Molecular Recognition between Aβ-Specific Single-Domain Antibody and Aβ Misfolded Aggregates
Abstract
1. Introduction
2. Materials and Methods
2.1. Gammabody Grafted with the C-Terminal 33–42 Aβ Residues
2.2. Misfolded Aβ Aggregates
2.3. Gammabody-Aβ Complexes
2.4. All-Atom MD Simulation Protocol
3. Results and Discussion
3.1. Structure and Dynamics of the Grafted Gammabodies
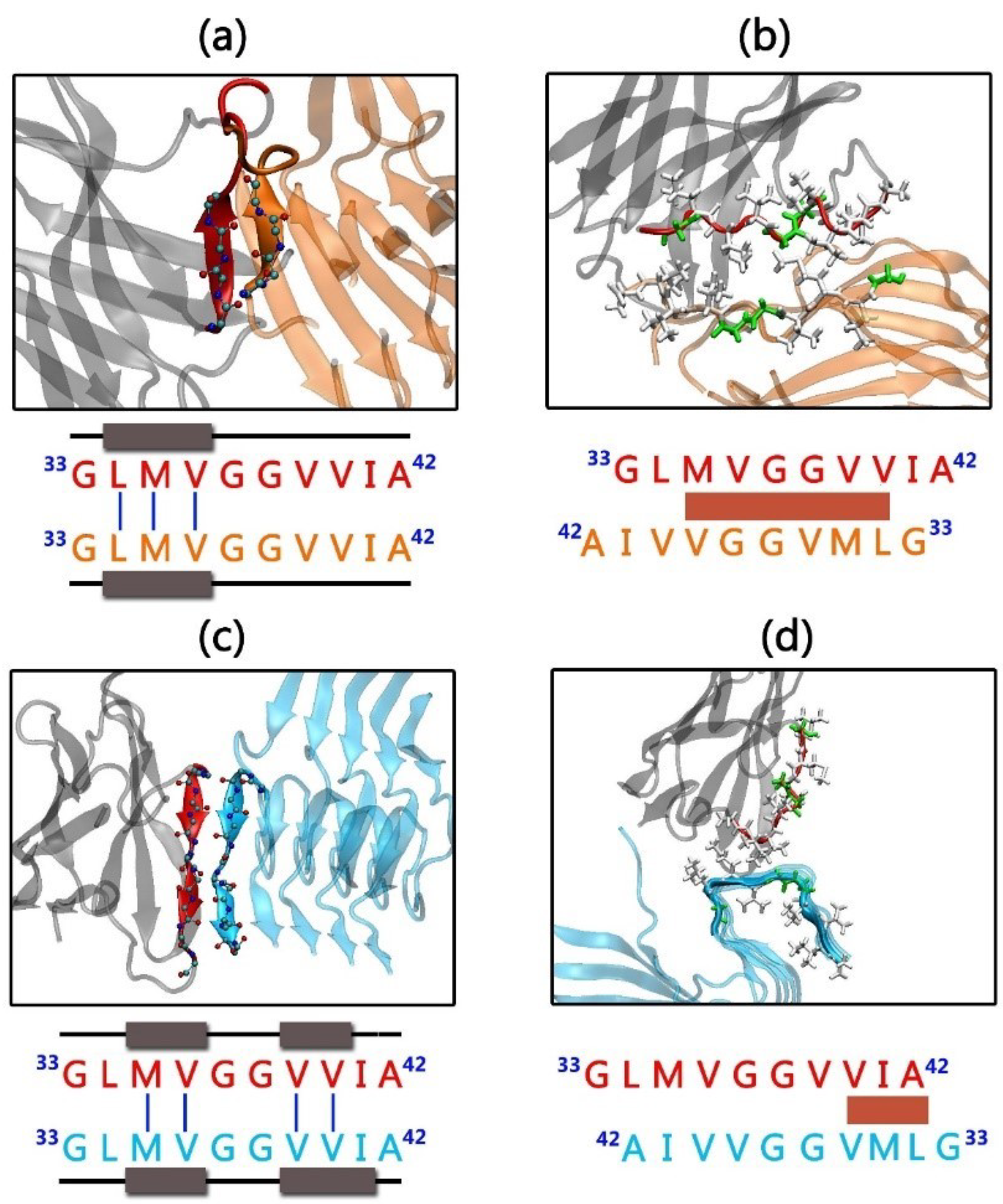
3.2. Gammabody-Aβ Recognitions
3.3. Interaction Energies between Gammabody and Aβ Aggregates
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cleary, J.P.; Walsh, D.M.; Hofmeister, J.J.; Shankar, G.M.; Kuskowski, M.A.; Selkoe, D.J.; Ashe, K.H. Natural oligomers of the amyloid-beta protein specifically disrupt cognitive function. Nat. Neurosci. 2005, 8, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Nussinov, R. Stabilities and conformations of Alzheimer’s beta -amyloid peptide oligomers (Abeta 16–22, Abeta 16–35, and Abeta 10–35): Sequence effects. Proc. Natl. Acad. Sci. USA 2002, 99, 14126–14131. [Google Scholar] [CrossRef] [PubMed]
- Stefani, M. Structural features and cytotoxicity of amyloid oligomers: Implications in Alzheimer’s disease and other diseases with amyloid deposits. Prog. Neurobiol. 2012, 99, 226–245. [Google Scholar] [CrossRef] [PubMed]
- Nasica-Labouze, J.; Nguyen, P.H.; Sterpone, F.; Berthoumieu, O.; Buchete, N.V.; Cote, S.; De Simone, A.; Doig, A.J.; Faller, P.; Garcia, A.; et al. Amyloid beta Protein and Alzheimer’s Disease: When Computer Simulations Complement Experimental Studies. Chem. Rev. 2015, 115, 3518–3563. [Google Scholar] [CrossRef] [PubMed]
- Middleton, C.T.; Marek, P.; Cao, P.; Chiu, C.C.; Singh, S.; Woys, A.M.; De Pablo, J.J.; Raleigh, D.P.; Zanni, M.T. Two-dimensional infrared spectroscopy reveals the complex behaviour of an amyloid fibril inhibitor. Nat. Chem. 2012, 4, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Miller, Y.; Ma, B.; Nussinov, R. Polymorphism in Alzheimer Abeta amyloid organization reflects conformational selection in a rugged energy landscape. Chem. Rev. 2010, 110, 4820–4838. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Nussinov, R. Selective molecular recognition in amyloid growth and transmission and cross-species barriers. J. Mol. Biol. 2012, 421, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Baumketner, A.; Shea, J.E. Folding landscapes of the Alzheimer amyloid-beta(12-28) peptide. J. Mol. Biol. 2006, 362, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Qi, R.; Wei, G.; Nussinov, R.; Ma, B. Self-aggregation and coaggregation of the p53 core fragment with its aggregation gatekeeper variant. Phys. Chem. Chem. Phys. 2016, 18, 8098–8107. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, A.; Balaji, P.V. Size, orientation and organization of oligomers that nucleate amyloid fibrils: Clues from MD simulations of pre-formed aggregates. BBA Proteins Proteom. 2012, 1824, 963–973. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, A. Insights into protein misfolding and amyloidogenesis. Phys. Chem. Chem. Phys. 2013, 15, 8867. [Google Scholar] [CrossRef] [PubMed]
- O’Nuallain, B.; Williams, A.D.; Westermark, P.; Wetzel, R. Seeding specificity in amyloid growth induced by heterologous fibrils. J. Biol. Chem. 2004, 279, 17490–17499. [Google Scholar] [CrossRef] [PubMed]
- Stefani, M. Biochemical and biophysical features of both oligomer/fibril and cell membrane in amyloid cytotoxicity. FEBS J. 2010, 277, 4602–4613. [Google Scholar] [CrossRef] [PubMed]
- Milanesi, L.; Sheynis, T.; Xue, W.F.; Orlova, E.V.; Hellewell, A.L.; Jelinek, R.; Hewitt, E.W.; Radford, S.E.; Saibil, H.R. Direct three-dimensional visualization of membrane disruption by amyloid fibrils. Proc. Natl. Acad. Sci. USA 2012, 109, 20455–20460. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Z.; Zhao, J.; Zheng, J. Molecular understanding of a potential functional link between antimicrobial and amyloid peptides. Soft Matter 2014, 10, 7425–7451. [Google Scholar] [CrossRef] [PubMed]
- Tofoleanu, F.; Brooks, B.R.; Buchete, N.V. Modulation of Alzheimer’s A beta Protofilament-Membrane Interactions by Lipid Headgroups. ACS Chem. Neurosci. 2015, 6, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Wang, Z.; Wang, D.; Wang, Z.; Martens, Y.A.; Wu, L.; Xu, Y.; Wang, K.; Li, J.; Huang, R.; et al. Amyloid-beta modulates microglial responses by binding to the triggering receptor expressed on myeloid cells 2 (TREM2). Mol. Neurodegener. 2018, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wu, X.; Li, X.; Jiang, L.L.; Gui, X.; Liu, Y.; Sun, Y.; Zhu, B.; Pina-Crespo, J.C.; Zhang, M.; et al. TREM2 Is a Receptor for beta-Amyloid that Mediates Microglial Function. Neuron 2018, 97, 1023.e7–1031.e7. [Google Scholar] [CrossRef] [PubMed]
- Yeh, F.L.; Hansen, D.V.; Sheng, M. TREM2, Microglia, and Neurodegenerative Diseases. Trends Mol. Med. 2017, 23, 512–533. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Qian, Z.; Wei, G. The inhibitory mechanism of a fullerene derivative against amyloid-beta peptide aggregation: An atomistic simulation study. Phys. Chem. Chem. Phys. 2016, 18, 12582–12591. [Google Scholar] [CrossRef] [PubMed]
- Bieschke, J.; Russ, J.; Friedrich, R.P.; Ehrnhoefer, D.E.; Wobst, H.; Neugebauer, K.; Wanker, E.E. EGCG remodels mature alpha-synuclein and amyloid-beta fibrils and reduces cellular toxicity. Proc. Natl. Acad. Sci. USA 2010, 107, 7710–7715. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.M.; Yu, X.; Patal, K.; Hu, R.D.; Chuang, S.; Zhang, G.; Zheng, J. Tanshinones Inhibit Amyloid Aggregation by Amyloid-beta Peptide, Disaggregate Amyloid Fibrils, and Protect Cultured Cells. ACS Chem Neurosci. 2013, 4, 1004–1015. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.S.; Lim, G.P.; Begum, A.N.; Ubeda, O.J.; Simmons, M.R.; Ambegaokar, S.S.; Chen, P.P.; Kayed, R.; Glabe, C.G.; Frautschy, S.A.; et al. Curcumin inhibits formation of amyloid beta oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J. Biol. Chem. 2005, 280, 5892–5901. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Men, P.; Kudo, W.; Perry, G.; Smith, M.A. Nanoparticle-chelator conjugates as inhibitors of amyloid-beta aggregation and neurotoxicity: A novel therapeutic approach for Alzheimer disease. Neurosci. Lett. 2009, 455, 187–190. [Google Scholar] [CrossRef] [PubMed]
- Cabaleiro-Lago, C.; Quinlan-Pluck, F.; Lynch, I.; Lindman, S.; Minogue, A.M.; Thulin, E.; Walsh, D.M.; Dawson, K.A.; Linse, S. Inhibition of Amyloid beta Protein Fibrillation by Polymeric Nanoparticles. J. Am. Chem. Soc. 2008, 130, 15437–15443. [Google Scholar] [CrossRef] [PubMed]
- Olmedo, I.; Araya, E.; Sanz, F.; Medina, E.; Arbiol, J.; Toledo, P.; Alvarez-Lueje, A.; Giralt, E.; Kogan, M.J. How changes in the sequence of the peptide CLPFFD-NH(2) can modify the conjugation and stability of gold nanoparticles and their affinity for beta-amyloid fibrils. Bioconj. Chem. 2008, 19, 1154–1163. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.D.; Zhang, M.Z.; Patel, K.; Wang, Q.M.; Chang, Y.; Gong, X.; Zhang, G.; Zheng, J. Cross-Sequence Interactions between Human and Rat Islet Amyloid Polypeptides. Langmuir 2014, 30, 5193–5201. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Z.; Hu, R.D.; Liang, G.Z.; Chang, Y.; Sun, Y.; Peng, Z.M.; Zheng, J. Structural and Energetic Insight into the Cross-Seeding Amyloid Assemblies of Human IAPP and Rat IAPP. J. Phys. Chem. B 2014, 118, 7026–7036. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Z.; Hu, R.D.; Chen, H.; Chang, Y.; Gong, X.; Liu, F.F.; Zheng, J. Interfacial interaction and lateral association of cross-seeding assemblies between hIAPP and rIAPP oligomers. Phys. Chem. Chem. Phys. 2015, 17, 10373–10382. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.D.; Zhang, M.Z.; Chen, H.; Jiang, B.B.; Zheng, J. Cross-Seeding Interaction between beta-Amyloid and Human Islet Amyloid Polypeptide. ACS Chem. Neurosci. 2015, 6, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Andreetto, E.; Yan, L.M.; Caporale, A.; Kapurniotu, A. Dissecting the Role of Single Regions of an IAPP Mimic and IAPP in Inhibition of A beta 40 Amyloid Formation and Cytotoxicity. ChemBioChem 2011, 12, 1313–1322. [Google Scholar] [CrossRef] [PubMed]
- Andreetto, E.; Yan, L.M.; Tatarek-Nossol, M.; Velkova, A.; Frank, R.; Kapurniotu, A. Identification of Hot Regions of the A beta-IAPP Interaction Interface as High-Affinity Binding Sites in both Cross- and Self-Association. Angew. Chem. Int. Ed. 2010, 49, 3081–3085. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.M.; Liang, G.Z.; Zhang, M.Z.; Zhao, J.; Patel, K.; Yu, X.; Zhao, C.; Ding, B.R.; Zhang, G.; Zhou, F.M.; et al. De Novo Design of Self-Assembled Hexapeptides as beta-Amyloid (A beta) Peptide Inhibitors. ACS Chem. Neurosci. 2014, 5, 972–981. [Google Scholar] [CrossRef] [PubMed]
- Sievers, S.A.; Karanicolas, J.; Chang, H.W.; Zhao, A.; Jiang, L.; Zirafi, O.; Stevens, J.T.; Munch, J.; Baker, D.; Eisenberg, D. Structure-based design of non-natural amino-acid inhibitors of amyloid fibril formation. Nature 2011, 475, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, Y.L.; Zhang, J.; Zhang, M.Z.; Zheng, J.; Teng, Y.; Liang, G.Z. A quantitative sequence-aggregation relationship predictor applied as identification of self-assembled hexapeptides. Chemom. Intell. Lab. 2015, 145, 7–16. [Google Scholar] [CrossRef]
- Kapurniotu, A.; Schmauder, A.; Tenidis, K. Structure-based design and study of non-amyloidogenic, double N-methylated IAPP amyloid core sequences as inhibitors of IAPP amyloid formation and cytotoxicity. J. Mol. Biol. 2002, 315, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.N.; Liu, C.; Zhao, M.L.; Eisenberg, D.; Nowick, J.S. Amyloid beta-sheet mimics that antagonize protein aggregation and reduce amyloid toxicity. Nat. Chem. 2012, 4, 927–933. [Google Scholar] [CrossRef] [PubMed]
- O’Nuallain, B.; Wetzel, R. Conformational Abs recognizing a generic amyloid fibril epitope. Proc. Natl. Acad. Sci. USA 2002, 99, 1485–1490. [Google Scholar] [CrossRef] [PubMed]
- Habicht, G.; Haupt, C.; Friedrich, R.P.; Hortschansky, P.; Sachse, C.; Meinhardt, J.; Wieligmann, K.; Gellermann, G.P.; Brodhum, M.; Gotz, J.; et al. Directed selection of a conformational antibody domain that prevents mature amyloid fibril formation by stabilizing A beta protofibrils. Proc. Natl. Acad. Sci. USA 2007, 104, 19232–19237. [Google Scholar] [CrossRef] [PubMed]
- Gardberg, A.S.; Dice, L.T.; Ou, S.; Rich, R.L.; Heimbrecht, E.; Ko, J.; Wetzel, R.; Myszka, D.G.; Patterson, P.H.; Dealwis, C. Molecular basis for passive immunotherapy of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2007, 104, 15659–15664. [Google Scholar] [CrossRef] [PubMed]
- Lambert, M.P.; Velasco, P.T.; Chang, L.; Viola, K.L.; Fernandez, S.; Lacor, P.N.; Khuon, D.; Gong, Y.S.; Bigio, E.H.; Shaw, P.; et al. Monoclonal antibodies that target pathological assemblies of A beta. J. Neurochem. 2007, 100, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Haupt, C.; Morgado, I.; Kumar, S.T.; Parthier, C.; Bereza, M.; Hortschansky, P.; Stubbs, M.T.; Horn, U.; Fandrich, M. Amyloid Fibril Recognition with the Conformational B10 Antibody Fragment Depends on Electrostatic Interactions. J. Mol. Biol. 2011, 405, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Morgado, I.; Wieligmann, K.; Bereza, M.; Ronicke, R.; Meinhardt, K.; Annamalai, K.; Baumann, M.; Wacker, J.; Hortschansky, P.; Malesevic, M.; et al. Molecular basis of beta-amyloid oligomer recognition with a conformational antibody fragment. Proc. Natl. Acad. Sci. USA 2012, 109, 12503–12508. [Google Scholar] [CrossRef] [PubMed]
- Dodel, R.; Hampel, H.; Depboylu, C.; Lin, S.Z.; Gao, F.; Shock, S.; Jackel, S.; Wei, W.; Buerger, K.; Hoft, C.; et al. Human antibodies against amyloid beta peptide: A potential treatment for Alzheimer’s disease. Ann. Neurol. 2002, 52, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Doody, R.S. Phase 3 Trials of Solanezumab for Mild-to-Moderate Alzheimer’s Disease (vol 370, pg 311, 2014). N. Engl. J. Med. 2014, 371, 584. [Google Scholar]
- Miles, L.A.; Crespi, G.A.; Doughty, L.; Parker, M.W. Bapineuzumab captures the N-terminus of the Alzheimer’s disease amyloid-beta peptide in a helical conformation. Sci. Rep. 2013, 3, 1302. [Google Scholar] [CrossRef] [PubMed]
- Bohrmann, B.; Baumann, K.; Benz, J.; Gerber, F.; Huber, W.; Knoflach, F.; Messer, J.; Oroszlan, K.; Rauchenberger, R.; Richter, W.F.; et al. Gantenerumab: A novel human anti-Abeta antibody demonstrates sustained cerebral amyloid-beta binding and elicits cell-mediated removal of human amyloid-beta. J. Alzheimers Dis. 2012, 28, 49–69. [Google Scholar] [CrossRef] [PubMed]
- La Porte, S.L.; Bollini, S.S.; Lanz, T.A.; Abdiche, Y.N.; Rusnak, A.S.; Ho, W.H.; Kobayashi, D.; Harrabi, O.; Pappas, D.; Mina, E.W.; et al. Structural Basis of C-terminal beta-Amyloid Peptide Binding by the Antibody Ponezumab for the Treatment of Alzheimer’s Disease. J. Mol. Biol. 2012, 421, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Sudduth, T.L.; Greenstein, A.; Wilcock, D.M. Intracranial Injection of Gammagard, a Human IVIg, Modulates the Inflammatory Response of the Brain and Lowers A beta in APP/PS1 Mice Along a Different Time Course than Anti-A beta Antibodies. J. Neurosci. 2013, 33, 9684–9692. [Google Scholar] [CrossRef] [PubMed]
- Ochs, H.D.; Pinciaro, P.J.; Grp, O.S. Octagam (R) 5%, an intravenous IgG product, is efficacious and well tolerated in subjects with primary immunodeficiency diseases. J. Clin. Immunol. 2004, 24, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Tayeb, H.O.; Murray, E.D.; Price, B.H.; Tarazi, F.I. Bapineuzumab and solanezumab for Alzheimer’s disease: Is the ‘amyloid cascade hypothesis’ still alive? Expert Opin. Biol. Ther. 2013, 13, 1075–1084. [Google Scholar] [CrossRef] [PubMed]
- Verdile, G.; Fuller, S.; Atwood, C.S.; Laws, S.M.; Gandy, S.E.; Martins, R.N. The role of beta amyloid in Alzheimer’s disease: Still a cause of everything or the only one who got caught? Pharmacol. Res. 2004, 50, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Rahman, K.S.; Chowdhury, E.U.; Sachse, K.; Kaltenboeck, B. Inadequate Reference Datasets Biased toward Short Non-epitopes Confound B-cell Epitope Prediction. J. Biol. Chem. 2016, 291, 14585–14599. [Google Scholar] [CrossRef] [PubMed]
- Ladiwala, A.R.A.; Bhattacharya, M.; Perchiacca, J.M.; Cao, P.; Raleigh, D.P.; Abedini, A.; Schmidt, A.M.; Varkey, J.; Langen, R.; Tessier, P.M. Rational design of potent domain antibody inhibitors of amyloid fibril assembly (vol 109, pg 19965, 2012). Proc. Natl. Acad. Sci. USA 2013, 110, 1560. [Google Scholar] [CrossRef]
- Perchiacca, J.M.; Ladiwala, A.R.A.; Bhattacharya, M.; Tessier, P.M. Structure-based design of conformation- and sequence-specific antibodies against amyloid beta. Proc. Natl. Acad. Sci. USA 2012, 109, 84–89. [Google Scholar] [CrossRef] [PubMed]
- Perchiacca, J.M.; Tessier, P.M. Engineering Aggregation-Resistant Antibodies. Annu. Rev. Chem. Biomol. 2012, 3, 263–286. [Google Scholar] [CrossRef] [PubMed]
- Bond, C.J.; Marsters, J.C.; Sidhu, S.S. Contributions of CDR3 to VHH domain stability and the design of monobody scaffolds for naive antibody libraries. J. Mol. Biol. 2003, 332, 643–655. [Google Scholar] [CrossRef]
- Barthelemy, P.A.; Raab, H.; Appleton, B.A.; Bond, C.J.; Wu, P.; Wiesmann, C.; Sidhu, S.S. Comprehensive analysis of the factors contributing to the stability and solubility of autonomous human V-H domains. J. Biol. Chem. 2008, 283, 3639–3654. [Google Scholar] [CrossRef] [PubMed]
- Perchiacca, J.M.; Lee, C.C.; Tessier, P.M. Optimal charged mutations in the complementarity-determining regions that prevent domain antibody aggregation are dependent on the antibody scaffold. Protein Eng. Des. Sel. 2014, 27, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Tiller, K.E.; Li, L.; Kumar, S.; Julian, M.C.; Garde, S.; Tessier, P.M. Arginine mutations in antibody complementarity-determining regions display context-dependent affinity/specificity trade-offs. J. Biol. Chem. 2017, 292, 16638–16652. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhu, D.; Zhu, J.; Nussinov, R.; Ma, B. Local and global anatomy of antibody-protein antigen recognition. J. Mol. Recognit. 2017. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Zhao, J.; Nussinov, R. Conformational selection in amyloid-based immunotherapy: Survey of crystal structures of antibody-amyloid complexes. Biochim. Biophys. Acta 2016, 1860 Pt B, 2672–2681. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Nussinov, R.; Ma, B. Mechanisms of recognition of Abeta monomer, oligomer, and fibril by homologous antibodies. J. Biol. Chem. 2017, 44, 18325–18343. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.Z.; Hu, R.D.; Chen, H.; Chang, Y.; Ma, J.; Liang, G.Z.; Mi, J.Y.; Wang, Y.R.; Zheng, J. Polymorphic cross-seeding amyloid assemblies of amyloid-beta and human islet amyloid polypeptide. Phys. Chem. Chem. Phys. 2015, 17, 23245–23256. [Google Scholar] [CrossRef] [PubMed]
- Luhrs, T.; Ritter, C.; Adrian, M.; Riek-Loher, D.; Bohrmann, B.; Dobeli, H.; Schubert, D.; Riek, R. 3D structure of Alzheimer’s amyloid-beta(1–42) fibrils. Proc. Natl. Acad. Sci. USA 2005, 102, 17342–17347. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Ma, B.; McElheny, D.; Parthasarathy, S.; Long, F.; Hoshi, M.; Nussinov, R.; Ishii, Y. Abeta(1–42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat. Struct. Mol. Biol. 2015, 22, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Jang, H.; Ma, B.; Tsai, C.J.; Nussinov, R. Modeling the Alzheimer Abeta17–42 fibril architecture: Tight intermolecular sheet-sheet association and intramolecular hydrated cavities. Biophys. J. 2007, 93, 3046–3057. [Google Scholar] [CrossRef] [PubMed]
- Miller, Y.; Ma, B.; Nussinov, R. Polymorphism of Alzheimer’s Abeta17–42 (p3) oligomers: The importance of the turn location and its conformation. Biophys. J. 2009, 97, 1168–1177. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.G.; Luo, Y.; Wei, G.H. A beta(16–22) Peptides Can Assemble into Ordered beta-Barrels and Bilayer beta-Sheets, while Substitution of Phenylalanine 19 by Tryptophan Increases the Population of Disordered Aggregates. J. Phys. Chem. B 2013, 117, 10149–10160. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kale, L.; Schulten, K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Bowers, M.T.; Shea, J.-E. Molecular structures of quiescently grown and brain-derived polymorphic fibrils of the Alzheimer amyloid Aβ9–40 peptide: A comparison to agitated fibrils. PLoS Comput. Biol. 2010, 6, e1000693. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hu, R.; Chen, H.; Gong, X.; Zhou, F.; Zhang, L.; Zheng, J. Polymorphic Associations and Structures of the Cross-Seeding of Abeta1–42 and hIAPP1-37 Polypeptides. J. Chem. Inf. Model. 2015, 55, 1628–1639. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Ma, B.; Chang, Y.; Nussinov, R. Molecular dynamics simulations of Alzheimer Abeta40 elongation and lateral association. Front. Biosci. 2008, 13, 3919–3930. [Google Scholar] [PubMed]
- Levine, Z.A.; Larini, L.; LaPointe, N.E.; Feinstein, S.C.; Shea, J.E. Regulation and aggregation of intrinsically disordered peptides. Proc. Natl. Acad. Sci. USA 2015, 112, 2758–2763. [Google Scholar] [CrossRef] [PubMed]
- Shea, J.E.; Urbanc, B. Insights into A beta Aggregation: A Molecular Dynamics Perspective. Curr. Top. Med. Chem. 2012, 12, 2596–2610. [Google Scholar] [CrossRef] [PubMed]







| System | ΔGgas (kcal/mol) | ΔGsol (kcal/mol) | TΔS (kcal/mol) | Etotal (kcal/mol) a | ΔGbinding b (kcal/mol) |
|---|---|---|---|---|---|
| 2U_Backbone | −363.7 ± 32.1 | 273.6 ± 30.7 | −26.0 | −1442.9 ± 49.0 | −64.2 ± 8.6 |
| 2U_SideChain | −283.9 ± 16.2 | 193.1 ± 15.1 | −25.9 | −1431.9 ± 45.4 | −64.8 ± 5.5 |
| 3S_Backbone | −117.6 ± 14.7 | 61.5 ± 13.8 | −26.1 | −1974.1 ± 48.0 | −30.0 ± 4.9 |
| 3S_SideChain | −94.3 ± 10.1 | 42.5 ± 6.4 | −26.1 | −1968.4 ± 50.2 | −25.7 ± 6.3 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Zheng, J.; Nussinov, R.; Ma, B. Molecular Recognition between Aβ-Specific Single-Domain Antibody and Aβ Misfolded Aggregates. Antibodies 2018, 7, 25. https://doi.org/10.3390/antib7030025
Zhang M, Zheng J, Nussinov R, Ma B. Molecular Recognition between Aβ-Specific Single-Domain Antibody and Aβ Misfolded Aggregates. Antibodies. 2018; 7(3):25. https://doi.org/10.3390/antib7030025
Chicago/Turabian StyleZhang, Mingzhen, Jie Zheng, Ruth Nussinov, and Buyong Ma. 2018. "Molecular Recognition between Aβ-Specific Single-Domain Antibody and Aβ Misfolded Aggregates" Antibodies 7, no. 3: 25. https://doi.org/10.3390/antib7030025
APA StyleZhang, M., Zheng, J., Nussinov, R., & Ma, B. (2018). Molecular Recognition between Aβ-Specific Single-Domain Antibody and Aβ Misfolded Aggregates. Antibodies, 7(3), 25. https://doi.org/10.3390/antib7030025






