1. Introduction and Preliminaries
A molecular graph (in chemical graph theory) is a simple graph (a graph that does not contain edges whose two ends are the same vertex, nor containing two edges with the same two ends) where the atoms of the molecule are represented as vertices and the bonds between them are the edges. A graph consists of a vertex set and an edge set . The number of edges incident to v of some vertex of a graph G is said to be its degree and is denoted by ; is the minimum degree of a graph, and is its maximum. We can represent an edge with vertices, , if it connects vertex u to vertex v.
Topological indices are a numerical quantity computed from the molecular graph of a chemical compound and typically remain the same under graph isomorphism. A topological index determined by means of using the degrees of the molecular graph is a family of indices that can be used to point out and model certain properties of chemical compounds; these indices are also known as degree-based topological indices [
1,
2,
3,
4]. In the same context, the M-polynomial plays a similar part; we can obtain closed-form formulas for degree-based topological indices from it [
5,
6,
7,
8,
9,
10].
There are some recent papers devoted to the computation of the M-polynomial of a given chemical graph. In particular, in [
11], the authors considered two infinite classes
and
of nanostar dendrimers and computed the Zagreb indices and Zagreb polynomials for these special nanostar dendrimers. In [
12], the author determined certain Zagreb polynomials of a special type of dendrimer nanostar
, and the Zagreb indices and Zagreb polynomials of the same dendrimer nanostars were computed in [
13]. The computation of some fifth multiplicative Zagreb indices of PAMAMdendrimers was reported in [
14]. The first and second reverse Zagreb indices, reverse hyper-Zagreb indices, and their polynomials of porphyrin, propyl ether imine, zinc porphyrin, and poly(ethylene amido amine) dendrimers were determined in [
15]. The computation of the M-polynomial and topological indices of the benzene ring embedded in a P-type surface network was given in [
16]. The M-polynomial and degree-based topological indices of polyhex nanotubes were given in [
9]. The main object of this paper is to derive the general form of the M-polynomial for two dendrimer nanostars
,
(
Figure 1A,B) and four types of nanotubes. Furthermore, we compute the first and second Zagreb index, the modified Zagreb index, the Randic index, and the symmetric division index for this family of molecules. We also give the graphical representation of the M-polynomial. A comparison between the obtained topological indices of these chemical graphs is outlined at the end.
2. Materials and Methods
Dendrimers are repetitively branching artificial star-shaped molecules synthesized from branched units called monomers using a nanoscale fabrication process; they have a well-defined structure with three major components: the core, branches, and end groups, where new branches are made from the core and are added recursively in steps. There are various applications of nanostar dendrimers for the formation of micro- and macro-capsules, chemical sensors, colored glasses, and modified electrodes. Due to these applications, these compounds have attracted much attention in both chemistry and mathematics alike.
Nanotubes are a nanoscale material that takes up a tube-like structure and can be made from various materials, organic or inorganic, such as carbon, boron, and silicon. CNT (Carbon Nanotubes) have a high thermal and electrical conductivity, while also being highly flexible and being good electron field emitters. They have various applications in energy storage, air and water filtration, fabrics, and thermal compounds and have attracted much attention from the scientific community.
Boron nanotubes are attracting much attention as well because of their remarkable properties such as structural stability, work function, transport properties, and electronic structure and are coming to challenge CNTs. In this article, we study the structures of two configurations of CNT, “zig-zag”
and “armchair”
(
Figure 2A,B), and two kinds of Boron nanotubes, triangular boron
and
-boron
(
Figure 3A,B).
As introduced by Grutman and Trinajstic, the first Zagreb index
and the second Zagreb index
are defined as follows [
17]:
It is well known that the first Zagreb index can be written as:
The modified Zagreb index
is defined to be:
, the general Randic index, is defined to be:
where
.
The symmetric division index,
, is defined to be:
Let
G be a simple molecular connected graph, and let
denote the degree of some vertex
. The vertex set
can be partitioned as follows:
where
. The edge set
can be partitioned into the following sets:
Then, the M-polynomial of a graph
G denoted as
is defined to be:
where
.
From the above polynomial, one can obtain numerous degree-based topological indices by differentiating and/or integrating with respect to
x and
y [
6]. The
Table 1 below lists some of the degree-based topological indices and how to derived them from the M-polynomial.
where
is defined to be:
3. Results
We found the M-polynomials of two special families of dendrimer nanostars denoted by and four types of nanotubes denoted by (). After that, we represented some of the M-polynomials as three-dimensional surfaces using Mathematica, then with the assistance of the above table, we found the topological indices of the aforementioned structures.
3.1. M-Polynomials of the Dendrimer Nanostars
The main objective of this subsection is to obtain the general closed form of the M-polynomial associated with the two special nanostar dendrimers ().
Theorem 1. Let be the molecular graph whose structure can be seen in Figure 1A. Then, its M-polynomial is: Proof. Studying the construction of
(see
Figure 1A), we can see that there are two partitions of the vertex set of
as:
and
. We also see that
can be partitioned as follows:
,
, and
. Thus, following from the definition, the M-polynomial of
is:
□
Theorem 2. Let be the molecular graph whose structure can be seen in Figure 1B. Then, its M-polynomial is: Proof. Studying the construction of
(see
Figure 2B), we can see that there are two partitions of the vertex set of
as:
and
. We also see that
can be partitioned as follows:
,
, and
. Thus, following straight from the definition, the M-polynomial of
is:
□
3.2. M-Polynomials of the Nanotubes
In this subsection, we will provide the general formula for the M-polynomial of the four nanotubes (
, where
), as shown in
Figure 2A,B and
Figure 3A,B. We also provide the graphical representation of these M-polynomials.
Theorem 3. Let be a molecular graph whose structure can be seen in Figure 2A. Then: Proof. Studying the construction of
, we can see that there are two partitions of the vertex set of
as:
and
. We also see that
can be partitioned as follows:
, and
Thus, following straight from the definition, the M-polynomial of
is:
□
Theorem 4. Let be a molecular graph whose structure can be seen in Figure 2B. Then: Proof. Studying the construction of
, we can see that there are two partitions of the vertex set of
:
and
. We also see that
can be partitioned as follows:
,
, and
Thus, the M-polynomial of
is:
□
Theorem 5. Let be a molecular graph whose structure can be seen in Figure 3A. Then: Proof. Studying the structure of
, we can see that there are two partitions of the vertex set of
:
and
. We also see that
can be partitioned as follows:
,
, and
. Thus, the M-polynomial of
is:
□
Theorem 6. Let be a molecular graph whose structure can be seen in Figure 3B. Then: Proof. Studying the construction of
, we can see that there are three partitions of the vertex set of
:
,
, and
. We also see that
can be partitioned as follows:
,
,
,
, and
. Thus, following straight from the definition, the M-polynomial of
is:
□
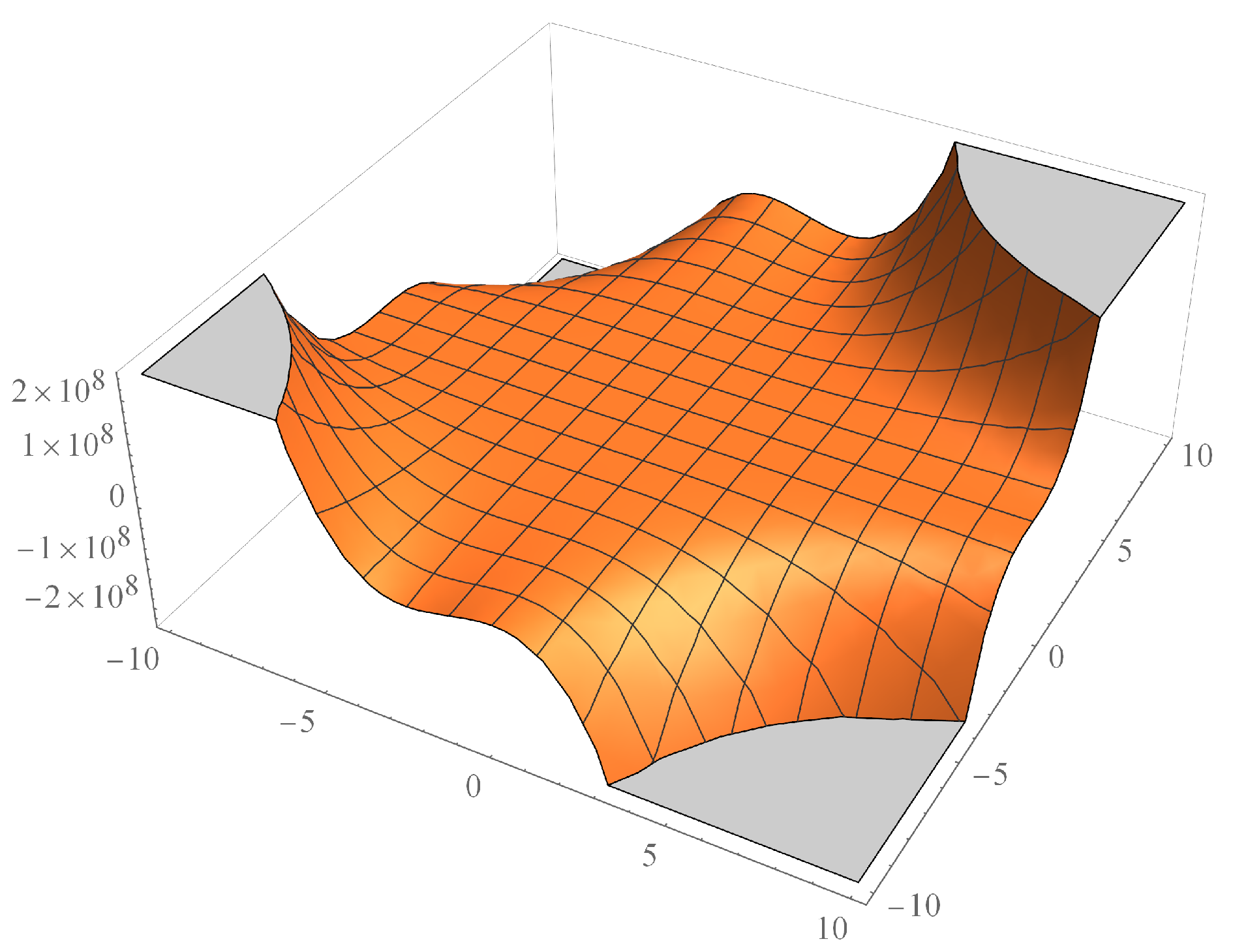
Now, we will give the 3D-plot of some of these M-polynomials with the help of Mathematica. In particulary, the 3d-plot of M-polynomial of dendrimer nanostar
is shown in
Figure 4 and for two nanotubes
and
in
Figure 5 and
Figure 6 respectively.
3.3. Degree-Based Topological Indices
In this subsection, by using the computed M-polynomials, some degree-based topological indices are determined for the aforementioned chemical graphs.
Theorem 7. Let . Then:
- 1
- 2
- 3
- 4
- 5
- 6
Proof. If
, then:
|
|
|
|
Using the results above and substituting:
□
Theorem 8. Let . Then:
- 1
- 2
- 3
- 4
- 5
- 6
Proof. If
, then:
Using the results above:
□
Theorem 9. Let . Then:
- 1
- 2
- 3
- 4
- 5
- 6
Proof. If
, then:
|
|
|
|
Using the results above:
□
Theorem 10. Let . Then:
- 1
- 2
- 3
- 4
- 5
- 6
Proof. If
m then:
Using the results above:
□
Using the same proving arguments, we have the following theorems for the remaining two nanotubes and .
Theorem 11. Let . Then:
- 1
- 2
- 3
- 4
- 5
- 6
Theorem 12. Let . Then:
- 1
- 2
- 3
- 4
- 5
- 6
4. Discussions
Topological indices are a numerical quantity computed from the molecular graph of a chemical compound and typically remain the same under graph isomorphism. They can be used to predict and model certain properties of chemical compounds; therefore, knowing the indices for these molecular graphs, we then understand the properties such as the chemical reactivity, physical features, biological activities, etc., of the associated molecule.
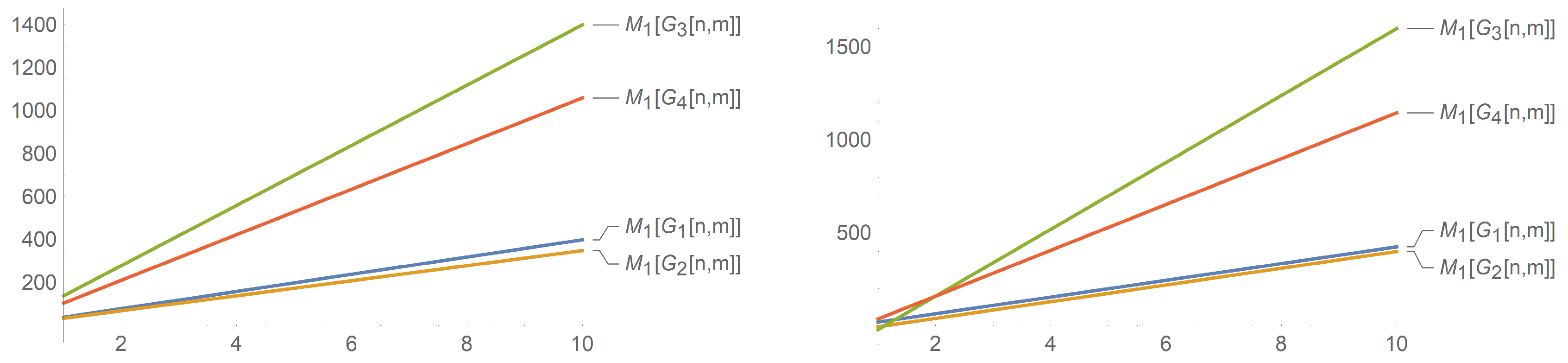
Having verified through experiments that the first Zagreb index is related to the total
-electron energy of the structure [
18,
19], hence a molecular graph having large values of
has a larger total
electron energy. Following from
Figure 7, we can find that both
and
have sharply rising total
-electron energy as values of
n increase and that
has a higher total
-electron energy as compared to
. Similarly, it follows from
Figure 8 that the total
-electron energy of
is greater than the other three nanotubes for
. It can also be observed that for a fixed value of
m, the total
-electron energies of
and
only differ from each other by some constant value.
Likewise, from the general Randic index, we can obtain the Randic index proposed by Randic by replacing
with −0.5. It has been observed by the chemist Randic in 1975 that the Randic index can be used in determining the physiochemical properties of alkanes; he observed that there is a good correlation between the Randic index and some physio-chemical properties of alkanes: boiling points, chromatography retention times, enthalpies of formation, surface areas, etc. In reference to
Figure 7, we could find that the Randic index of
and
both sharply increase with
n and that
is much greater than
. Observing
Figure 9, we find that
for
. The Randic index for
is greater than the rest of the three nanotubes if
, and the three nanotubes have Randic indices that steadily increase past
for all
.
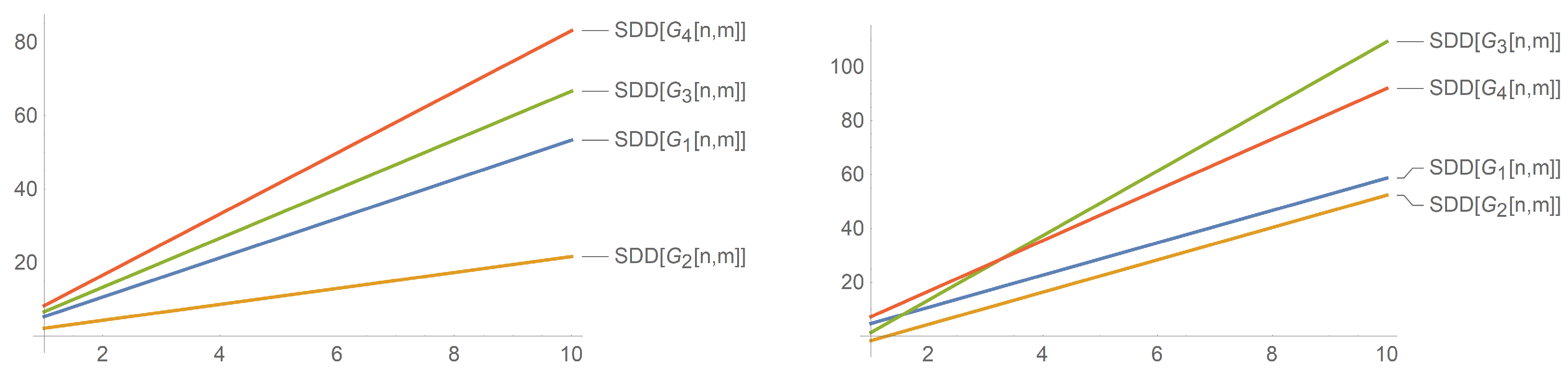
On the testing sets supplied by the International Academy of Mathematical Chemistry, it showed good predictive properties. It can also decently predict the total surface area for polychlorobiphenyls. Therefore, from
Figure 8, we note that the symmetric division index of
rises sharply with
being much greater than
. Similarly, we can also take note in reference to
Figure 10 that
has the largest index among all four nanotubes for
.
The above graphs for the nanotubes show a correlation of , , and with n and m. It can be seen that the indices have a linear correlation with n and m. This contrasts the correlation of the indices of and with n as the graphs are exponential and quickly rise. We only give an analysis for these three indices as the other indices ( and ) are similar and can be obtained from the Randic index with specific values of ( and , respectively), and therefore, the correlation between the indices and n (and m) does not change.
5. Conclusions
We computed the M-polynomial of two classes of nanostar dendrimers and four nanotubes and used the M-polynomials to obtain a wide list of indices for our molecular graphs. By analyzing our results, we found that were better at achieving larger values of the calculated topological indices in comparison to and likewise that were better at achieving larger values of these indices than the other three nanotubes. For future prospects, one can compute the M-polynomial for other types of nanostar dendrimers or nanotubes. For example, single-walled inorganic aluminosilicate and aluminogermanate nanotubes of a periodic structure and precise dimensions can be investigated related to the current study.