Figure 1.
3D stack sample decomposition/composition.
Figure 1.
3D stack sample decomposition/composition.
Figure 2.
(a) Individual slices for sample 6 - „2013-01-23-col0-8_GC1“. (b) 3D view of sample 6 - „2013-01-23-col0-8_GC1“.
Figure 2.
(a) Individual slices for sample 6 - „2013-01-23-col0-8_GC1“. (b) 3D view of sample 6 - „2013-01-23-col0-8_GC1“.
Figure 3.
(a) Individual slices for sample 6 - „2013-01-23-col0-8_GC1“. (b) 3D view of sample 6 - „2013-01-23-col0-8_GC1“.
Figure 3.
(a) Individual slices for sample 6 - „2013-01-23-col0-8_GC1“. (b) 3D view of sample 6 - „2013-01-23-col0-8_GC1“.
Figure 4.
MATLAB plot—CNN architecture.
Figure 4.
MATLAB plot—CNN architecture.
Figure 5.
Diagram for the voter operator.
Figure 5.
Diagram for the voter operator.
Figure 6.
Graphical demonstration of the median operator. Suppose that there are five generated CNNs: (a) Each of CNNs made semantic segmentation for a slice, with grey pixels being background label, and white pixels being foreground (nucleus) label. (b) After sorting resulting segmentation slices, median slice can be selected.
Figure 6.
Graphical demonstration of the median operator. Suppose that there are five generated CNNs: (a) Each of CNNs made semantic segmentation for a slice, with grey pixels being background label, and white pixels being foreground (nucleus) label. (b) After sorting resulting segmentation slices, median slice can be selected.
Figure 7.
Let ground truth image (GTI) be defined as referenced slice, and segmented image (SI) as segmented slice calculated from semantic segmentation of CNN. White value represents foreground (nucleus) label, and black value represents background label. If the GTI (white (+)) pixel goes into SI (white (+)) pixel, that is TP. If the GTI (white (+)) pixel goes into SI (black (-)) pixel, that is FN. If the GTI (black (-)) pixel goes into SI (white (+)) pixel that is FP. If the GTI (black (-)) pixel goes into SI (black (-)) pixel, that is TN.
Figure 7.
Let ground truth image (GTI) be defined as referenced slice, and segmented image (SI) as segmented slice calculated from semantic segmentation of CNN. White value represents foreground (nucleus) label, and black value represents background label. If the GTI (white (+)) pixel goes into SI (white (+)) pixel, that is TP. If the GTI (white (+)) pixel goes into SI (black (-)) pixel, that is FN. If the GTI (black (-)) pixel goes into SI (white (+)) pixel that is FP. If the GTI (black (-)) pixel goes into SI (black (-)) pixel, that is TN.
Figure 8.
Generic diagram of the complete process.
Figure 8.
Generic diagram of the complete process.
Figure 9.
Generic diagram of GGTI DL variants 1A−2B.
Figure 9.
Generic diagram of GGTI DL variants 1A−2B.
Figure 10.
Generic diagram of GGTI DL variants 3A and AB.
Figure 10.
Generic diagram of GGTI DL variants 3A and AB.
Figure 11.
(a) Training block for variant 1A. (b) Training block for variant 1B. (c) Training block for variant 2A and 3A. (d) Training block for variant 2B and 3B.
Figure 11.
(a) Training block for variant 1A. (b) Training block for variant 1B. (c) Training block for variant 2A and 3A. (d) Training block for variant 2B and 3B.
Figure 12.
Diagram of software framework. It presents software implementation of modules, as well as inputs and outputs.
Figure 12.
Diagram of software framework. It presents software implementation of modules, as well as inputs and outputs.
Figure 13.
(a) Global GUI contains links to all other important GUI elements, as well as some other additional options and helpers. (b) GUI for MGTI—Module 1. (c) GUI for GGTI AP7—Module 2. (d) GUI (in usage) for variants 1A, 1B, 2A and 2B—Module 3.
Figure 13.
(a) Global GUI contains links to all other important GUI elements, as well as some other additional options and helpers. (b) GUI for MGTI—Module 1. (c) GUI for GGTI AP7—Module 2. (d) GUI (in usage) for variants 1A, 1B, 2A and 2B—Module 3.
Figure 14.
(a) GUI for GGTI DL variants 3A and 3B—Module 3. (b) Module 4 GUI offers load and preview of referenced, segmented and original slice, as well as export to MAT/TIFF data types, etc. (c) The second Module 4 GUI offers calculation of all metrics. (d) The GUI for batch 3D segmentation and evaluation.
Figure 14.
(a) GUI for GGTI DL variants 3A and 3B—Module 3. (b) Module 4 GUI offers load and preview of referenced, segmented and original slice, as well as export to MAT/TIFF data types, etc. (c) The second Module 4 GUI offers calculation of all metrics. (d) The GUI for batch 3D segmentation and evaluation.
Figure 21.
(a) Sample image/slice of BBBC039 dataset. (b) Sample image/slice of BBBC035 dataset.
Figure 21.
(a) Sample image/slice of BBBC039 dataset. (b) Sample image/slice of BBBC035 dataset.
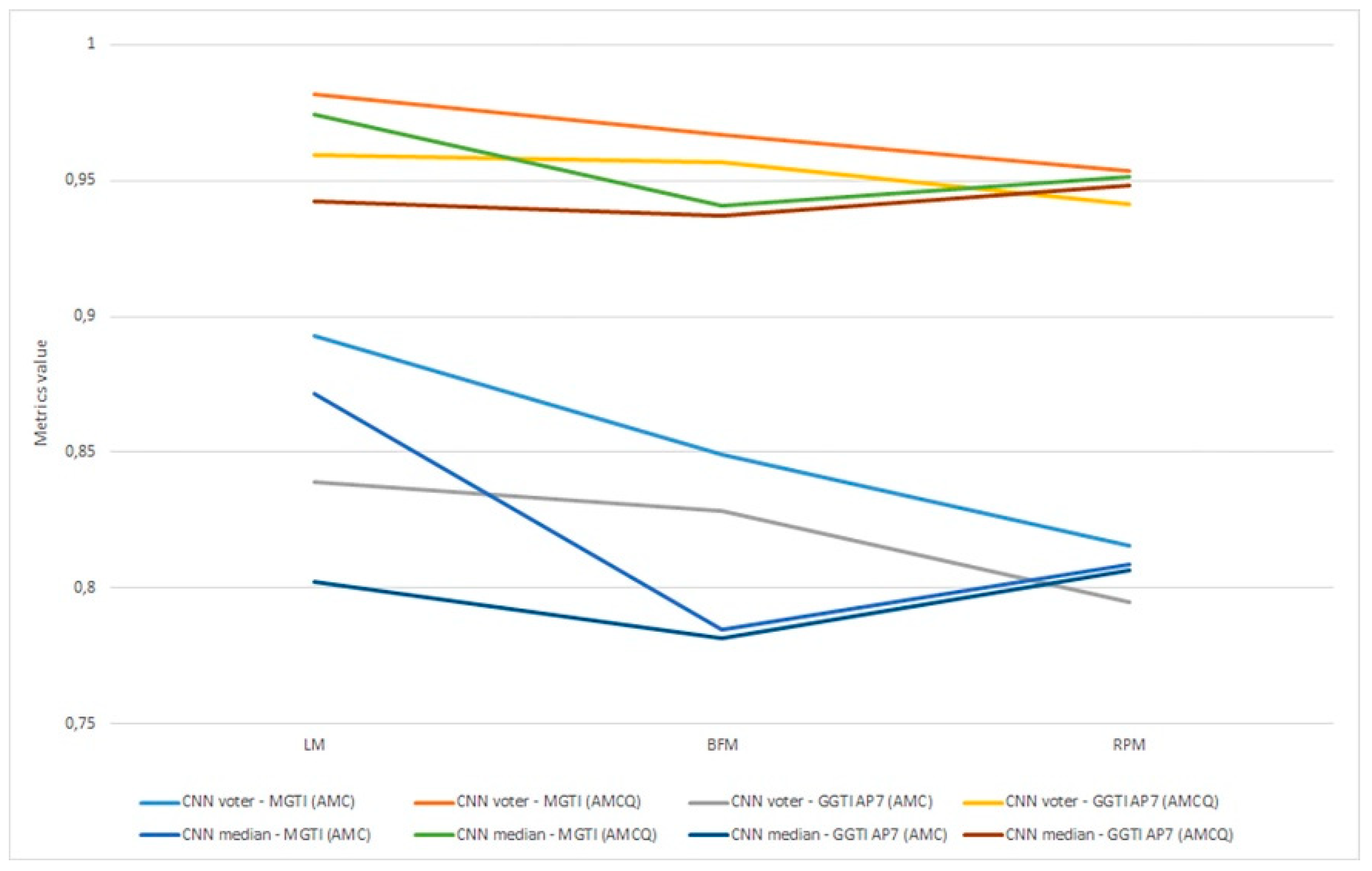
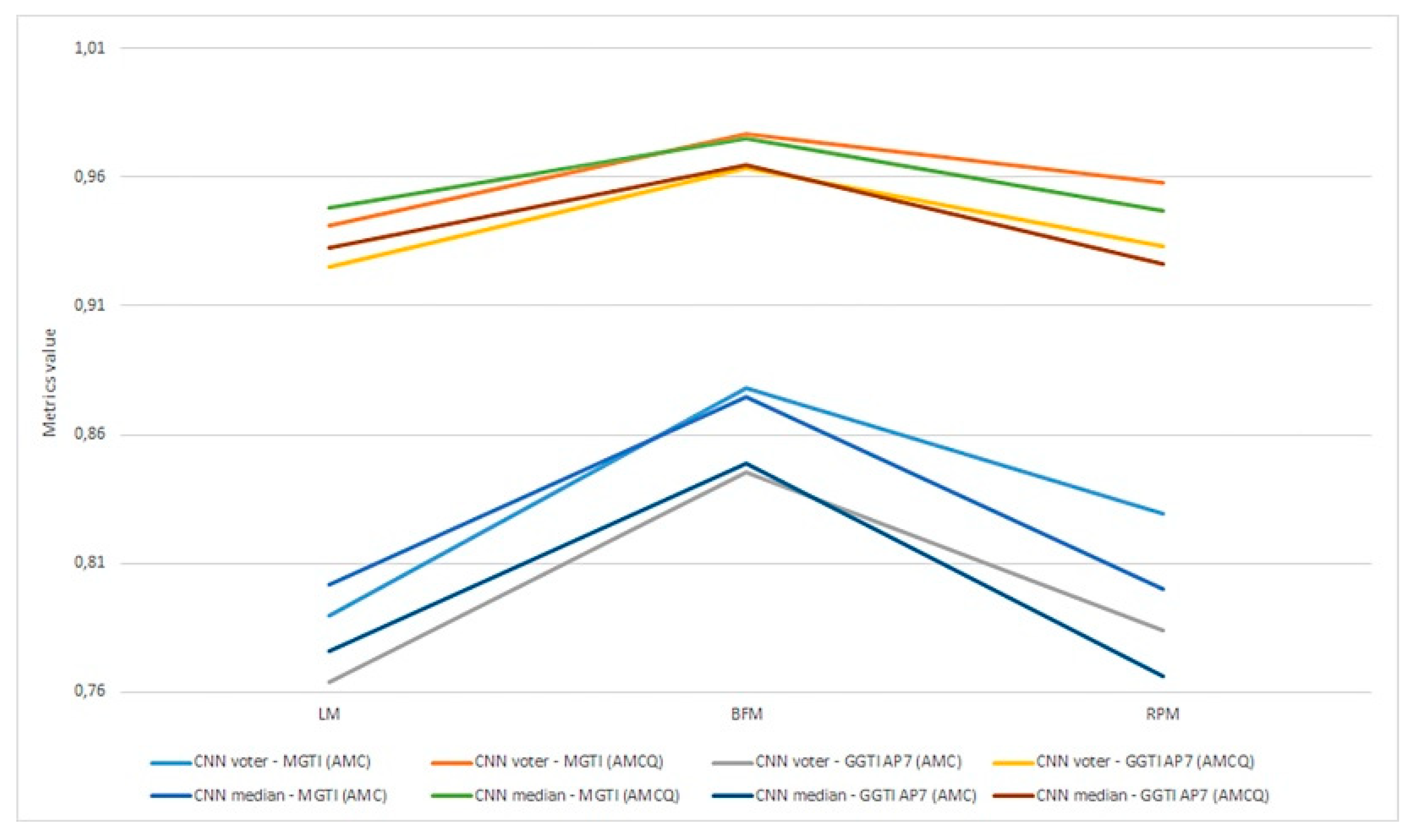
Figure 22.
(a) Experiment 1-6, results for LPM method and metrics. (b) Experiment 1-6, results for BFM method and metrics. (c) Experiment 1–6, results for RPM method and metrics.
Figure 22.
(a) Experiment 1-6, results for LPM method and metrics. (b) Experiment 1-6, results for BFM method and metrics. (c) Experiment 1–6, results for RPM method and metrics.
Figure 23.
(a) Variant 1A—segmented slice of MGTI and LM CNN voter. (b) Variant 1B—segmented slices of MGTI and LM CNN voter.
Figure 23.
(a) Variant 1A—segmented slice of MGTI and LM CNN voter. (b) Variant 1B—segmented slices of MGTI and LM CNN voter.
Figure 24.
(a) Variant 2A—segmented slices of MGTI and LM CNN voter. (b) Variant 2B—segmented slices of MGTI and LM CNN median.
Figure 24.
(a) Variant 2A—segmented slices of MGTI and LM CNN voter. (b) Variant 2B—segmented slices of MGTI and LM CNN median.
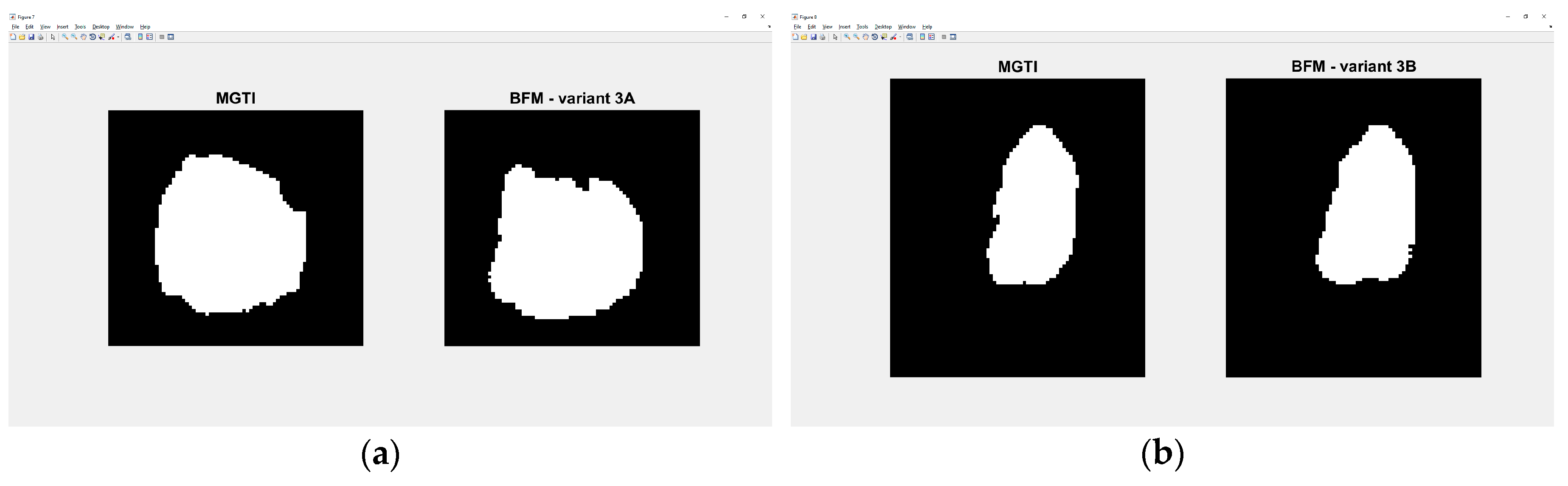
Figure 25.
(a) Variant 3A—segmented slice of MGTI and BFM CNN voter. (b) Variant 3B—segmented slices of MGTI and BFM CNN voter.
Figure 25.
(a) Variant 3A—segmented slice of MGTI and BFM CNN voter. (b) Variant 3B—segmented slices of MGTI and BFM CNN voter.
Figure 26.
Experiment 7, results for six 3D stack samples and AMCQ metrics.
Figure 26.
Experiment 7, results for six 3D stack samples and AMCQ metrics.
Figure 27.
Segmentation of first and last five slices of Arabidopsis thaliana sample 1. GGTI AP7 is not having controlled segmentation over darker slices of samples, while GGTI DL gives controlled and reliable segmentation.
Figure 27.
Segmentation of first and last five slices of Arabidopsis thaliana sample 1. GGTI AP7 is not having controlled segmentation over darker slices of samples, while GGTI DL gives controlled and reliable segmentation.
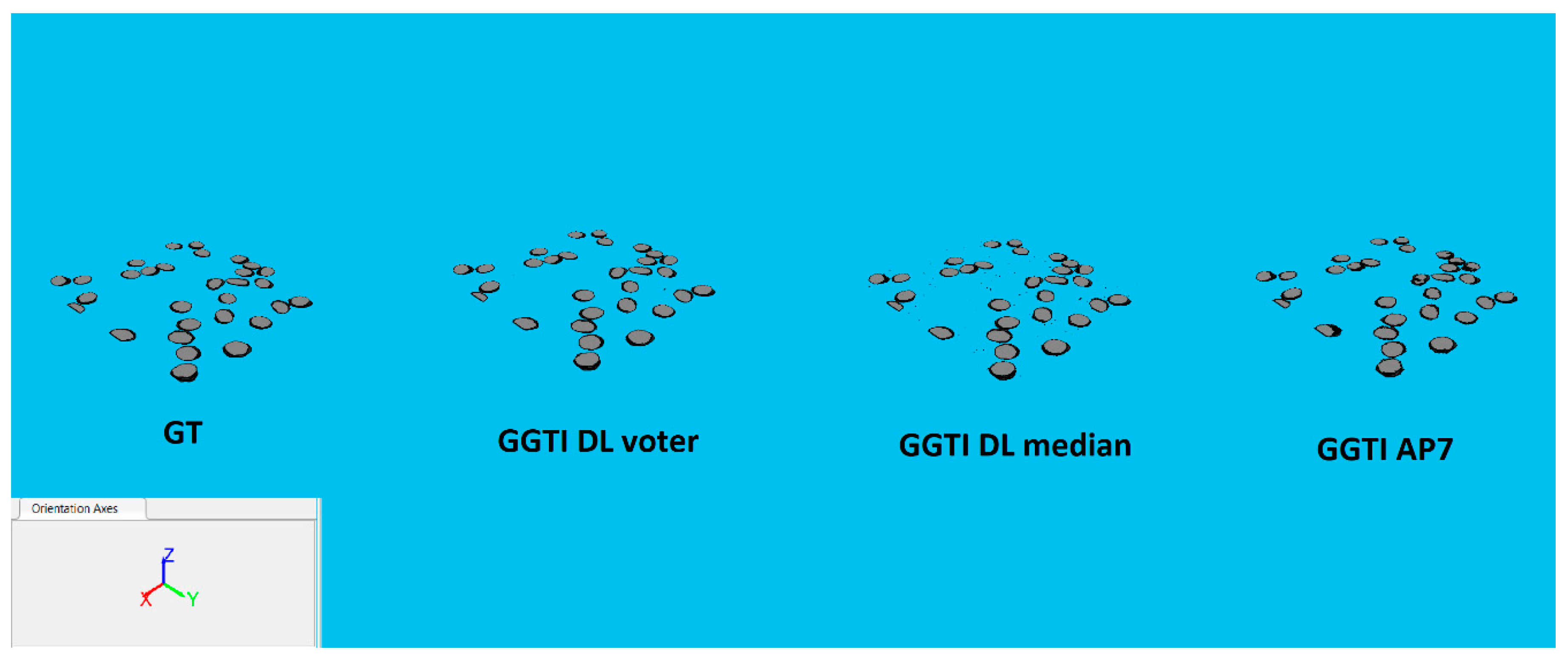
Figure 28.
3D segmentations of sample 4: MGTI, GGTI DL CNN voter, GGTI DL CNN median and GGTI AP7.
Figure 28.
3D segmentations of sample 4: MGTI, GGTI DL CNN voter, GGTI DL CNN median and GGTI AP7.
Figure 29.
Experiment 8, results for all generalization variants and AMCQ metrics.
Figure 29.
Experiment 8, results for all generalization variants and AMCQ metrics.
Figure 30.
3D segmentations of BBBC039 testing sample: GT, GGTI DL CNN voter, GGTI DL CNN median and GGTI AP7.
Figure 30.
3D segmentations of BBBC039 testing sample: GT, GGTI DL CNN voter, GGTI DL CNN median and GGTI AP7.
Figure 31.
3D segmentations of BBBC035 testing sample: GT, GGTI DL CNN voter, GGTI DL CNN median and GGTI AP7.
Figure 31.
3D segmentations of BBBC035 testing sample: GT, GGTI DL CNN voter, GGTI DL CNN median and GGTI AP7.
Table 1.
Preview of variants 1A–3B.
Table 1.
Preview of variants 1A–3B.
| Variant | Training | Usage of Pre-Trained CNNs | Semantic Segmentation—Validation |
|---|
| Number of Slices Per 3D Stack Sample | Number of 3D Stack Samples |
|---|
| Variant 1A | 1 | 1 | No | Validation slice belongs to training 3D stack sample, different than training slice. |
| Variant 1B | 1 | >1 | No | Validation slice belongs to 3D stack sample that has not been used in training. |
| Variant 2A | >1 | 1 | No | Validation slice belongs to training 3D stack sample, different than training slices. |
| Variant 2B | >1 | >1 | No | Validation slice belongs to 3D stack sample that has not been used in training. |
| Variant 3A | >1 | 1 | Yes—from variant 2A | Validation slice belongs to training 3D stack sample, different than training slices. |
| Variant 3B | >1 | >1 | Yes—from variant 2B | Validation slice belongs to 3D stack sample that has not been used in training. |
Table 2.
Results for variant 1A—CNN voter compared to MGTI.
Table 2.
Results for variant 1A—CNN voter compared to MGTI.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.88078 | 0.83361 | 0.84929 |
| AMCQ | 0.97683 | 0.95812 | 0.96392 |
Table 3.
Results for variant 1A—CNN voter compared to GGTI AP7.
Table 3.
Results for variant 1A—CNN voter compared to GGTI AP7.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.82300 | 0.76498 | 0.76699 |
| AMCQ | 0.95221 | 0.92090 | 0.92143 |
Table 4.
Results for variant 1A—CNN median compared to MGTI.
Table 4.
Results for variant 1A—CNN median compared to MGTI.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.84017 | 0.84468 | 0.84664 |
| AMCQ | 0.96353 | 0.96340 | 0.96325 |
Table 5.
Results for variant 1A—CNN median compared to GGTI AP7.
Table 5.
Results for variant 1A—CNN median compared to GGTI AP7.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.81129 | 0.78347 | 0.76811 |
| AMCQ | 0.95034 | 0.93191 | 0.92203 |
Table 6.
Results for variant 1B—CNN voter compared to MGTI.
Table 6.
Results for variant 1B—CNN voter compared to MGTI.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.87835 | 0.87316 | 0.87280 |
| AMCQ | 0.97628 | 0.97484 | 0.97476 |
Table 7.
Results for variant 1B—CNN voter compared to GGTI AP7.
Table 7.
Results for variant 1B—CNN voter compared to GGTI AP7.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.85212 | 0.84450 | 0.84727 |
| AMCQ | 0.96493 | 0.96239 | 0.96397 |
Table 8.
Results for variant 1B—CNN median compared to MGTI.
Table 8.
Results for variant 1B—CNN median compared to MGTI.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.87857 | 0.86307 | 0.86438 |
| AMCQ | 0.97639 | 0.97083 | 0.97116 |
Table 9.
Results for variant 1B—CNN median compared to GGTI AP7.
Table 9.
Results for variant 1B—CNN median compared to GGTI AP7.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.85351 | 0.84903 | 0.85025 |
| AMCQ | 0.96549 | 0.96472 | 0.96522 |
Table 10.
Results for variant 2A—CNN voter compared to MGTI.
Table 10.
Results for variant 2A—CNN voter compared to MGTI.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.89308 | 0.84920 | 0.81585 |
| AMCQ | 0.98159 | 0.96668 | 0.95337 |
Table 11.
Results for variant 2A—CNN voter compared to GGTI AP7.
Table 11.
Results for variant 2A—CNN voter compared to GGTI AP7.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.83880 | 0.82822 | 0.79492 |
| AMCQ | 0.95949 | 0.95694 | 0.94158 |
Table 12.
Results for variant 2A—CNN median compared to MGTI.
Table 12.
Results for variant 2A—CNN median compared to MGTI.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.87130 | 0.78491 | 0.80887 |
| AMCQ | 0.97429 | 0.94065 | 0.95123 |
Table 13.
Results for variant 2A—CNN median compared to GGTI AP7.
Table 13.
Results for variant 2A—CNN median compared to GGTI AP7.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.802365 | 0.78129 | 0.80637 |
| AMCQ | 0.942518 | 0.93690 | 0.94822 |
Table 14.
Results for variant 2B—CNN voter compared to MGTI.
Table 14.
Results for variant 2B—CNN voter compared to MGTI.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.88564 | 0.87881 | 0.87008 |
| AMCQ | 0.97879 | 0.97616 | 0.97308 |
Table 15.
Results for variant 2B—CNN voter compared to GGTI AP7.
Table 15.
Results for variant 2B—CNN voter compared to GGTI AP7.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.85552 | 0.84212 | 0.83191 |
| AMCQ | 0.96629 | 0.96000 | 0.95552 |
Table 16.
Results for variant 2B—CNN median compared to MGTI.
Table 16.
Results for variant 2B—CNN median compared to MGTI.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.88728 | 0.87827 | 0.86879 |
| AMCQ | 0.97912 | 0.97564 | 0.97283 |
Table 17.
Results for variant 2B—CNN median compared to GGTI AP7.
Table 17.
Results for variant 2B—CNN median compared to GGTI AP7.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.86313 | 0.84981 | 0.83226 |
| AMCQ | 0.96959 | 0.96298 | 0.95602 |
Table 18.
Results for variant 3A—CNN voter compared to MGTI.
Table 18.
Results for variant 3A—CNN voter compared to MGTI.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.78964 | 0.87792 | 0.82941 |
| AMCQ | 0.94142 | 0.97669 | 0.95796 |
Table 19.
Results for variant 3A—CNN voter compared to GGTI AP7.
Table 19.
Results for variant 3A—CNN voter compared to GGTI AP7.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.76392 | 0.84530 | 0.78378 |
| AMCQ | 0.92526 | 0.96347 | 0.93344 |
Table 20.
Results for variant 3A—CNN median compared to MGTI.
Table 20.
Results for variant 3A—CNN median compared to MGTI.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.80183 | 0.87481 | 0.80027 |
| AMCQ | 0.94786 | 0.97493 | 0.94680 |
Table 21.
Results for variant 3A—CNN median compared to GGTI AP7.
Table 21.
Results for variant 3A—CNN median compared to GGTI AP7.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.77626 | 0.84862 | 0.76622 |
| AMCQ | 0.93260 | 0.96478 | 0.92621 |
Table 22.
Results for variant 3B—CNN voter compared to MGTI.
Table 22.
Results for variant 3B—CNN voter compared to MGTI.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.92281 | 0.93390 | 0.90248 |
| AMCQ | 0.98783 | 0.99064 | 0.98234 |
Table 23.
Results for variant 3B—CNN voter compared to GGTI AP7.
Table 23.
Results for variant 3B—CNN voter compared to GGTI AP7.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.88202 | 0.88944 | 0.86098 |
| AMCQ | 0.97356 | 0.97536 | 0.96547 |
Table 24.
Results for variant 3B—CNN median compared to MGTI.
Table 24.
Results for variant 3B—CNN median compared to MGTI.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.91094 | 0.91695 | 0.89490 |
| AMCQ | 0.98550 | 0.98607 | 0.97895 |
Table 25.
Results for variant 3B—CNN median compared to GGTI AP7.
Table 25.
Results for variant 3B—CNN median compared to GGTI AP7.
| Metrics | LM | BFM | RPM |
|---|
| AMC | 0.88326 | 0.87748 | 0.86304 |
| AMCQ | 0.97570 | 0.97187 | 0.96556 |
Table 26.
Results for Sample 1—2012-12-21-crwn12-008_GC1 batch 3D segmentation - CNN voter, CNN median, GGTI AP7 and NucleusJ compared to MGTI.
Table 26.
Results for Sample 1—2012-12-21-crwn12-008_GC1 batch 3D segmentation - CNN voter, CNN median, GGTI AP7 and NucleusJ compared to MGTI.
| Metrics | GGTI DL Voter Compared to MGTI | GGTI DL Median Compared to MGTI | GGTI AP7 Compared to MGTI | NucleusJ Compared to MGTI |
|---|
| AMC | 0.79218 | 0.79983 | 0.85271 | 0.89196 |
| AMCQ | 0.93450 | 0.94039 | 0.96455 | 0.97612 |
Table 27.
Results for Sample 2—2012-12-21-crwn12-009_GC1 batch 3D segmentation - CNN voter, CNN median, GGTI AP7 and NucleusJ compared to MGTI.
Table 27.
Results for Sample 2—2012-12-21-crwn12-009_GC1 batch 3D segmentation - CNN voter, CNN median, GGTI AP7 and NucleusJ compared to MGTI.
| Metrics | GGTI DL Voter Compared to MGTI | GGTI DL Median Compared to MGTI | GGTI AP7 Compared to MGTI | NucleusJ Compared to MGTI |
|---|
| AMC | 0.80584 | 0.80520 | 0.93701 | 0.91979 |
| AMCQ | 0.94448 | 0.94438 | 0.99165 | 0.98655 |
Table 28.
Results for Sample 3—2012-12-21-crwn12-009_GC2 batch 3D segmentation - CNN voter, CNN median, GGTI AP7 and NucleusJ compared to MGTI.
Table 28.
Results for Sample 3—2012-12-21-crwn12-009_GC2 batch 3D segmentation - CNN voter, CNN median, GGTI AP7 and NucleusJ compared to MGTI.
| Metrics | GGTI DL Voter Compared to MGTI | GGTI DL Median Compared to MGTI | GGTI AP7 Compared to MGTI | NucleusJ Compared to MGTI |
|---|
| AMC | 0.84245 | 0.83838 | 0.92516 | 0.91983 |
| AMCQ | 0.96110 | 0.95991 | 0.98907 | 0.98760 |
Table 29.
Results for Sample 4—2013-01-23-col0-3_GC1 batch 3D segmentation - CNN voter, CNN median, GGTI AP7 and NucleusJ compared to MGTI.
Table 29.
Results for Sample 4—2013-01-23-col0-3_GC1 batch 3D segmentation - CNN voter, CNN median, GGTI AP7 and NucleusJ compared to MGTI.
| Metrics | GGTI DL Voter Compared to MGTI | GGTI DL Median Compared to MGTI | GGTI AP7 Compared to MGTI | NucleusJ Compared to MGTI |
|---|
| AMC | 0.89753 | 0.88032 | 0.85723 | 0.81302 |
| AMCQ | 0.98200 | 0.97724 | 0.96604 | 0.94573 |
Table 30.
Results for Sample 5—2013-01-23-col0-3_GC2 batch 3D segmentation - CNN voter, CNN median, GGTI AP7 and NucleusJ compared to MGTI.
Table 30.
Results for Sample 5—2013-01-23-col0-3_GC2 batch 3D segmentation - CNN voter, CNN median, GGTI AP7 and NucleusJ compared to MGTI.
| Metrics | GGTI DL Voter Compared to MGTI | GGTI DL Median Compared to MGTI | GGTI AP7 Compared to MGTI | NucleusJ Compared to MGTI |
|---|
| AMC | 0.87893 | 0.86141 | 0.89951 | 0.84845 |
| AMCQ | 0.97182 | 0.96666 | 0.98087 | 0.95708 |
Table 31.
Results for Sample 6—2013-01-23-col0-8_GC1 batch 3D segmentation - CNN voter, CNN median, GGTI AP7 and NucleusJ compared to MGTI.
Table 31.
Results for Sample 6—2013-01-23-col0-8_GC1 batch 3D segmentation - CNN voter, CNN median, GGTI AP7 and NucleusJ compared to MGTI.
| Metrics | GGTI DL Voter Compared to MGTI | GGTI DL Median Compared to MGTI | GGTI AP7 Compared to MGTI | NucleusJ Compared to MGTI |
|---|
| AMC | 0.91628 | 0.88757 | 0.92987 | 0.89224 |
| AMCQ | 0.98525 | 0.97637 | 0.98928 | 0.97586 |
Table 32.
Variant 1G—Results for dataset: BBBC039—Nuclei of U2OS cells in a chemical screen; GGTI DL/GGTI AP7 compared to GT.
Table 32.
Variant 1G—Results for dataset: BBBC039—Nuclei of U2OS cells in a chemical screen; GGTI DL/GGTI AP7 compared to GT.
| Metrics | GGTI DL Voter Compared to GT | GGTI DL Median Compared to GT | GGTI AP7 Compared to GT |
|---|
| AMC | 0.91455 | 0.91182 | 0.94410 |
| AMCQ | 0.98568 | 0.98517 | 0.99341 |
Table 33.
Variant 1G—Results for dataset: BBBC035—Simulated nuclei of HL60 cells; GGTI DL/GGTI AP7 compared to GT.
Table 33.
Variant 1G—Results for dataset: BBBC035—Simulated nuclei of HL60 cells; GGTI DL/GGTI AP7 compared to GT.
| Metrics | GGTI DL Voter Compared to GT | GGTI DL Median Compared to GT | GGTI AP7 Compared to GT |
|---|
| AMC | 0.94451 | 0.93626 | 0.92042 |
| AMCQ | 0.99172 | 0.98970 | 0.98569 |
Table 34.
Variant 2G—Results for dataset: BBBC039—Nuclei of U2OS cells in a chemical screen; GGTI DL/GGTI AP7 compared to GT.
Table 34.
Variant 2G—Results for dataset: BBBC039—Nuclei of U2OS cells in a chemical screen; GGTI DL/GGTI AP7 compared to GT.
| Metrics | GGTI DL Voter Compared to GT | GGTI DL Median Compared to GT | GGTI AP7 Compared to GT |
|---|
| AMC | 0.95688 | 0.95536 | 0.94410 |
| AMCQ | 0.99616 | 0.99596 | 0.99341 |
Table 35.
Variant 2G—Results for dataset: BBBC035—Simulated nuclei of HL60 cells; GGTI DL/GGTI AP7 compared to GT.
Table 35.
Variant 2G—Results for dataset: BBBC035—Simulated nuclei of HL60 cells; GGTI DL/GGTI AP7 compared to GT.
| Metrics | GGTI DL Voter Compared to GT | GGTI DL Median Compared to GT | GGTI AP7 Compared to GT |
|---|
| AMC | 0.93432 | 0.93133 | 0.92042 |
| AMCQ | 0.98891 | 0.98830 | 0.98569 |
Table 36.
Variant 3G—Results for dataset: BBBC039—Nuclei of U2OS cells in a chemical screen; GGTI DL/GGTI AP7 compared to GT.
Table 36.
Variant 3G—Results for dataset: BBBC039—Nuclei of U2OS cells in a chemical screen; GGTI DL/GGTI AP7 compared to GT.
| Metrics | GGTI DL Voter Compared to GT | GGTI DL Median Compared to GT | GGTI AP7 Compared to GT |
|---|
| AMC | 0.94754 | 0.94666 | 0.94410 |
| AMCQ | 0.99444 | 0.99431 | 0.99341 |
Table 37.
Variant 3G—Results for dataset: BBBC035—Simulated nuclei of HL60 cells; GGTI DL/GGTI AP7 compared to GT.
Table 37.
Variant 3G—Results for dataset: BBBC035—Simulated nuclei of HL60 cells; GGTI DL/GGTI AP7 compared to GT.
| Metrics | GGTI DL Voter Compared to GT | GGTI DL Median Compared to GT | GGTI AP7 Compared to GT |
|---|
| AMC | 0.91323 | 0.91395 | 0.92042 |
| AMCQ | 0.98376 | 0.98392 | 0.98569 |