-induced anaphylaxis by the prompt administration of sugammadex suggesting that encapsulation of the drug somehow reverses the ongoing anaphylactic events [reviewed in 5 and 39].
Drug carrier systems such as the cyclodextrans, dendrimers, vesicles, nanoparticles, micelles, hydrogels and soluble polymers are being introduced to improve drug delivery, stability and solubility, reduce toxicity and irritation, mask taste and increase microbial stability etc. This has the possibility of producing changes in immunological recognition including an increase in the allergenicity of a drug or conferring allergenicity on a previously 'non-allergenic' molecule. This possibility of altered allergenic properties should not be overlooked in preclinical drug safety assessments or by allergists and dermatologists [89].
IgE antibodies that react with polyamines via recognition of primary amine groups (especially in the form of di- or higher primary amines), detected in the sera of some patients allergic to a number of different drugs remains a conundrum since there appears to be no data or even hints on the origin of the antibodies or their clinical significance if any [47]. Further investigations of the incidences of these antibodies in normal non-allergic subjects and in patients allergic to drugs, inhalants, food and venoms may help to shed some light.
While the development of an immunoassay for the detection of IgE antibodies to thiopentone in the sera of patients allergic to the induction agent is a valuable diagnostic aid [49,52,53], the complexity of findings and the necessary controls when the imprecisely defined drug-solid phase is used with sera from patients allergic to NMBDs, make the assay somewhat problematic [48,51]. Nevertheless, identification of the barbiturate IgE-binding structures is a significant addition to the slowly accumulating list of allergenic determinants on ‘small’ molecules. It is also apparent that with the almost total replacement of thiopentone by drugs such as propofol, the need for, and application of, the thiopentone antibody test has fallen dramatically.
There are some puzzling, if not intriguing, aspects of results obtained so far with immunoassays designed to detect IgE antibodies to quinolone antibacterial drugs. From the limited chemical approaches applied, it seems that the important allergenic structures of the 4-quinolone and 1,8-naphthyridine groups of drugs are located on the side of the molecules containing the 3-carboxy substituent [5,55] but the finding of an occasional serum from apparently normal, healthy, non-allergic subjects that react positively and apparently specifically in the assay is hard to explain. Considered together with the variable skin test results obtained by a number of different investigators [56,57,58,59] there seems to be still a lot to learn about reactions provoked by the quinolone group of drugs.
The capacity of chlorhexidine to sensitize patients via contact with skin and mucous membranes and its generally unanticipated and 'hidden' nature in a variety of medicaments and other products makes this drug a warning and a model for other widely used contact agents that are generally not considered to represent a sensitizing problem. Results so far indicate that the IgE antibody response to chlorhexidine when used as an antiseptic is fairly homogeneous with essentially the whole molecule complementary to the IgE antibody combining sites [69]. The chlorhexidine molecule contains identical structures at each end raising the possibility that the free, unbound molecule, like NMBDs [5,32], may be able to bridge combining sites of complementary and adjacent IgE molecules on the mast cell surface. The terminal chlorophenyl groups are separated by 16 atoms and the IgE-reactive chlorguanide groups are bridged by a six carbon chain. The distances involved and the flexibility of the chlorhexidine molecule suggest that cross-linking at the mast cell surface is possible.
While the existing immunoassays for the detection of IgE to aspirin and pyrazolone NSAIDs appear to be useful potential tests for routine diagnostic application, few if any immunoassays have been introduced and well studied for many other important drugs including the clinically valuable antibiotics such as macrolides, tetracyclines and rifamycins. This is not necessarily a serious problem or indeed surprising since most drugs are only rarely implicated in type I hypersensitivity reactions and the absence of tests is almost certainly a reflection of the absence of research borne out of a lack of need. Almost any drug has probably provoked an immediate allergic reaction, including anaphylaxis, in at least one patient in the world at some time, but this is not necessarily reason enough for the development of a specific immunoassay for the detection of IgE antibodies in such rare instances. However, from the viewpoint of those interested in building up a picture or library of the fine structural features of IgE antibody-binding determinants on drug molecules administered to humans and involved in type I hypersensitivity reactions, every bit of structural information that helps to define a drug determinant is of interest.
Acknowledgments
I thank Nghia H. Pham for his help in the preparation of the figures and tables.
Conflicts of Interest
The author declares no conflict of interest.
References
- Kabat, E.A.; Mayer, M.M. Experimental Immunochemistry, 2nd ed.; Charles C. Thomas: Springfield, IL, USA, 1961. [Google Scholar]
- Landsteiner, K. The Specificity of Serological Reactions; Dover: New York, NY, USA, 1962. [Google Scholar]
- Trier, N.H.; Hansen, P.R.; Houen, G. Production and characterization of peptide antibodies. Methods 2012, 56, 136–144. [Google Scholar] [CrossRef]
- Baldo, B.A. Immunochemical perspectives of allergy: In the steps of Karl Landsteiner. In Molecular Approaches to the Study of Allergens. Monographs in Allergy; Karger: Basel, Switzerland, 1990; Volume 28. [Google Scholar]
- Baldo, B.A.; Pham, N.H. Drug Allergy: Clinical Aspects, Diagnosis, Mechanisms, Structure-Activity Relationships; Springer: New York, NY, USA, 2013. [Google Scholar]
- Kaliner, M.; Austen, K.F. Immunological release of chemical mediators from human tissues. Ann. Rev. Pharmacol. 1975, 15, 177–189. [Google Scholar] [CrossRef]
- Gould, H.J.; Sutton, B.J.; Beavil, A.J.; Beavil, R.L.; McCloskey, N.; Coker, H.A.; Fear, D.; Smurthwaite, L. The biology of IgE and the basis of allergic disease. Annu. Rev. Immunol. 2003, 21, 579–628. [Google Scholar] [CrossRef]
- Ogawa, Y.; Grant, J.A. Mediators of anaphylaxis. Immunol. Allergy Clin. North. Am. 2007, 27, 249–260. [Google Scholar] [CrossRef]
- Fisher, M.M.; Baldo, B.A. Acute anaphylactic reactions. Med. J. Aust. 1988, 149, 34–38. [Google Scholar]
- Mendelson, L.M. Adverse reactions to β-lactam antibiotics. Immunol. Allergy Clin. North. Am. 1998, 18, 745–757. [Google Scholar] [CrossRef]
- Salkind, A.R.; Cuddy, P.G.; Foxworth, J.W. The rational clinical examination. Is this patient allergic to penicillin? An evidence-based analysis of the likelihood of penicillin allergy. JAMA 2001, 285, 2498–2505. [Google Scholar] [CrossRef]
- Dewdney, J.M. Immunology of the antibiotics. In The Antigens; Sela, M., Ed.; Academic Press: New York, NY, USA, 1977; Volume 4, pp. 73–228. [Google Scholar]
- Levine, B.B. Immunochemical mechanisms of drug allergy. In Textbook of Immunopathology, 2nd ed.; Miescher, P.A., Muller-Eberhard, H.J., Eds.; Grune and Stratton: New York, NY, USA, 1976; Volume 2, pp. 403–419. [Google Scholar]
- Parker, C.W. Drug allergy. In Clinical Immunology; Parker, C.W., Ed.; Saunders: Philadelphia, PA, USA, 1980; Volume 2, pp. 1219–1260. [Google Scholar]
- Levine, B.B.; Redmond, A.P. Minor haptenic determinant-specific reagins of penicillin hypersensitivity in man. Int. Arch. Allergy Appl. Immunol. 1969, 35, 445–455. [Google Scholar] [CrossRef]
- Harle, D.G.; Baldo, B.A. Identification of penicillin allergenic determinants that bind IgE antibodies in the sera of subjects with penicillin allergy. Mol. Immunol. 1990, 27, 1063–1071. [Google Scholar] [CrossRef]
- Baldo, B.A.; Pham, N.H.; Weiner, J. Detection and side-chain specificity of IgE antibodies to flucloxacillin in allergic subjects. J. Mol. Recognit. 1995, 8, 171–177. [Google Scholar] [CrossRef]
- Zhao, Z.; Baldo, B.A.; Baumgart, K.W.; Mallon, D.F.J. Fine structural recognition specificities of IgE antibodies distinguishing amoxicilloyl and amoxicillanyl determinants in allergic subjects. J. Mol. Recognit. 2001, 14, 300–307. [Google Scholar] [CrossRef]
- Zhao, Z.; Baldo, B.A.; Rimmer, J. β-Lactam allergenic determinants: fine structural recognition of a cross-reacting determinant on benzylpenicillin and cephalothin. Clin. Exp. Allergy 2002, 32, 1644–1650. [Google Scholar] [CrossRef]
- Harle, D.G.; Baldo, B.A. Drugs as allergens: An immunoassay for detecting IgE antibodies to cephalosporins. Int. Arch. Allergy Appl. Immunol. 1990, 92, 439–444. [Google Scholar] [CrossRef]
- Hamilton-Miller, J.M.T.; Newton, G.G.F.; Abraham, E.P. Products of aminolysis and enzymic hydrolysis of the cephalosporins. Biochem. J. 1970, 116, 371–384. [Google Scholar]
- Faraci, W.S.; Pratt, R.F. Elimination of a good leaving group from the 3'-position of a cephalosporin need not be concerted with β-lactam ring opening: TEM-2 β-lactamase-catalysed hydrolysis of pyridine -2-azo-4'-(N',N'-dimethylaniline) cephalosporin (PADAC) and of cephaloridine. J. Am. Chem. Soc. 1984, 106, 1489–1490. [Google Scholar] [CrossRef]
- Pham, N.H.; Baldo, B.A. β-Lactam drug allergens: Fine structural recognition patterns of cephalosporin- reactive IgE antibodies. J. Mol. Recognit. 1996, 9, 287–296. [Google Scholar] [CrossRef]
- Harle, D.G.; Baldo, B.A.; Wells, J.V. Drugs as allergens: Detection and combining site specificities of IgE antibodies to sulfamethoxazole. Mol. Immunol. 1988, 25, 1347–1354. [Google Scholar] [CrossRef]
- Harle, D.G.; Baldo, B.A.; Smal, M.A.; Van Nunen, S.A. An immunoassay for the detection of IgE antibodies to trimethoprim in the sera of allergic patients. Clin. Allergy 1987, 17, 209–216. [Google Scholar] [CrossRef]
- Smal, M.A.; Baldo, B.A.; Harle, D.G. Drugs as allergens: The molecular basis of IgE binding to trimethoprim. Allergy 1988, 43, 184–191. [Google Scholar] [CrossRef]
- Pham, N.H.; Baldo, B.A.; Manfredi, M.; Zerboni, R. Fine structural specificity differences of trimethoprim allergenic determinants. Clin. Exp. Allergy 1996, 26, 1155–1160. [Google Scholar] [CrossRef]
- Fisher, M.M.; Baldo, B.A. Anaphylaxis during anaesthesia: Current aspects of diagnosis and prevention. Eur. J. Anaesthesiol. 1994, 11, 263–284. [Google Scholar]
- Mertes, P.M.; Alla, F.; Trechot, P.; Auroy, Y.; Jougla, E. Anaphylaxis during anesthesia in France: An 8-year national survey. J. Allergy Clin. Immunol. 2011, 128, 366–373. [Google Scholar] [CrossRef]
- Fisher, M.M.; Munro, I. Life-threatening anaphylactoid reactions to muscle relaxants. Anesth. Analg. 1983, 62, 559–564. [Google Scholar]
- Baldo, B.A.; Fisher, M.M. Substituted ammonium ions as allergenic determinants in drug allergy. Nature 1983, 306, 262–264. [Google Scholar] [CrossRef]
- Baldo, B.A.; Fisher, M.M. Anaphylaxis to muscle relaxant drugs: Cross-reactivity and molecular basis of binding of IgE antibodies detected by radioimmunoassay. Mol. Immunol. 1983, 20, 1393–1400. [Google Scholar] [CrossRef]
- Baldo, B.A.; Fisher, M.M. Mechanisms of IgE-dependent anaphylaxis to anaesthetic drugs. Ann. Fr. Anesth. Reanim. 1993, 12, 131–140. [Google Scholar] [CrossRef]
- Baldo, B.A.; Fisher, M.M.; Pham, N.H. On the origin and specificity of antibodies to neuromuscular blocking (muscle relaxant) drugs: an immunochemical perspective. Clin. Exp. Allergy 2009, 39, 325–344. [Google Scholar]
- Didier, A.; Cador, D.; Bongrand, P.; Furstoss, R.; Fourneron, P.; Senft, M.; Philip-Joet, F.; Charpin, D.; Charpin, J.; Vervloet, D. Role of quaternary ammonium ion determinants in allergy to muscle relaxants. J. Allergy Clin. Immunol. 1987, 79, 578–584. [Google Scholar] [CrossRef]
- Floorvaag, E.; Johansson, S.G.O.; Oman, H.; Venemalm, L.; Degerbeck, F.; Dybendal, T.; Lundberg, M. Prevalence of IgE antibodies to morphine. Relation to the high and low incidences of NMBA anaphylaxis in Norway and Sweden, respectively. Acta Anaesthesiol. Scand. 2005, 49, 437–444. [Google Scholar] [CrossRef]
- Baldo, B.A.; Fisher, M.M. Detection of serum IgE antibodies that react with alcuronium and tubocurarine after lifethreatening reactions to muscle-relaxant drugs. Anaesth. Intens. Care 1983, 11, 194–197. [Google Scholar]
- Harle, D.G.; Baldo, B.A.; Fisher, M.M. Assays for, and cross-reactivities of, IgE antibodies to the muscle relaxants gallamine, decamethonium and succinylcholine (suxamethonium). J. Immunol. Methods 1985, 78, 293–305. [Google Scholar] [CrossRef]
- Baldo, B.A.; McDonnell, N.J.; Pham, N.H. The cyclodextrin sugammadex and anaphylaxis to rocuronium: Is rocuronium still potentially allergenic in the inclusion complex form? Mini Rev. Med. Chem. 2012, 12, 701–712. [Google Scholar] [CrossRef]
- Ebo, D.G.; Venemalm, L.; Bridts, C.H.; Degerbeck, F.; Hagberg, H.; De Clerck, L.S.; Stevens, W.J. Immunoblobulin E antibodies to rocuronium. A new diagnostic tool. Anesthesiology 2007, 107, 253–259. [Google Scholar] [CrossRef]
- Allen, S.J.; Gallagher, A.; Paxton, L.D. Anaphylaxis to rocuronium. Anaesthesia. 2000, 55, 1223–1224. [Google Scholar] [CrossRef]
- Heier, T.; Guttormsen, A.B. Anaphylactic reactions during induction of anaesthesia using rocuronium for muscle relaxation: A report including 3 cases. Acta. Anesthesiol. Scand. 2000, 44, 775–781. [Google Scholar] [CrossRef]
- Neal, S.M.; Manthri, P.R.; Gadiyar, V.; Wildsmith, J.A.W. Histaminoid reactions associated with rocuronium. Br. J. Anaesth. 2000, 84, 108–111. [Google Scholar] [CrossRef]
- Rose, M.; Fisher, M.M. Rocuronium: high risk for anapylaxis. Br. J. Anaesth. 2001, 86, 678–682. [Google Scholar] [CrossRef]
- Bhananker, S.M.; O’Donnell, J.T.; Salemi, J.R.; Bishop, M.J. The risk of anaphylactic reactions to rocuronium in the United States is comparable to that of vecuronium: An analysis of food and drug administration reporting of adverse events. Anesth. Analg. 2005, 101, 819–822. [Google Scholar] [CrossRef]
- Johansson, S.G.O.; Oman, H.; Degerbeck, F.; Tunelli, J.; Florvaag, E.; Nopp, A. Anaphylaxis to atracurium—a non-QAI-dependent reaction? Acta. Anaesthesiol. Scand. 2012, 56, 262–263. [Google Scholar]
- Zhao, Z.; Baldo, B.A.; O’Brien, R.M.; Plomley, R.F. Reaction with, and fine structural recognition of polyamines by human IgE antibodies. Mol. Immunol. 2000, 37, 233–240. [Google Scholar] [CrossRef]
- Baldo, B.A.; Fisher, M.M.; Harle, D.G. Allergy to thiopentone. Clin. Rev. Allergy 1991, 9, 295–307. [Google Scholar]
- Harle, D.G.; Baldo, B.A.; Smal, M.A.; Wajon, P.; Fisher, M.M. Detection of thiopentone-reactive IgE antibodies following anaphylactoid reactions during anaesthesia. Clin. Allergy 1986, 16, 493–498. [Google Scholar] [CrossRef]
- Harle, D.G.; Baldo, B.A.; Smal, M.A.; Fisher, M.M. Drugs as allergens: The molecular basis of IgE binding to thiopentone. Int. Arch. Allergy Appl. Immunol. 1987, 84, 277–283. [Google Scholar] [CrossRef]
- Harle, D.G.; Baldo, B.A.; Fisher, M.M. The molecular basis of IgE antibody binding to thiopentone. Binding of IgE from thiopentone-allergic and non-allergic subjects. Mol. Immunol. 1990, 27, 853–858. [Google Scholar] [CrossRef]
- Fisher, M.M.; Ross, J.D.; Harle, D.G.; Baldo, B.A. Anaphylaxis to thiopentone: An unusual outbreak in a single hospital. Anaesth. Intens. Care 1989, 17, 361–365. [Google Scholar]
- Fisher, M.M.; Baldo, B.A.; Silbert, B.S. Anaphylaxis during anesthesia: Use of radioimmunoassays to determine etiology and drugs responsible in fatal cases. Anesthesiology 1991, 75, 1112–1115. [Google Scholar] [CrossRef]
- Baldo, B.A.; Fisher, M.M. Diagnosis of IgE-dependent anaphylaxis to neuromuscular blocking drugs, thiopentone and opioids. Ann. Fr. Anesth. Reanim. 1993, 12, 173–181. [Google Scholar] [CrossRef]
- Baldo, B.A.; Pham, N.H.; Zhao, Z. Chemistry of drug allergenicity. Curr. Opin. Allergy Clin. Immunol. 2001, 1, 327–335. [Google Scholar]
- Vieluf, D.; Russwurm, R.; Przybilla, B.; Ring, J. Anaphylactic reactions to quinolones. J. Allergy Clin. Immunol. 1991, 87, 228. [Google Scholar]
- Colas, C.; Pola, J.; Zapata, C.; Dominguez, A.; Gimeno, A.; Duce, F. Quinolones hypersensitivity: Clinical and immunological aspects. J. Allergy Clin. Immunol. 1993, 91, 365. [Google Scholar]
- Dávila, I.; Diez, M.L.; Quirce, S.; Fraj, J.; de La Hoz, B.; Lázaro, M. Cross-reactivity between quinolones. Report of three cases. Allergy 1993, 48, 388–390. [Google Scholar] [CrossRef]
- Diaz, M.V.; Labairu, T.L.; del Pozo Gil, M.D.; Sarramián, A.B.; Mahave, I.G. In vivo diagnostic tests in adverse reactions to quinolones. J. Invest. Allergol. Clin. Immunol. 2007, 17, 393–398. [Google Scholar]
- Manfredi, M.; Severino, M.; Testi, S.; Macchia, D.; Ermini, G.; Pichler, W.J.; Campi, P. Detection of specific IgE to quinolones. J. Allergy Clin. Immunol. 2004, 113, 155–160. [Google Scholar] [CrossRef]
- Aranda, A.; Mayorga, C.; Ariza, A.; Doña, I.; Rosado, A.; Blanca-Lopez, N.; Andreu, I.; Torres, M.J. In vitro evaluation of IgE-mediated hypersensitivity to quinolones. Allergy 2011, 66, 247–254. [Google Scholar] [CrossRef]
- Rouzaire, P.; Nosbaum, A.; Mullet, C.; Diot, N.; Dubost, R.; Bienvenu, F.; Guilloux, L.; Piriou, V.; Bienvenu, J.; Bérard, F. Immediate allergic hypersensitivity to quinolones associates with neuromuscular blocking agent sensitization. J. Allergy Clin. Immunol. In Pract. 2013, 1, 273–279. [Google Scholar] [CrossRef]
- Pham, N.H. Immunochemical and diagnostic approaches to drug allergy. Ph.D. thesis, University of Sydney, Sydney, Australia, 1999. [Google Scholar]
- Pham, N.H.; Baldo, B.A.; Puy, R.M. Studies on the mechanism of multiple drug allergies. Structural basis of drug recognition. J. Immunoass. Immunoch. 2001, 22, 47–73. [Google Scholar] [CrossRef]
- Baldo, B.A.; Pham, N.H. On the question of the association between immediate hypersensitivity to quinolones and neuromuscular blocking drug sensitization. J. Allergy Clin. Immunol. In Pract. 2013, 1, 709–710. [Google Scholar] [CrossRef]
- Okano, M.; Nomura, M.; Hata, S.; Okada, N.; Sato, K.; Kitano, Y.; Tashiro, M.; Yoshimoto, Y.; Hama, R.; Aoki, T. Anaphylactic symptoms due to chlorhexidine gluconate. Arch. Dermatol. 1989, 125, 50–52. [Google Scholar] [CrossRef]
- Lim, K.S.; Kam, P.C.A. Chlorhexidine–pharmacology and clinical applications. Anaesth. Intens. Care 2008, 36, 502–512. [Google Scholar]
- Ohtoshi, T.; Yamauchi, N.; Tadokoro, K.; Miyachi, S.; Suzuki, S.; Miyamoto, T.; Muranaka, M. IgE antibody-mediated shock reaction caused by topical application of chlorhexidine. Clin. Allergy 1986, 16, 155–161. [Google Scholar] [CrossRef]
- Pham, N.H.; Weiner, J.M.; Reisner, G.S.; Baldo, B.A. Anaphylaxis to chlorhexidine. Case report. Implication of immunoglobulin E antibodies and identification of an allergenic determinant. Clin. Exp. Allergy 2000, 30, 1001–1007. [Google Scholar] [CrossRef]
- Baldo, B.A.; Pham, N.H. Histamine-releasing and allergenic properties of opioid analgesic drugs: Resolving the two. Anaesth. Intens. Care 2012, 40, 216–235. [Google Scholar]
- Harle, D.G.; Baldo, B.A.; Coroneos, N.J.; Fisher, M.M. Anaphylaxis following administration of papaveretum. Case report: Implication of IgE antibodies that react with morphine and codeine, and identification of an allergenic determinant. Anesthesiology 1989, 71, 489–494. [Google Scholar] [CrossRef]
- Zhu, D.; Becker, W.M.; Schulz, K.H.; Schubeler, K.; Schlaak, M. The presence of specific IgE to salicyloyl and O-methylsalicyloyl in aspirin-sensitive patients. Asian Pac. J. Allergy Immunol. 1992, 10, 25–32. [Google Scholar]
- Zhu, D.; Becker, W.M.; Schulz, K.H.; Schubeler, K.; Schlaak, M. Detection of IgE antibodies specific for 1-phenyl-2,3-dimethyl-3-pyrazoline-5-one by RAST: A serological diagnostic method for sensitivity to pyrazoline drugs. Asian Pac. J. Allergy Immunol. 1992, 10, 95–101. [Google Scholar]
- Himly, M.; Jahn-Schmid, B.; Pittertschatscher, K.; Bohle, B.; Grubmayr, K.; Ferreira, F.; Ebner, H.; Ebner, C. IgE-mediated immediate-type hypersensitivity to the pyrazolone drug propyphenazone. J. Allergy. Clin. Immunol. 2003, 111, 882–888. [Google Scholar] [CrossRef]
- Davies, R.J.; Pepys, J. Asthma due to inhaled chemical agents: The macrolide antibiotic spiramycin. Clin. Allergy 1975, 1, 99–107. [Google Scholar] [CrossRef]
- Pascual, C.; Crespo, J.F.; Quiralte, J.; Lopez, C.; Wheeler, G.; Martin-Esteban, M. In vitro detection of specific IgE antibodies to erythromycin. J. Allergy Clin. Immunol. 1995, 95, 668–671. [Google Scholar] [CrossRef]
- Jorro, G.; Morales, C.; Brasó, J.V.; Peláez, A. Anaphylaxis to erythromycin. Ann. Allergy Asthma Immunol. 1996, 77, 456–458. [Google Scholar] [CrossRef]
- Martinez, E.; Collazos, J.; Mayo, J. Hypersensitivity reactions to rifampin. Pathogenetic mechanisms, clinical manifestations, management strategies, and review of the anaphylactic-like reactions. Medicine 1999, 78, 361–369. [Google Scholar] [CrossRef]
- Magnan, A.; Venemalm, L.; Porro, F.; Vervloet, D. Anaphylactic reaction to rifamycin SV: Presence of specific IgE antibodies. J. Allergy Clin. Immunol. 1999, 103, 954–956. [Google Scholar] [CrossRef]
- Bassi, L.; Di Bernardino, L.; Silvestri, L.G. IgE antibodies in patients allergic to rifampicin. Int. Arch. Allergy Appl. Immunol. 1976, 51, 390–394. [Google Scholar] [CrossRef]
- Antonicelli, L.; Micucci, C.; Bilò, M.B.; Manfredi, M.; Valentini, M.; Campi, P. IgE-mediated reactions to rifaximin and rifamycin SV and cross-reactivity among rifamycins. Allergy 2009, 64, 1232–1233. [Google Scholar] [CrossRef]
- Baldo, B.A.; Zhao, Z.; Pham, N.H. Antibiotic allergy: Immunochemical and clinical considerations. Curr. Allergy Asthma Rep. 2008, 8, 49–55. [Google Scholar] [CrossRef]
- Montañez, M.I.; Mayorga, C.; Torres, M.J.; Ariza, A.; Blanca, M.; Perez-Inestrosa, E. Synthetic approach to gain insight into antigenic determinants of cephalosporins: In vitro studies of chemical structure-IgE molecular recognition relationships. Chem. Res. Toxicol. 2011, 24, 706–717. [Google Scholar] [CrossRef]
- Romano, A.; Viola, M.; Guéant-Rodriguez, R.-M.; Gaeta, F.; Valluzzi, R.; Guéant, J.L. Brief communication: Tolerability of menopenem in patients with IgE-mediated hypersensitivity to penicillins. Ann. Intern. Med. 2007, 146, 266–269. [Google Scholar] [CrossRef]
- Cribb, A.E.; Miller, M.; Leeder, J.S.; Hill, J.; Spielberg, S.P. Reactions of the nitroso and hydroxylamine metabolites of sulfamethoxazole with reduced glutathione. Implications for idiosyncratic toxicity. Drug Metab. Dispos. 1991, 19, 900–906. [Google Scholar]
- Cribb, A.E.; Lee, B.L.; Trepanier, L.A.; Spielberg, S.P. Adverse reactions to sulfonamide and sulfonamide-trimethoprim antimicrobials: Clinical syndromes and pathogenesis. Adverse Drug React. Toxicol. Rev. 1996, 15, 9–50. [Google Scholar]
- Adams, J.M.; Bennett, D.J.; Bom, A.; Clark, J.K.; Feilden, H.; Hutchinson, E.J.; Palin, R.; Prosser, A.; Rees, D.C.; Rosair, G.M.; et al. Cyclodextrin-derived host molecules as reversal agents for the neuromuscular blocker rocuronium bromide: Synthesis and structure-activity relationships. J. Med. Chem. 2002, 45, 1806–1816. [Google Scholar] [CrossRef]
- Zhang, M.-Q. Drug-specific cyclodextrins: The future of rapid neuromuscular block reversal? Drugs Future 2003, 28, 347–354. [Google Scholar] [CrossRef]
- Baldo, B.A.; McDonnell, N.J.; Pham, N.H. Drug-specific cyclodextrins with emphasis on sugammadex, the neuromuscular blocker rocuronium and perioperative anaphylaxis: Implications for drug allergy. Clin. Exp. Allergy 2011, 41, 1663–1678. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
