Docking of Platinum Compounds on Cube Rhombellane Functionalized Homeomorphs
Abstract
:1. Introduction
2. Methods
2.1. Docking Procedure
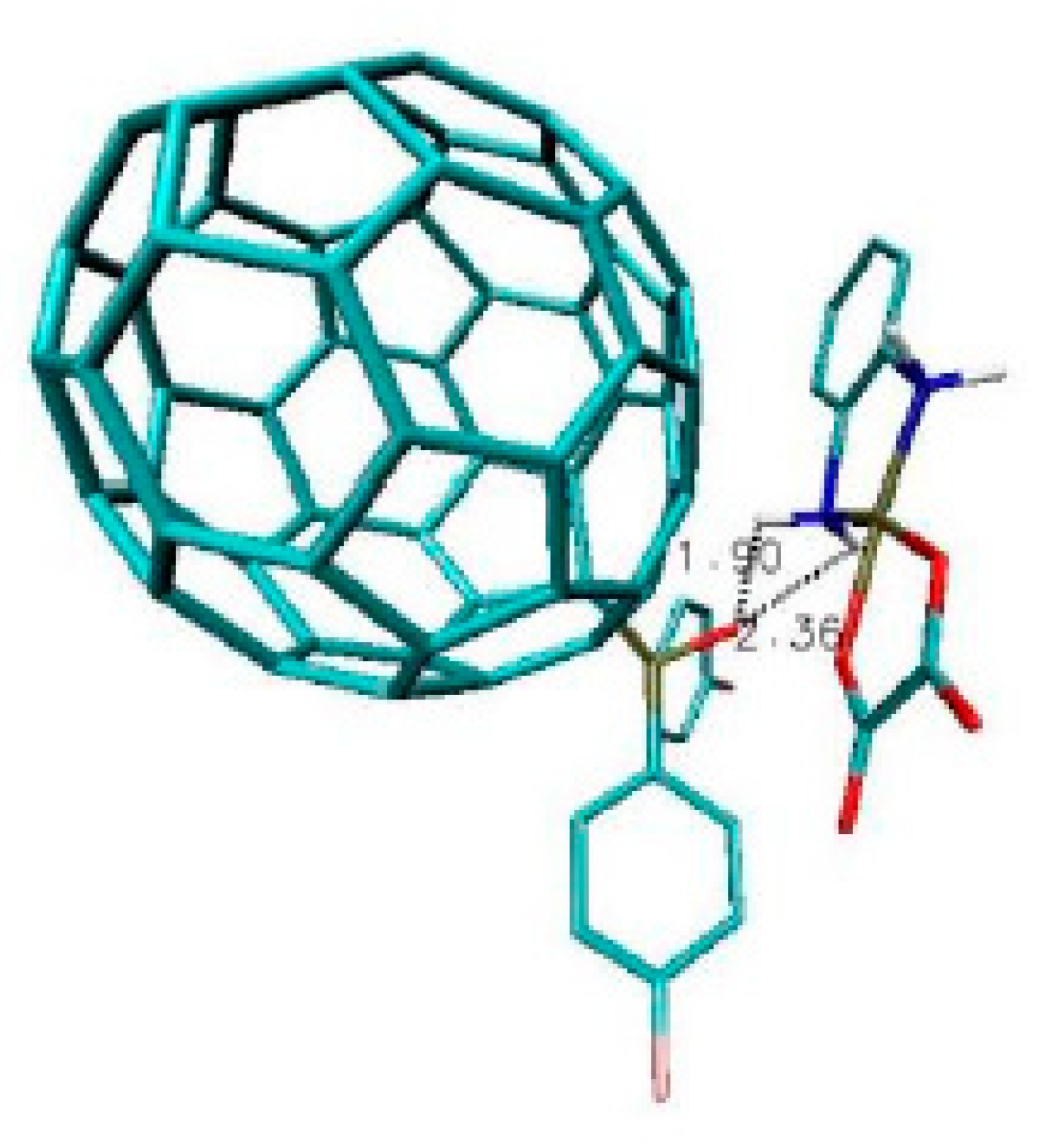
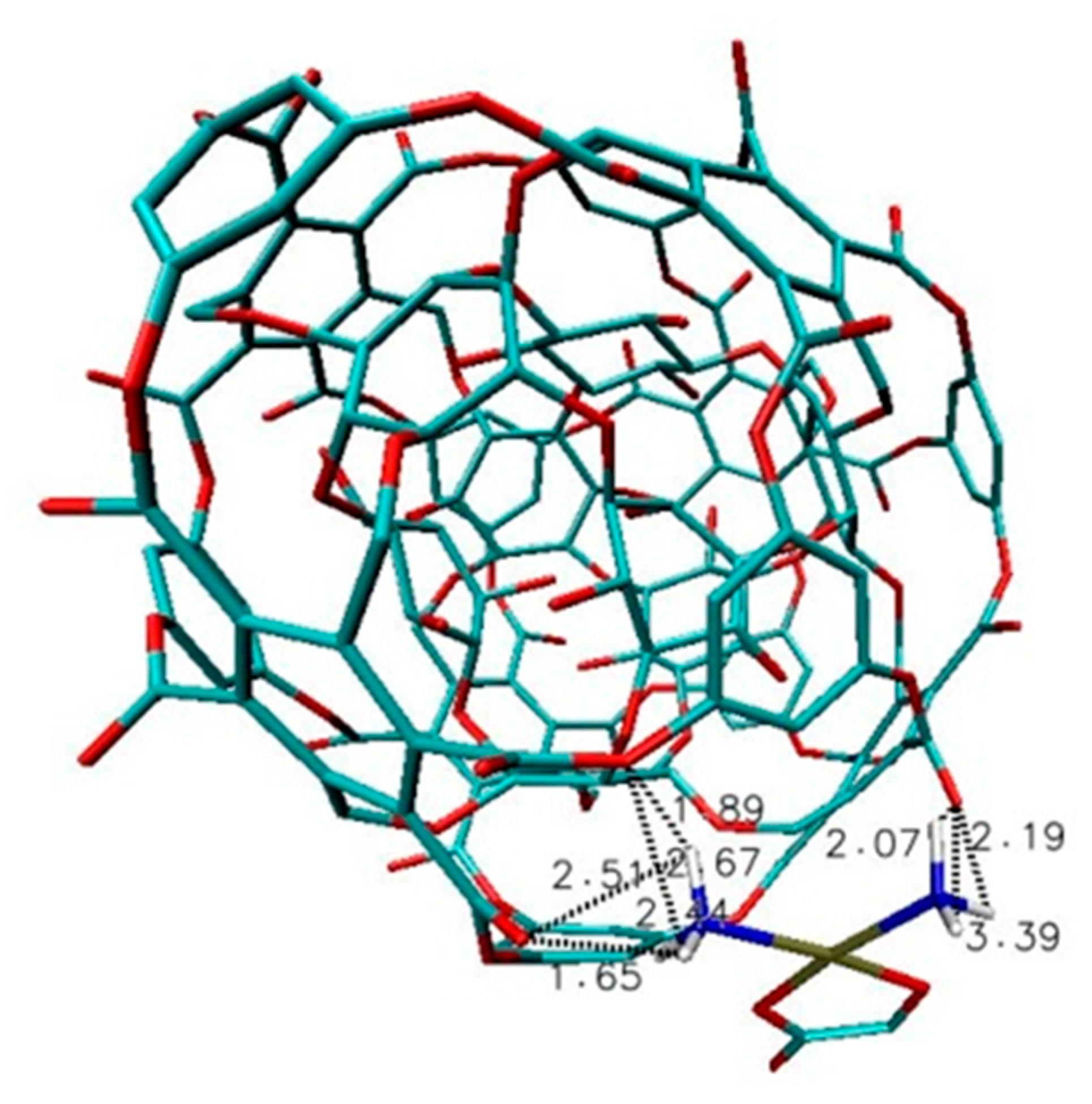
2.2. Results and Discussion
3. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rosenberg, B.; Vancamp, L.; Krigas, T. Inhibition of cell division in Escherichia coli by electrolysis products from a platinum electrode. Nature 1965, 205, 698–699. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, B.; Vancamp, L.; Trosko, J.E.; Mansour, V.H. Platinum compounds: A new class of potent antitumour agents. Nature 1969, 222, 385–386. [Google Scholar] [CrossRef] [PubMed]
- Carozzi, V.; Chiorazzi, A.; Canta, A.; Oggioni, N.; Gilardini, A.; Rodriguez-Menendez, V.; Avezza, F.; Crippa, L.; Ceresa, C.; Nicolini, G.; et al. Effect of the chronic combined administration of cisplatin and paclitaxel in a rat model of peripheral neurotoxicity. Eur. J. Cancer 2009, 45, 656–665. [Google Scholar] [CrossRef] [PubMed]
- Mcwhinney, S.R.; Goldberg, R.M.; Mcleod, H.L. Platinum neurotoxicity pharmacogenetics. Mol. Cancer Ther. 2009, 8, 10–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brabec, V.; Kasparova, J. Modifications of DNA by platinum complexes. Relation to resistance of tumors to platinum antitumor drugs. Drug Resist. Updat. 2005, 8, 131–146. [Google Scholar] [CrossRef]
- Hah, S.S.; Stivers, K.M.; De Vere White, R.W.; Henderson, P.T. Kinetics of carboplatin-DNA binding in genomic DNA and bladder cancer cells as determined by accelerator mass spectrometry. Chem. Res. Toxicol. 2006, 19, 622–626. [Google Scholar] [CrossRef] [Green Version]
- Szefler, B.; Czeleń, P.; Szczepanik, A.; Cysewski, P. Does affinity of cisplatin to B-Vitamins impair the therapeutic effect in the case of patient with lung cancer consuming carrot or beet juice. Anti-Cancer Agents Med. Chem. 2019, 19. [Google Scholar] [CrossRef]
- Szefler, B. Nanotechnology, from quantum mechanical calculations up to drug delivery. Int. J. Nanomed. 2018, 13, 6143–6176. [Google Scholar] [CrossRef] [Green Version]
- Szefler, B.; Diudea, M.V.; Grudziński, I.P. Nature of polyethyleneimine-glucose oxidase interactions. Stud. Univ. Babes-Bolyai Chem. 2016, 61, 249–260. [Google Scholar]
- Szefler, B.; Diudea, M.; Putz, M.V.; Grudziński, I.P. Molecular dynamic studies of the complex polyethylenimine and glucose oxidase. Int. J. Mol. Sci. 2016, 17, 1796. [Google Scholar] [CrossRef] [Green Version]
- Szefler, B.; Czeleń, P. Docking of cisplatin on fullerene derivatives and some cube rhombellane functionalized homeomorphs. Symmetry 2019, 11, 874. [Google Scholar] [CrossRef] [Green Version]
- Szefler, B.; Czeleń, P. Docking of polyethylenimines derivatives on cube rhombellane functionalized homeomorphs. Symmetry 2019, 11, 1048. [Google Scholar] [CrossRef] [Green Version]
- Czeleń, P.; Szefler, B. The immobilization of oxindole derivatives with use of cube rhombellane homeomorphs. Symmetry 2019, 11, 900. [Google Scholar] [CrossRef] [Green Version]
- Pauling, L.; Wheland, G.W. The nature of the chemical bond. V. The quantum mechanical calculation of the resonance energy of benzene and naphthalene and the hydrocarbon free radicals. J. Chem. Phys. 1933, 1, 362–374. [Google Scholar] [CrossRef] [Green Version]
- Daudel, R.; Lefebre, R.; Moser, C. Quantum Chemistry; Interscience: New York, NY, USA, 1959. [Google Scholar]
- Diudea, M.V.; Lungu, C.N.; Nagy, C.L. Cube-rhombellane related structures: A drug perspective. Molecules 2018, 23, 2533. [Google Scholar] [CrossRef] [Green Version]
- Nagy, K. Computational Exploration of Functionalized Rhombellanes: Building Blocks and Double-Shell Structures. Symmetry 2020, 12, 343. [Google Scholar] [CrossRef] [Green Version]
- Pop, R.; Medeleanu, M.; Diudea, M.V.; Szefler, B.; Cioslowski, J. Fullerenes patched by flowers. Cent. Eur. J. Chem. 2013, 11, 527–534. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision, A.1; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Randić, M. Aromaticity of polycyclic conjugated hydrocarbons. Chem. Rev. 2003, 103, 3449–3605. [Google Scholar] [CrossRef]
- Diudea, M.V.; Nagy, C.L. Periodic Nanostructures; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Pauling, L. The Nature of the Chemical Bond University, Vol. 260; Cornell University Press: Ithaca, NY, USA, 1960. [Google Scholar]
- Gomes, J.A.N.F.; Mallion, R.B. Aromaticity and ring currents. Chem. Rev. 2001, 101, 1349–1383. [Google Scholar] [CrossRef]
- Cyrański, M.K.; Krygowski, T.M.; Katritzky, A.R.; Schleyer, P.V.R. To what extent can aromaticity be defined uniquely. J. Org. Chem. 2002, 67, 1333–1338. [Google Scholar] [CrossRef]
- Chen, Z.; Wannere, C.S.; Crominboeuf, C.; Puchta, R.; Schleyer, R.V.P. Nucleus-independent chemical shifts (NICS) as an aromaticity criterion. Chem. Rev. 2005, 105, 3842–3888. [Google Scholar] [CrossRef] [PubMed]
- Nagy, K.; Szefler, B.; Nagy, C.L. Computational Exploration of FunctionalizedRhombellanes: Building Blocks andDouble-Shell Structures. Symmetry 2020, 12, 343. [Google Scholar] [CrossRef] [Green Version]
- Pop, R.; Janežić, D. Interaction of indomethacin with functionalized rhombellanes. Croat. Chem. Acta 2019, 4, 1–7. [Google Scholar]
- PubChem. Available online: https://pubchem.ncbi.nlm.nih.gov/ (accessed on 19 March 2020).
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. Autodock4 and AutoDockTools4: Automated docking with selective receptor flexiblity. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoichet, B.K.; Kuntz, I.D.; Bodian, D.L. Molecular docking using shape descriptors. J. Comput. Chem. 2004, 13, 380–397. [Google Scholar] [CrossRef]
- Autodock. Available online: http://autodock.1369657.n2.nabble.com/ADL-Parameters-for-docking-with-metal-ions-in-receptor-td2505649.html (accessed on 6 November 2019).
- Abagyan, R.; Totrov, M. High-throughput docking for lead generation. Curr. Opin. Chem. Biol. 2001, 5, 375. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Nguyen, T.H.; Pham, N.H.; Huy, N.T.; Bay, M.V.; Pham, M.Q.; Nam, P.C.; Vu, V.V.; Ngo, S.T. Autodock vina adopts more accurate binding poses but autodock4 forms better binding affinity. J. Chem. Inf. Model 2020, 60, 204–211. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
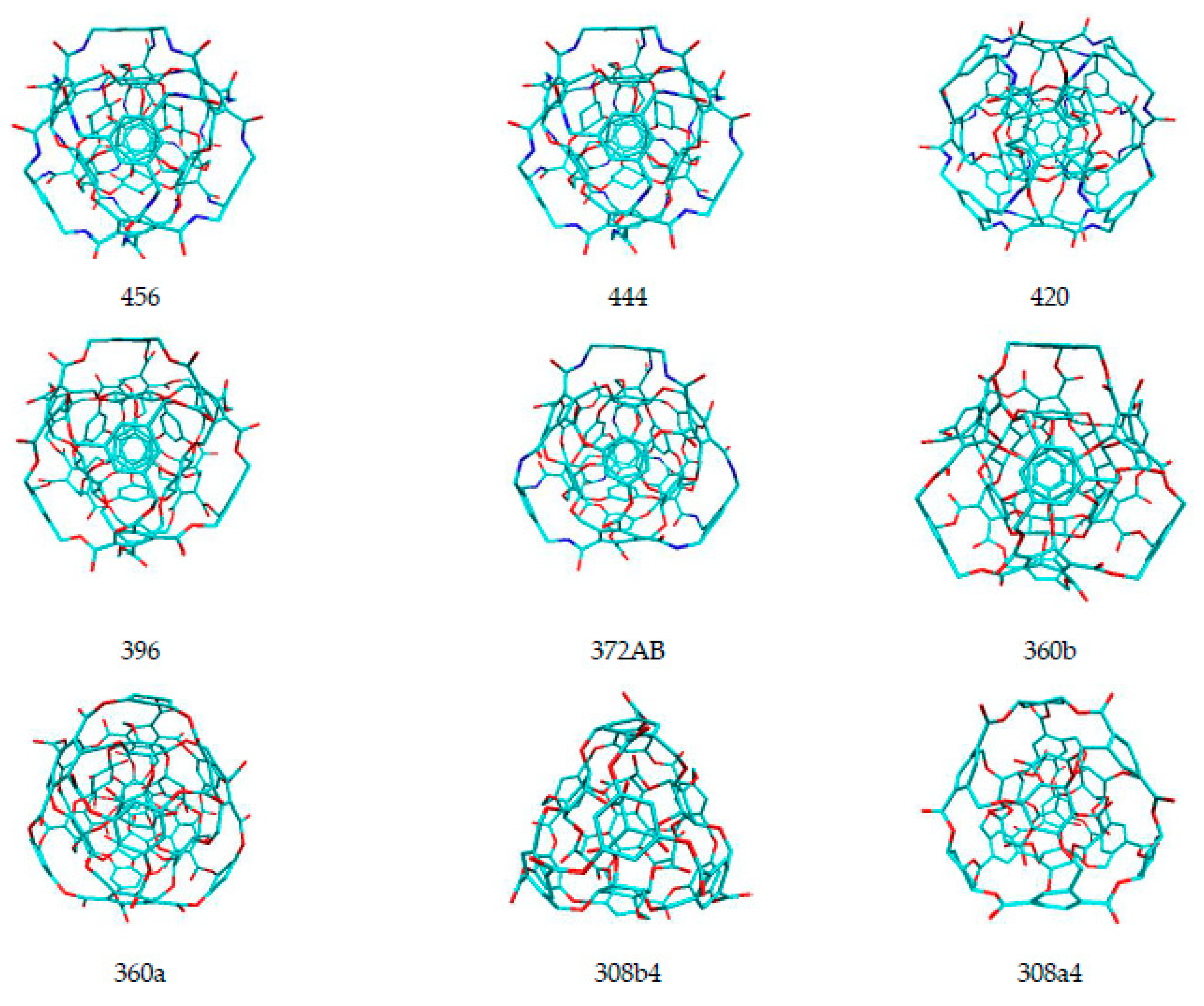
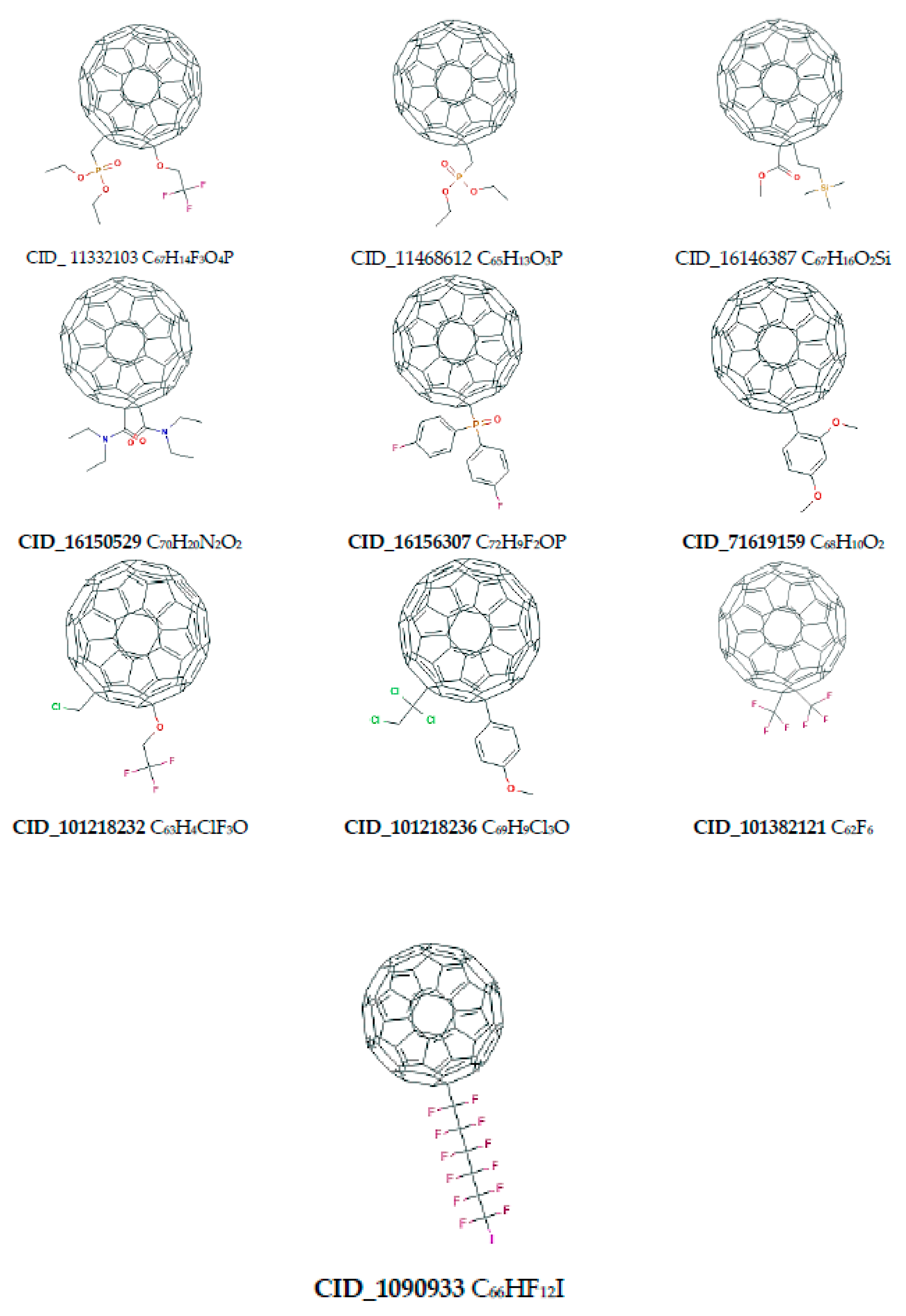



| Name of Nanostructures Title | Binding Energy (kcal/mol) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CID_71619159 | −4.45 | −4.45 | −4.45 | −4.45 | −4.45 | −4.45 | −4.45 | −4.45 | −4.45 | −4.45 |
| CID_53469305 | −4.31 | −4.31 | −4.31 | −4.3 | −4.3 | −4.29 | −4.29 | −4.29 | −4.29 | −4.29 |
| CID_16156307 | −4.98 | −4.98 | −4.98 | −4.98 | −4.98 | −4.98 | −4.98 | −4.97 | −4.82 | −4.81 |
| CID_16146387 | −4.17 | −4.12 | −4.12 | −4.12 | −4.12 | −4.12 | −4.11 | −4.11 | −4.11 | −4.11 |
| CID_11468612 | −4.48 | −4.48 | −4.48 | −4.48 | −4.48 | −4.48 | −4.48 | −4.48 | −4.48 | −4.48 |
| CID_11332103 | −4.45 | −4.45 | −4.45 | −4.45 | −4.45 | −4.45 | −4.45 | −4.45 | −4.45 | −4.45 |
| CID_10909337 | −3.66 | −3.64 | −3.63 | −3.63 | −3.63 | −3.63 | −3.62 | −3.62 | −3.6 | −3.59 |
| CID_101218236 | −4.47 | −4.47 | −4.47 | −4.47 | −4.47 | −4.47 | −4.47 | −4.47 | −4.47 | −4.23 |
| CID_101218232 | −4.37 | −4.37 | −4.37 | −4.37 | −4.37 | −4.37 | −4.37 | −4.37 | −4.37 | −4.37 |
| 456 | −3.8 | −3.8 | −3.79 | −3.79 | −3.79 | −3.79 | −3.79 | −3.79 | −3.79 | −3.75 |
| 444 | −3.82 | −3.82 | −3.82 | −3.82 | −3.82 | −3.82 | −3.82 | −3.81 | −3.82 | −3.81 |
| 420 | −3.87 | −3.87 | −3.87 | −3.87 | −3.87 | −3.87 | −3.87 | −3.86 | −3.86 | −3.85 |
| 396 | −4.02 | −4.02 | −4.02 | −4.02 | −4.02 | −4.02 | −4.01 | −4 | −3.99 | −3.98 |
| 372AB | −4.08 | −4.08 | −4.08 | −4.08 | −4.08 | −4.08 | −4.07 | −4.05 | −3.95 | −3.95 |
| 360b | −4.18 | −4.18 | −4.17 | −4.17 | −4.17 | −4.17 | −4.16 | −4.16 | −4.16 | −4.16 |
| 360a | −4.06 | −4.06 | −4.06 | −4.06 | −3.98 | −3.98 | −3.96 | −3.96 | −3.96 | −3.96 |
| 308b4 | −4.6 | −4.6 | −4.6 | −4.6 | −4.6 | −4.6 | −4.6 | −4.6 | −4.6 | −4.6 |
| 308a4 | −4.66 | −4.66 | −4.66 | −4.66 | −4.66 | −4.66 | −4.66 | −4.66 | −4.49 | −4.48 |
| Name of Nanostructures Title | Binding Energy (kcal/mol) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CID_101218232 | −4.97 | −4.96 | −4.96 | −4.94 | −4.95 | −4.94 | −4.94 | −4.93 | −4.9 | −4.83 |
| CID_101218236 | −4.67 | −4.67 | −4.67 | −4.67 | −4.67 | −4.65 | −4.63 | −4.52 | −4.52 | −4.38 |
| CID_10909337_C | −4.06 | −4.05 | −4.05 | −3.93 | −4.01 | −4 | −4 | −3.93 | −3.88 | −3.85 |
| CID_11332103 | −4.82 | −4.82 | −4.82 | −4.82 | −4.82 | −4.77 | −4.52 | −4.45 | −4.4 | −4.4 |
| CID_11468612 | −4.93 | −4.93 | −4.93 | −4.93 | −4.93 | −4.93 | −4.93 | −4.92 | −4.9 | −4.89 |
| CID_16146387 | −4.62 | −4.61 | −4.61 | −4.6 | −4.59 | −4.59 | 4.58 | 4.56 | 4.54 | −4.47 |
| CID_16150529 | −4.48 | −4.47 | −4.45 | −4.45 | −4.45 | −4.45 | −4.44 | −4.44 | −4.43 | −4.39 |
| CID_16156307 | −5.85 | −5.85 | −5.84 | −5.83 | −5.36 | −5.36 | −5.36 | −5.36 | −5.35 | −5.35 |
| CID_53469305 | −4.33 | −4.11 | −4.19 | −4.17 | −4.13 | −4.13 | −4.13 | −4.12 | −4.12 | −4.12 |
| CID_71619159 | −4.62 | −4.62 | −4.62 | −4.62 | −4.61 | −4.61 | −4.61 | −4.61 | −4.59 | −4.55 |
| 308a4 | −4.76 | −4.74 | −4.43 | −4.3 | −4.2 | −4.16 | −4.14 | −4.14 | −4.14 | −4.11 |
| 308b4 | −4.43 | −4.17 | −4.17 | −4.17 | −4.16 | −4.16 | −4.11 | −4.12 | −4.11 | −4.04 |
| 360a | −4.41 | −4.39 | −4.03 | −4 | −4.15 | −4.07 | −3.93 | −3.93 | −3.92 | −3.9 |
| 360b | −3.87 | −3.83 | −3.83 | −3.82 | −3.81 | −3.82 | −3.81 | −3.81 | −3.8 | −3.77 |
| 372AB | −3.93 | −3.93 | −3.92 | −3.9 | −3.9 | −3.89 | −3.89 | −3.89 | −3.88 | −3.85 |
| 396 | −4.51 | −4.5 | −4.49 | −4.43 | −4.28 | −4.08 | −4.03 | −3.91 | −3.9 | −3.89 |
| 420 | −4.44 | −4.4 | −4.35 | −4.38 | −4.34 | −4.32 | −4.17 | −3.84 | −3.84 | −3.82 |
| 444 | −3.95 | −3.95 | −3.93 | −3.93 | −3.93 | −3.85 | −3.87 | −3.83 | −3.76 | −3.73 |
| 456 | −3.83 | −3.81 | −3.81 | −3.81 | −3.82 | −3.82 | −3.79 | −3.76 | −3.74 | −3.65 |
| Name of Nanostructures Title | Binding Energy (kcal/mol) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CID_10909337 | −3.41 | −3.41 | −3.41 | −3.41 | −3.41 | −3.41 | −3.41 | −3.41 | −3.41 | −3.41 |
| CID_11332103 | −3.79 | −3.78 | −3.78 | −3.78 | −3.78 | −3.77 | −3.77 | −3.78 | −3.77 | −3.76 |
| CID_11468612 | −3.7 | −3.7 | −3.7 | −3.7 | −3.69 | −3.69 | −3.69 | −3.69 | −3.69 | −3.69 |
| CID_16146387 | −3.64 | −3.64 | −3.64 | −3.64 | −3.64 | −3.64 | −3.64 | −3.64 | −3.64 | −3.64 |
| CID_16150529 | −3.55 | −3.55 | −3.55 | −3.55 | −3.54 | −3.54 | −3.53 | −3.53 | −3.53 | −3.52 |
| CID_16156307 | −4.61 | −4.61 | −4.61 | −4.61 | −4.6 | −4.6 | −4.6 | −4.6 | −4.6 | −4.6 |
| CID_53469305 | −3.56 | −3.56 | −3.56 | −3.56 | −3.55 | −3.55 | −3.55 | −3.55 | −3.55 | −3.51 |
| CID_71618962 | −3.17 | −3.17 | −3.17 | −3.17 | −3.17 | −3.17 | −3.16 | −3.16 | −3.16 | −3.16 |
| CID_71619055 | −3.94 | −3.94 | −3.94 | −3.94 | −3.94 | −3.94 | −3.94 | −3.94 | −3.94 | −3.94 |
| CID_71619159 | −3.76 | −3.76 | −3.76 | −3.76 | −3.76 | −3.76 | −3.76 | −3.73 | −3.73 | −3.73 |
| CID_101218232 | −3.82 | −3.82 | −3.82 | −3.81 | −3.81 | −3.81 | −3.81 | −3.8 | −3.81 | −3.8 |
| CID_101218236 | −3.39 | −3.39 | −3.39 | −3.39 | −3.39 | −3.39 | −3.37 | −3.37 | −3.37 | −3.36 |
| CID_101266715 | −3.44 | −3.44 | −3.43 | −3.43 | −3.43 | −3.43 | −3.43 | −3.42 | −3.42 | −3.42 |
| CID_101382121 | −2.59 | −2.59 | −2.59 | −2.57 | −2.56 | −2.56 | −2.56 | −2.56 | −2.56 | −2.56 |
| 456 | −3.65 | −3.64 | −3.63 | −3.63 | −3.43 | −3.41 | −3.39 | −3.39 | −3.39 | −3.37 |
| 444 | −3.66 | −3.43 | −3.41 | −3.41 | −3.41 | −3.41 | −3.38 | −3.37 | −3.37 | −3.36 |
| 420 | −3.51 | −3.5 | −3.48 | −3.48 | −3.5 | −3.5 | −3.49 | −3.49 | −3.49 | −3.49 |
| 396 | −3.59 | −3.59 | −3.58 | −3.58 | −3.58 | −3.57 | −3.57 | −3.56 | −3.56 | −3.54 |
| 372AB | −3.85 | −3.84 | −3.84 | −3.83 | −3.78 | −3.78 | −3.78 | −3.78 | −3.78 | −3.77 |
| 360b | −3.65 | −3.65 | −3.64 | −3.64 | −3.64 | −3.63 | −3.63 | −3.62 | −3.61 | −3.6 |
| 360a | −4.05 | −4.05 | −4.05 | −4.04 | −4.04 | −4.01 | −4 | −4 | −4 | −4 |
| 308b4 | −4.08 | −4.08 | −4.08 | −4.08 | −4.08 | −4.07 | −4.06 | −4.06 | −4.06 | −4.07 |
| 308a4 | −4 | −4 | −4 | −4 | −4 | −4 | −3.99 | −3.99 | −3.99 | −3.99 |
| Name of Nanostructures Title Binding Energy (kcal/mol) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CID_10909337 | −1.3 | −1.29 | −1.29 | −1.29 | −1.29 | −1.28 | −1.28 | −1.24 | −1.26 | −1.24 |
| CID_11332103 | −2.81 | −2.81 | −2.8 | −2.77 | −2.76 | −2.75 | −2.62 | −2.65 | −2.61 | −2.45 |
| CID_11468612 | −2.88 | −2.83 | −2.82 | −2.81 | −2.79 | −2.77 | −2.75 | −2.75 | −2.74 | −2.74 |
| CID_16146387 | −2.31 | −2.29 | −2.27 | −2.26 | −2.24 | −2.24 | −2.23 | −2.21 | −2.12 | −2.12 |
| CID_16150529 | −2.26 | −2.26 | −2.25 | −2.25 | −2.24 | −2.21 | −2.23 | −2.23 | −2.14 | −2.14 |
| CID_16156307 | −4.14 | −3.96 | −3.95 | −3.86 | −3.81 | −3.75 | −3.64 | −3.59 | −3.4 | −3.38 |
| CID_53469305 | −2.26 | −2.07 | −2.23 | −2.2 | −2.18 | −2.18 | −2.18 | −2.16 | −2.11 | −2.08 |
| CID_71618962 | −2.2 | −2.19 | −2.18 | −2.17 | −2.17 | −2.17 | −2.16 | −2.16 | −2.15 | −2.14 |
| CID_71619055 | −2.4 | −2.4 | −2.4 | −2.39 | −2.39 | −2.39 | −2.39 | −2.38 | −2.37 | −2.36 |
| CID_71619159 | −2.24 | −2.24 | −2.24 | −2.24 | −2.24 | −2.23 | −2.23 | −2.22 | −2.19 | −2.17 |
| CID_101218232 | −3.04 | −3.01 | −2.99 | −2.95 | −2.97 | −2.97 | −2.97 | −2.97 | −2.96 | −2.93 |
| CID_101218236 | −3.09 | −3.08 | −3.08 | −3.05 | −3.01 | −3.01 | −3.01 | −3.01 | −3 | −3 |
| CID_101266715 | −1.59 | −1.59 | −1.59 | −1.59 | −1.59 | −1.58 | −1.54 | −1.53 | −1.52 | −1.52 |
| CID_101382121 | −1.12 | −1.11 | −1.11 | −1.1 | −1.1 | −1.1 | −1.09 | −1.09 | −1.08 | −1.03 |
| 456 | −2.96 | −2.94 | −2.94 | −2.82 | −2.79 | −2.89 | −2.85 | −2.78 | −2.73 | −2.7 |
| 444 | −2.9 | −2.8 | −2.68 | −2.87 | −2.77 | −2.81 | −2.79 | −2.78 | −2.69 | −2.61 |
| 420 | −2.57 | −2.57 | −2.5 | −2.57 | −2.57 | −2.51 | −2.5 | −2.57 | −2.51 | −2.45 |
| 396 | −2.75 | −2.61 | −2.58 | −2.56 | −2.58 | −2.52 | −2.57 | −2.54 | −2.53 | −2.39 |
| 372AB | −2.61 | −2.61 | −2.56 | −2.61 | −2.51 | −2.58 | −2.49 | −2.47 | −2.48 | −2.45 |
| 360b | −2.58 | −2.56 | −2.56 | −2.53 | −2.56 | −2.54 | −2.53 | −2.52 | −2.5 | −2.5 |
| 360a | −3.02 | −3.02 | −2.94 | −2.85 | −2.84 | −2.88 | −2.8 | −2.73 | −2.73 | −2.56 |
| 308b4 | −2.72 | −2.54 | −2.53 | −2.67 | −2.64 | −2.41 | −2.59 | −2.56 | −2.45 | −2.36 |
| 308a4 | −2.79 | −2.69 | −2.64 | −2.68 | −2.66 | −2.63 | −2.55 | −2.41 | −2.4 | −2.29 |
| Name of Nanostructures | MAX Binding Energy | MIN Binding Energy | AVERAGE | SD | Binding Constant [Kmax] |
|---|---|---|---|---|---|
| CID_71619159 | −4.45 | −4.45 | −4.45 | 9 × 10−16 | 1827.7 |
| CID_53469305 | −4.31 | −4.29 | −4.298 | 9 × 10−3 | 1443.0 |
| CID_16156307 | −4.98 | 4.81 | −4.946 | 7 × 10−2 | 4470.8 |
| CID_16146387 | −4.17 | −4.11 | −4.121 | 2 × 10−2 | 1139.3 |
| CID_11468612 | −4.48 | −4.48 | −4.48 | 9 × 10−16 | 1922.6 |
| CID_11332103 | −4.45 | −4.45 | −4.45 | 9 × 10−16 | 1827.7 |
| CID_10909337 | −3.66 | −3.59 | −3.625 | 2 × 10−2 | 481.7 |
| CID_101218236 | −4.47 | −4.23 | −4.446 | 7 × 10−2 | 1890.4 |
| CID_101218232 | −4.37 | −4.37 | −4.37 | 9 × 10−16 | 1596.8 |
| 456 | −3.8 | −3.75 | −3.788 | 1 × 10−2 | 610.2 |
| 444 | −3.82 | −3.81 | −3.818 | 4 × 10−3 | 631.1 |
| 420 | −3.87 | −3.85 | −3.866 | 7 × 10−3 | 686.7 |
| 396 | −4.02 | −3.98 | −4.01 | 1 × 10−2 | 884.5 |
| 372AB | −4.08 | −3.95 | −4.05 | 5 × 10−2 | 978.8 |
| 360b | −4.18 | −4.16 | −4.168 | 7 × 10−3 | 1158.7 |
| 360a | −4.06 | −3.96 | −4.004 | 5 × 10−2 | 946.3 |
| 308b4 | −4.6 | −4.6 | −4.6 | 9 × 10−16 | 2354.2 |
| 308a4 | −4.66 | −4.48 | −4.625 | 7 × 10−2 | 2605.1 |
| Name of Nanostructures | MAX Binding Energy | MIN Binding Energy | AVERAGE | SD | Binding Constant [Kmax] |
|---|---|---|---|---|---|
| CID_101218232 | −4.97 | −4.83 | −4.932 | 0.038678 | 4396.0 |
| CID_101218236 | −4.67 | −4.38 | −4.605 | 0.09426 | 2649.4 |
| CID_10909337_C | −4.06 | −3.85 | −3.976 | 0.070456 | 946.3 |
| CID_11332103 | −4.82 | −4.4 | −4.664 | 0.184076 | 3412.8 |
| CID_11468612 | −4.93 | −4.89 | −4.922 | 0.014 | 4109.0 |
| CID_16146387 | −4.62 | −4.47 | −4.577 | 0.042438 | 2435.0 |
| CID_16150529 | −4.48 | −4.39 | −4.445 | 0.022913 | 1922.6 |
| CID_16156307 | −5.85 | −5.35 | −5.551 | 0.238095 | 19413.6 |
| CID_53469305 | −4.33 | −4.11 | −4.155 | 0.062968 | 1492.6 |
| CID_71619159 | −4.62 | −4.55 | −4.606 | 0.020591 | 2435.0 |
| 308a4 | −4.76 | −4.11 | −4.312 | 0.23731 | 3084.1 |
| 308b4 | −4.43 | −4.04 | −4.164 | 0.096974 | 1767.0 |
| 360a | −4.41 | −3.9 | −4.073 | 0.179279 | 1708.3 |
| 360b | −3.87 | −3.77 | −3.817 | 0.024104 | 686.7 |
| 372AB | −3.93 | −3.85 | −3.898 | 0.023152 | 759.9 |
| 396 | −4.51 | −3.89 | −4.202 | 0.253567 | 2022.4 |
| 420 | −4.44 | −3.82 | −4.19 | 0.242899 | 1797.1 |
| 444 | −3.95 | −3.73 | −3.873 | 0.075637 | 785.9 |
| 456 | −3.83 | −3.65 | −3.784 | 0.052192 | 641.8 |
| Name of Nanostructures | MAX Binding Energy | MIN Binding Energy | AVERAGE | SD | Binding Constant [Kmax] |
|---|---|---|---|---|---|
| CID_10909337 | −3.41 | −3.41 | −3.41 | 0.00 | 315.9 |
| CID_11332103 | −3.79 | −3.76 | −3.78 | 0.01 | 599.9 |
| CID_11468612 | −3.70 | −3.69 | −3.69 | 0.00 | 515.4 |
| CID_16146387 | −3.64 | −3.64 | −3.64 | 0.00 | 465.8 |
| CID_16150529 | −3.55 | −3.52 | −3.54 | 0.01 | 400.1 |
| CID_16156307 | −4.61 | −4.60 | −4.60 | 0.00 | 2394.3 |
| CID_53469305 | −3.56 | −3.51 | −3.55 | 0.01 | 406.9 |
| CID_71618962 | −3.17 | −3.16 | −3.17 | 0.00 | 210.7 |
| CID_71619055 | −3.94 | −3.94 | −3.94 | 0.00 | 772.8 |
| CID_71619159 | −3.76 | −3.73 | −3.75 | 0.01 | 570.3 |
| CID_101218232 | −3.82 | −3.80 | −3.81 | 0.01 | 631.1 |
| CID_101218236 | −3.39 | −3.36 | −3.38 | 0.01 | 305.4 |
| CID_101266715 | −3.44 | −3.42 | −3.43 | 0.01 | 332.3 |
| CID_101382121 | −2.59 | −2.56 | −2.57 | 0.01 | 79.2 |
| 456 | −3.65 | −3.37 | −3.49 | 0.12 | 473.7 |
| 444 | −3.66 | −3.36 | −3.42 | 0.08 | 481.7 |
| 420 | −3.51 | −3.48 | −3.49 | 0.01 | 374.0 |
| 396 | −3.59 | −3.54 | −3.57 | 0.01 | 428.1 |
| 372AB | −3.85 | −3.77 | −3.80 | 0.03 | 663.9 |
| 360b | −3.65 | −3.60 | −3.63 | 0.02 | 473.7 |
| 360a | −4.05 | −4.00 | −4.02 | 0.02 | 930.4 |
| 308b4 | −4.08 | −4.06 | −4.07 | 0.01 | 978.8 |
| 308a4 | −4.00 | −3.99 | −4.00 | 0.00 | 855.1 |
| Name of Nanostructures | MAX Binding Energy | MIN Binding Energy | AVERAGE | SD | Binding Constant [Kmax] |
|---|---|---|---|---|---|
| CID_10909337 | −1.30 | −1.24 | −1.28 | 0.02 | 9.0 |
| CID_11332103 | −2.81 | −2.45 | −2.70 | 0.11 | 114.8 |
| CID_11468612 | −2.88 | −2.74 | −2.79 | 0.04 | 129.1 |
| CID_16146387 | −2.31 | −2.12 | −2.23 | 0.06 | 49.3 |
| CID_16150529 | −2.26 | −2.14 | −2.22 | 0.04 | 45.4 |
| CID_16156307 | −4.14 | −3.38 | −3.75 | 0.23 | 1083.1 |
| CID_53469305 | −2.26 | −2.07 | −2.17 | 0.06 | 45.4 |
| CID_71618962 | −2.20 | −2.14 | −2.17 | 0.02 | 41.0 |
| CID_71619055 | −2.40 | −2.36 | −2.39 | 0.01 | 57.4 |
| CID_71619159 | −2.24 | −2.17 | −2.22 | 0.02 | 43.8 |
| CID_101218232 | −3.04 | −2.93 | −2.98 | 0.03 | 169.2 |
| CID_101218236 | −3.09 | −3.00 | −3.03 | 0.03 | 184.1 |
| CID_101266715 | −1.59 | −1.52 | −1.56 | 0.03 | 14.6 |
| CID_101382121 | −1.12 | −1.03 | −1.09 | 0.02 | 6.6 |
| 456 | −2.96 | −2.70 | −2.84 | 0.09 | 147.8 |
| 444 | −2.90 | −2.61 | −2.77 | 0.08 | 133.6 |
| 420 | −2.57 | −2.45 | −2.53 | 0.04 | 76.5 |
| 396 | −2.75 | −2.39 | −2.56 | 0.08 | 103.7 |
| 372AB | −2.61 | −2.45 | −2.54 | 0.06 | 81.9 |
| 360b | −2.58 | −2.50 | −2.54 | 0.03 | 77.8 |
| 360a | −3.02 | −2.56 | −2.84 | 0.13 | 163.6 |
| 308b4 | −2.72 | −2.36 | −2.55 | 0.11 | 98.6 |
| 308a4 | −2.79 | −2.29 | −2.57 | 0.15 | 110.9 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szefler, B.; Czeleń, P. Docking of Platinum Compounds on Cube Rhombellane Functionalized Homeomorphs. Symmetry 2020, 12, 749. https://doi.org/10.3390/sym12050749
Szefler B, Czeleń P. Docking of Platinum Compounds on Cube Rhombellane Functionalized Homeomorphs. Symmetry. 2020; 12(5):749. https://doi.org/10.3390/sym12050749
Chicago/Turabian StyleSzefler, Beata, and Przemysław Czeleń. 2020. "Docking of Platinum Compounds on Cube Rhombellane Functionalized Homeomorphs" Symmetry 12, no. 5: 749. https://doi.org/10.3390/sym12050749
APA StyleSzefler, B., & Czeleń, P. (2020). Docking of Platinum Compounds on Cube Rhombellane Functionalized Homeomorphs. Symmetry, 12(5), 749. https://doi.org/10.3390/sym12050749






