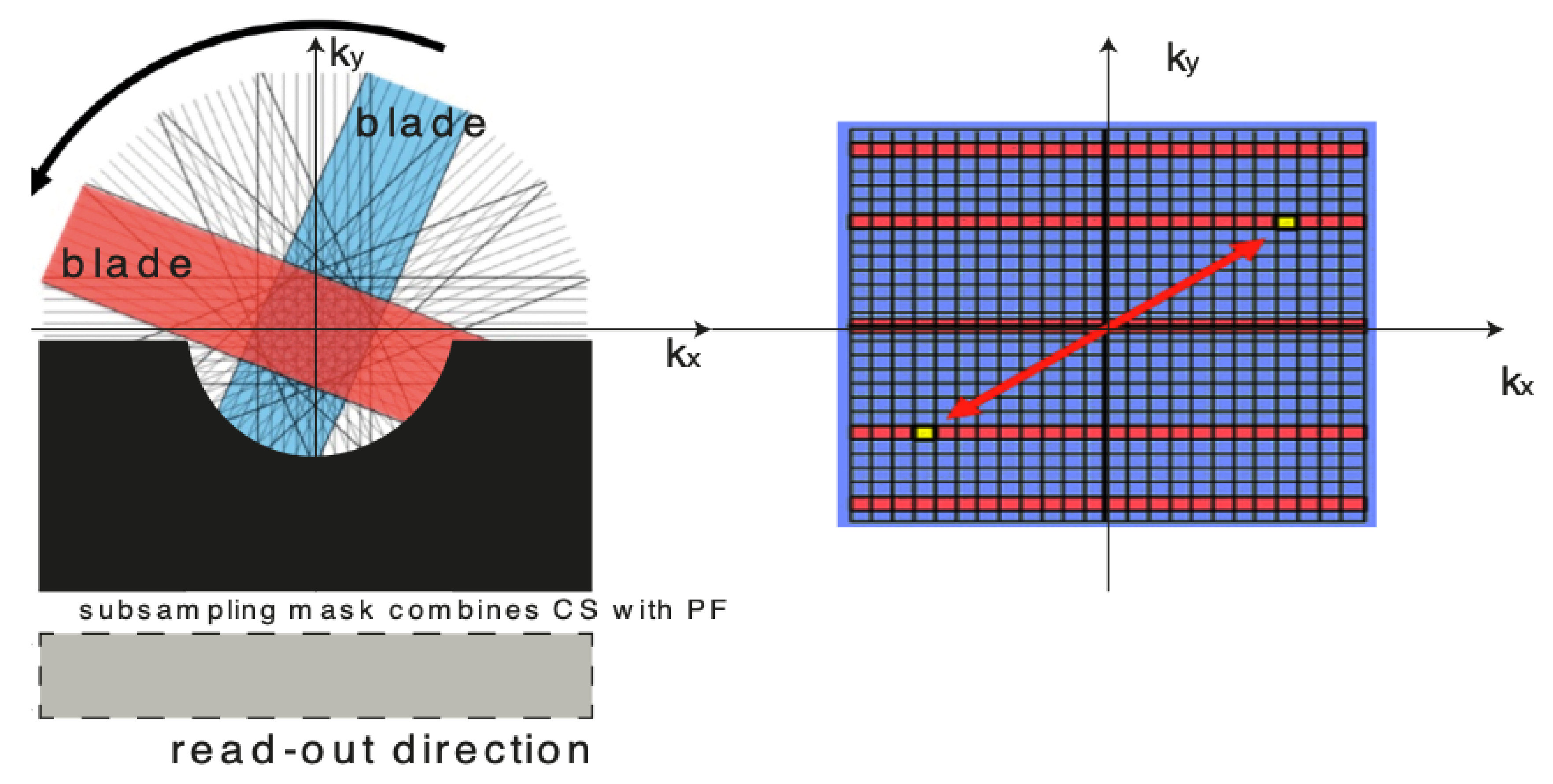
For over nearly two decades, SR procedures have effectively been exploited to enhance the spatial resolution of diagnostic images after k-spaces data are collected, thus making the doctor’s diagnosis easier. The variety of applications and methods has grown ever since, especially in the MRI modality, exposing the interest of the community to such post- processing. MRI, CT, PET and hybrid techniques are still suffering from insufficient spatial resolution, contrast issues, visual noise scattering and blurring produced by motion artefacts. These underlying issues can lead to problems in identifying abnormalities. Reducing scanning time is a serious challenge for many medical imaging techniques. Compressed Sensing (CS) theory delivers an appealing framework to address this inconvenience since it provides theoretical guarantees on the reconstruction of sparse signals by projection on a low dimensional linear subspace. Numerous adjustments here are presented and proven to be efficient for enhancing MRI spatial resolution while reducing acquisition lengths. The proposed technique minimises artefacts produced by highly sparse data, even in the influence of misregistration artefacts. In order to expedite the process of k-space filling and enhance spatial resolution, the algorithm combines several sub-techniques, that is, compressive-sensing, Poisson Disc sampling and Partial Fourier with SR technique. This combination allowed for improving both image definition and time consumption. Furthermore, improved upper frequencies provides better edge delineation. The proposed algorithm can be directly implemented to MR scanners without any hardware modifications. Moreover, it has been proven that the implemented technique produces enhanced and sharper shapes. It really minimises the risk of misdiagnosis. The financial aspects in healthcare must be addressed and sufficient value to all participants are supposed to justify their use. Apart from enhancing the spatial resolution, this technique can be useful in addressing misregistration issues. Phantom-based studies as well in-vivo experiments have proven to be successful in reducing examination time. The achieved results show an improvement of definition and readability of the MR images, see
Figure 5,
Figure 6,
Figure 7,
Figure 8,
Figure 9,
Figure 10,
Figure 11,
Figure 12,
Figure 13,
Figure 14,
Figure 15,
Figure 16,
Figure 17,
Figure 18,
Figure 19,
Figure 20,
Figure 21,
Figure 22 and
Figure 23. The techniques used to identify abnormalities that are claimed to be potentially malignant or pre-malignant expose a higher capability to spot them due to the improved image quality parameters. Furthermore, the achievements have been validated by PSNR and IEM metrics [
45] that can measure medical image quality with great competence. In more extended studies numerous scanning techniques were investigated. The proposed algorithm has been compared with SENSE, GRAPPA as well as unmodified PROPELLER. Despite reasonable results achieved using competitive algorithms, the proposed algorithms offered the shortest scanning time, see
Table 7. In order to get a quantitative assessment, the peak signal-to-noise ratio (PSNR) and the mean absolute error (MAE) have been utilised. Both the smaller MAE or higher PSNR confirmed robustness of the applied algorithm. All the most commonly applied image quality metrics, including the IEM, confirmed the weight of the results. The proposed algorithm was compared with 4 state-of-the-art SRRs: Non-Rigid Multi-Modal 3D Medical Image Registration Based on Foveated Modality Independent Neighbourhood Descriptor [
45], Residual dense network for image super-resolution [
15], Enhanced deep residual networks for single image super-resolution [
14] and Image super-resolution using very deep residual channel attention networks [
16]. L1-cost regularisation is applied to learn the nets of the following procedures—Enhanced deep residual networks for single image super-resolution, Residual dense network for image super-resolution and Image super-resolution using very deep residual channel attention networks. The compressively-sensed, Super-Resolution images have achieved the highest IEM values [
46]. it is obvious to see that the compression ratios highly affect the PSNR scores (see
Table 1,
Table 2,
Table 3,
Table 4,
Table 5,
Table 6,
Table 7,
Table 8,
Table 9,
Table 10,
Table 11,
Table 12,
Table 13,
Table 14,
Table 15,
Table 16,
Table 17 and
Table 18). It is worth being underlined that satisfying results occurred for halved sampling spaces. The mean PSNR and IEM values are used to compare the results. Each simulation was run N = 100 times. The signed rank test was applied to all the image quality metrics to verify if the null hypothesis that the central tendency of the difference was zero at numerous acceleration rates. All the statistical tests were performed using The R Project for Statistical Computing, see
Figure 10,
Figure 11,
Figure 12,
Figure 13,
Figure 14,
Figure 15,
Figure 16,
Figure 17,
Figure 18,
Figure 19,
Figure 20,
Figure 21,
Figure 22 and
Figure 23. A separate group t-student‘s test was conducted in order to compare the PSNR mean scores between 2 seperate sets, with a paired
t-test. The significance test, Student’s
t-test was carried out on the PSNR and are found that the probability validates the method’s performance. The results are exposed in
Table 1,
Table 2,
Table 3,
Table 4,
Table 5,
Table 6,
Table 7,
Table 8,
Table 9,
Table 10,
Table 11,
Table 12,
Table 13,
Table 14,
Table 15,
Table 16,
Table 17,
Table 18 and
Table 19. The achieved p-values have proven high statistical significance. Future research will be concentrated on testing competitive solutions which operate using Discrete Shearlet Transform because of its ability to be a good candidate for MRI sparse sampling. Presently, the algorithm is being validated via testing using DW-MRI/PET scanners.
Table 7.
Comparison of the scanning parameters
| Scanning Pattern | TR | TE | FOV | Voxel (mm) | Total Scan Duration (s) | p |
|---|
| PROPELLER | 1200 | 180 | 290 | 0.96/0.96/1.00 | 360 | 0.159 |
| SENSE | 1200 | 180 | 290 | 0.96/0.96/1.00 | 353 | 0.226 |
| GRAPPA | 1200 | 180 | 290 | 0.96/0.96/1.00 | 320 | 0.136 |
| the proposed algorithm | 1200 | 180 | 290 | 0.96/0.96/1.00 | 112 | 0.103 |
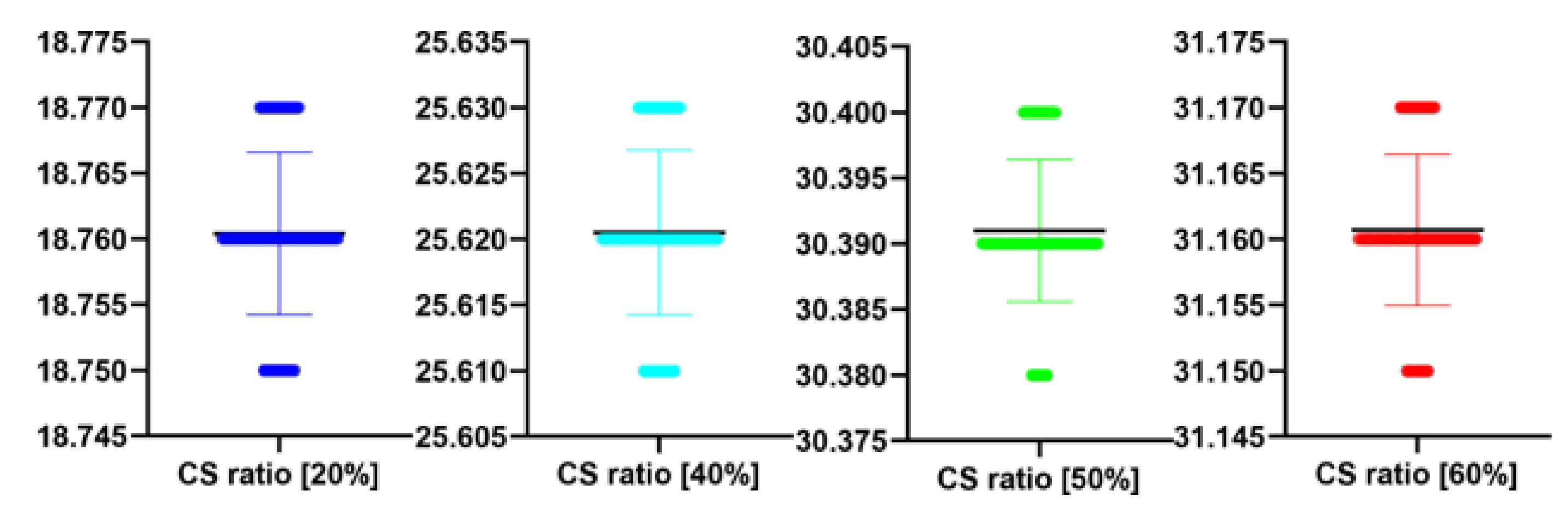
Table 8.
The performance of the proposed algorithm at different CS ratio. MAE abbreviation stands for Mean Average Error.
| CS Ratio [%] | PSNR [dB] | MAE | IEM |
|---|
| 20 | 18.76 | 19.82 | 1.79 |
| 40 | 25.62 | 18.01 | 1.88 |
| 50 | 30.39 | 15.66 | 3.99 |
| 60 | 31.16 | 14.20 | 4.31 |
Table 9.
Stats of the IEM metrics for
Figure 12. M denotes mean of observed IEMs
| Reconstruction Method | N | M | SD | t | p |
|---|
| Image reconstructed from partially sampled | | | | | |
| PROPELLER blade | 100 | 1.56 | 0.01 | 1.133 | 0.260 |
| no motion corrected regular sampling scheme | | | | | |
| (with no downsampling applied) | 100 | 3.12 | 0.01 | 0.294 | 0.070 |
| B-spline cubic interpolation /image registration applied/ | 100 | 2.62 | 0.01 | 1.842 | 0.061 |
| Non-Rigid Multi-Modal 3D Medical Image Registration | | | | | |
| Based on Foveated Modality Independent Neighbourhood Descriptor | 100 | 2.11 | 0.01 | 0.183 | 0.275 |
| Enhanced deep residual networks for single image super-resolution | 100 | 2.32 | 0.01 | −0.327 | 0.436 |
| Image super-resolution using very deep residual channel | | | | | |
| attention networks | 100 | 2.15 | 0.01 | −0.366 | 0.412 |
| Residual dense network for image SR | 100 | 2.43 | 0.01 | −0.412 | 0.432 |
| the proposed method | 100 | 2.82 | 0.01 | −0.51 | 0.551 |
| Image reconstructed from partially sampled | | | | | |
| PROPELLER blade | 100 | 3.89 | 0.01 | −0.371 | 0.202 |
Table 10.
The performance of the proposed algorithm at different CS ratio for
Figure 12. MAE abbreviation stands for Mean Average Error.
| CS Ratio [%] | PSNR [dB] | MAE | IEM |
|---|
| 20 | 18.44 | 20.01 | 1.75 |
| 40 | 28.42 | 18.03 | 1.92 |
| 50 | 36.22 | 16.05 | 3.89 |
| 60 | 38.11 | 14.55 | 4.31 |
Table 11.
Stats of the PSNR metrics for
Figure 12. M denotes mean of observed PSNRs.
| CS Quality [%] | N | M | SD | t(99) | p |
|---|
| 20 | 100 | 18.44 | 0.01 | 0.139 | 0.889 |
| 40 | 100 | 28.42 | 0.01 | 0.728 | 0.469 |
| 50 | 100 | 36.22 | 0.01 | 1.789 | 0.077 |
| 60 | 100 | 38.11 | 0.01 | 1.830 | 0.070 |
Table 12.
Stats of the IEM metrics for
Figure 12. M denotes mean of observed IEMs
| CS Quality [%] | N | M | SD | t(99) | p |
|---|
| 20 | 100 | 1.75 | 0.01 | 1.000 | 0.329 |
| 40 | 100 | 1.92 | 0.01 | −0.865 | 0.389 |
| 50 | 100 | 3.89 | 0.01 | −0.672 | 0.503 |
| 60 | 100 | 4.31 | 0.01 | 0.961 | 0.339 |
Table 13.
Stats of the IEM metrics for
Figure 17. M denotes mean of observed IEMs.
| Reconstruction Method | N | M | SD | t | p |
|---|
| Image reconstructed from partially sampled | | | | | |
| PROPELLER blade | 100 | 1.56 | 0.01 | 1.133 | 0.260 |
| no motion corrected regular sampling scheme | | | | | |
| (with no downsampling applied) | 100 | 3.12 | 0.01 | 0.294 | 0.770 |
| B-spline cubic interpolation /image registration applied/ | 100 | 2.62 | 0.01 | 1.822 | 0.021 |
| Non-Rigid Multi-Modal 3D Medical Image Registration | | | | | |
| Based on Foveated Modality Independent Neighbourhood Descriptor | 100 | 2.10 | 0.01 | 0.123 | 0.225 |
| Enhanced deep residual networks for single image super-resolution | 100 | 2.31 | 0.01 | −0.317 | 0.426 |
| Image super-resolution using very deep residual channel | | | | | |
| attention networks | 100 | 2.12 | 0.01 | −0.346 | 0.442 |
| Residual dense network for image SR | 100 | 2.49 | 0.01 | −0.462 | 0.412 |
| the proposed method | 100 | 2.80 | 0.01 | −0.511 | 0.521 |
| Image reconstructed from partially sampled | | | | | |
| PROPELLER blade | 100 | 3.89 | 0.01 | −0.371 | 0.202 |
Table 14.
The performance of the proposed algorithm at different CS ratio. MAE abbreviation stands for Mean Average Error.
| CS Ratio [%] | PSNR [dB] | MAE | IEM |
|---|
| 20 | 19.31 | 20.01 | 1.67 |
| 40 | 24.14 | 18.23 | 1.91 |
| 50 | 29.28 | 16.02 | 3.68 |
| 60 | 31.08 | 15.65 | 4.19 |
Table 15.
Stats of the PSNR metrics for
Figure 17. M denotes mean of observed PSNRs.
| CS Quality [%] | N | M | SD | t(99) | p |
|---|
| 20 | 100 | 19.31 | 0.01 | 0.542 | 0.589 |
| 40 | 100 | 24.14 | 0.01 | −0.713 | 0.478 |
| 50 | 100 | 29.28 | 0.01 | −1.044 | 0.299 |
| 60 | 100 | 31.08 | 0.01 | −1.021 | 0.310 |
Table 16.
Stats of the IEM metrics for
Figure 17. M denotes mean of observed IEMs.
| CS Quality [%] | N | M | SD | t(99) | p |
|---|
| 20 | 100 | 1.67 | 0.01 | −0.575 | 0.566 |
| 40 | 100 | 1.91 | 0.01 | −0.134 | 0.894 |
| 50 | 100 | 3.68 | 0.01 | 0.542 | 0.589 |
| 60 | 100 | 4.19 | 0.01 | 0.588 | 0.558 |
Table 17.
Stats of the PSNR metrics for
Figure 22. M denotes mean of observed PSNRs. MAE abbreviation stands for Mean Average Error.
| Reconstruction/Sampling Algorithm | N | M | MAE | SD | t(99) | p |
|---|
| the PROPELLER sampling reconstruction | 100 | 29.84 | 18.22 | 0.01 | −1.881 | 0.063 |
| the proposed algorithm | 100 | 34.58 | 15..01 | 0.00 | 1.149 | 0.253 |
Table 18.
Stats of the IEM metrics for
Figure 22. M denotes mean of observed IEMs
| Reconstruction/Sampling Algorithm | N | M | SD | t(99) | p |
|---|
| the PROPELLER sampling reconstruction | 100 | 2.04 | 0.01 | 0.139 | 0.889 |
| the proposed algorithm | 100 | 3.64 | 0.00 | −1.228 | 0.222 |
Table 19.
The performance of the proposed algorithm at different CS ratio. MAE abbreviation stands for Mean Average Error.
| CS Ratio [%] | PSNR [dB] | MAE | IEM |
|---|
| 20 | 21.04 | 20.22 | 1.88 |
| 40 | 27.45 | 18.23 | 1.92 |
| 50 | 34.58 | 15.45 | 3.64 |
| 60 | 36.01 | 15.01 | 4.17 |