Deep Neural Network Compression for Plant Disease Recognition
Abstract
:1. Introduction
2. Related Works
2.1. Pruning
2.1.1. Pruning Granularity
2.1.2. Pruning Strategies
2.2. Knowledge Distillation
2.3. Quantization
2.4. Lightweight Networks
2.5. DL Architectures for Plant Disease Detection
3. Methodology
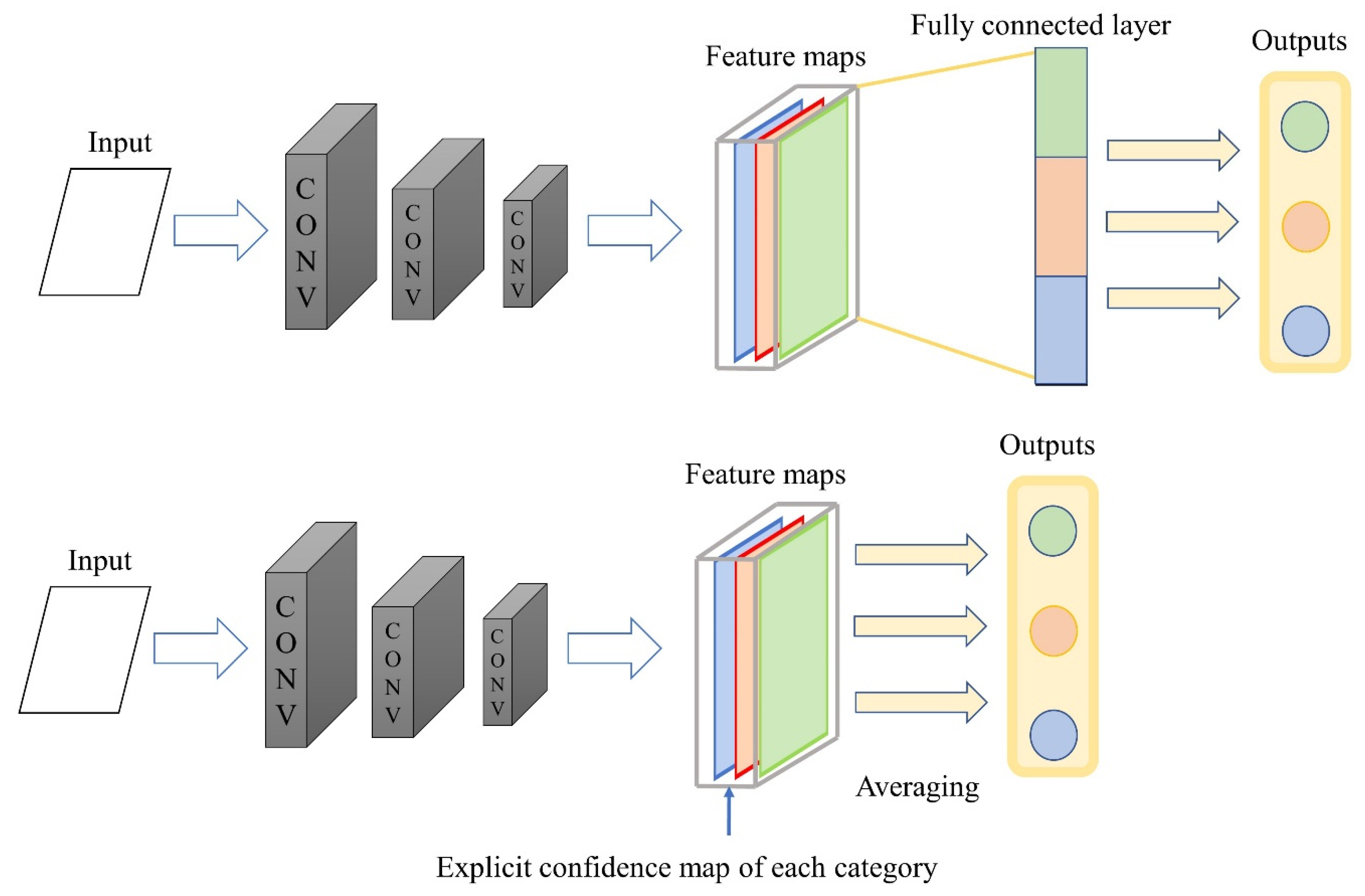
3.1. Lightweight on Fully Connected Layers
3.2. Iterative Pruning with Knowledge Distillation
3.2.1. Iterative Pruning
| Algorithm 1. Framework of iterative pruning with knowledge distillation |
| Initialization |
| 1: Convolution layer index to prune: ; |
| 2: Student model pruning rate at step : ; |
| 3: Student model final pruning rate at step : ; |
| Pretraining |
| 4: Training lightweight model ; |
| 5: Pretrained student model : ; |
| Iterative pruning |
| 6: |
| 7: Pruning ( ) filters of with feature map ranks; |
| Knowledge distillation |
| 8: Retraining model with knowledge distillation with ; |
| 9: If () then Goto Iterative pruning |
| Final model |
| 10: Return student model ; |
3.2.2. Retraining
3.3. Fixed-Point Quantization
4. Experimental Evaluation
4.1. Experimental Settings
4.1.1. Datasets
4.1.2. Configurations
4.2. Experimental Results
4.2.1. Filter Pruning
4.2.2. Quantization
4.3. Comparsion with Lightweight Model
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chetlur, S.; Woolley, C.; Vandermersch, P.; Cohen, J.; Tran, J.; Catanzaro, B.; Shelhamer, E. Cudnn: Efficient primitives for deep learning. arXiv 2014, arXiv:1410.0759. Available online: https://arxiv.org/abs/1410.0759 (accessed on 3 May 2021).
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Young, T.; Hazarika, D.; Poria, S.; Cambria, E. Recent trends in deep learning based natural language processing. IEEE Comput. Intell. Mag. 2018, 13, 55–75. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Advances in Neural Information Processing Systems; NIPS: Long Beac, CA, USA, 2017; pp. 5998–6008. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; Van Der Laak, J.A.; Van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [Green Version]
- Girshick, R. Fast r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 13–16 December 2015; pp. 1440–1448. [Google Scholar]
- Noh, H.; Hong, S.; Han, B. Learning deconvolution network for semantic segmentation. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 13–16 December 2015; pp. 1520–1528. [Google Scholar]
- Jia, X.; Gavves, E.; Fernando, B.; Tuytelaars, T. Guiding the long-short term memory model for image caption generation. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 13–16 December 2015; pp. 2407–2415. [Google Scholar]
- Rahnemoonfar, M.; Sheppard, C. Deep count: Fruit counting based on deep simulated learning. Sensors 2017, 17, 905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, H.; Chen, C. Fast implementation of real-time fruit detection in apple orchards using deep learning. Comput. Electron. Agric. 2020, 168, 105108. [Google Scholar] [CrossRef]
- Xiong, J.; Yu, D.; Liu, S.; Shu, L.; Wang, X.; Liu, Z.J.E. A review of plant phenotypic image recognition technology based on deep learning. Electronics 2021, 10, 81. [Google Scholar] [CrossRef]
- Kamilaris, A.; Prenafeta-Boldú, F.X. Deep learning in agriculture: A survey. Comput. Electron. Agric. 2018, 147, 70–90. [Google Scholar] [CrossRef] [Green Version]
- Mortensen, A.K.; Dyrmann, M.; Karstoft, H.; Jørgensen, R.N.; Gislum, R. Semantic segmentation of mixed crops using deep convolutional neural network. In Proceedings of the CIGR-AgEng Conference, Aarhus, Denmark, 26–29 June 2016; pp. 1–6. [Google Scholar]
- Hasan, R.I.; Yusuf, S.M.; Alzubaidi, L. Review of the state of the art of deep learning for plant diseases: A broad analysis and discussion. Plants 2020, 9, 1302. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- LeCun, Y.; Bottou, L.; Bengio, Y.; Haffner, P. Gradient-based learning applied to document recognition. Proc. IEEE 1998, 86, 2278–2324. [Google Scholar] [CrossRef] [Green Version]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. Available online: https://arxiv.org/abs/1409.1556 (accessed on 3 May 2021).
- Zhang, C.; Patras, P.; Haddadi, H. Deep learning in mobile and wireless networking: A survey. IEEE Commun. Surv. Tutorials 2019, 21, 2224–2287. [Google Scholar] [CrossRef] [Green Version]
- McCool, C.; Perez, T.; Upcroft, B. Mixtures of lightweight deep convolutional neural networks: Applied to agricultural robotics. IEEE Robot. Autom. Lett. 2017, 2, 1344–1351. [Google Scholar] [CrossRef]
- Gonzalez-Huitron, V.; León-Borges, J.A.; Rodriguez-Mata, A.; Amabilis-Sosa, L.E.; Ramírez-Pereda, B.; Rodriguez, H. Disease detection in tomato leaves via CNN with lightweight architectures implemented in Raspberry Pi 4. Comput. Electron. Agric. 2021, 181, 105951. [Google Scholar] [CrossRef]
- Chen, J.; Wang, W.; Zhang, D.; Zeb, A.; Nanehkaran, Y.A. Attention embedded lightweight network for maize disease recognition. Plant Pathol. 2021, 70, 630–642. [Google Scholar] [CrossRef]
- Tang, Z.; Yang, J.; Li, Z.; Qi, F. Grape disease image classification based on lightweight convolution neural networks and channelwise attention. Comput. Electron. Agric. 2020, 178, 105735. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, D.; Nanehkaran, Y.A. Identifying plant diseases using deep transfer learning and enhanced lightweight network. Multimed. Tools Appl. 2020, 79, 31497–31515. [Google Scholar] [CrossRef]
- Choudhary, T.; Mishra, V.; Goswami, A.; Sarangapani, J. A comprehensive survey on model compression and acceleration. Artif. Intell. Rev. 2020, 53, 5113–5155. [Google Scholar] [CrossRef]
- Hughes, D.; Salathé, M. An open access repository of images on plant health to enable the development of mobile disease diagnostics. arXiv 2015, arXiv:1511.08060. Available online: https://arxiv.org/abs/1511.08060 (accessed on 3 May 2021).
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Denker, J.S.; Solla, S.A. Optimal brain damage. In Advances in Neural Information Processing Systems; NIPS: Morgan Kaufmann, CA, USA, 1990; pp. 598–605. Available online: https://dl.acm.org/doi/10.5555/109230.109298 (accessed on 3 May 2021).
- Denil, M.; Shakibi, B.; Dinh, L.; Ranzato, M.A.; De Freitas, N. Predicting parameters in deep learning. arXiv 2013, arXiv:1306.0543. Available online: https://arxiv.org/abs/1306.0543 (accessed on 3 May 2021).
- Hassibi, B.; Stork, D.G. Second Order Derivatives for Network Pruning: Optimal Brain Surgeon. In Advances in Neural Information Processing Systems 5; NIPS: Morgan Kaufmann, CA, USA, 1993; pp. 164–171. [Google Scholar]
- Han, S.; Pool, J.; Tran, J.; Dally, W.J. Learning both weights and connections for efficient neural networks. arXiv 2015, arXiv:1506.02626. Available online: https://arxiv.org/abs/1506.02626 (accessed on 3 May 2021).
- Han, S.; Liu, X.; Mao, H.; Pu, J.; Pedram, A.; Horowitz, M.A.; Dally, W.J. EIE: Efficient inference engine on compressed deep neural network. ACM SIGARCH Comput. Archit. News 2016, 44, 243–254. [Google Scholar] [CrossRef]
- Li, H.; Kadav, A.; Durdanovic, I.; Samet, H.; Graf, H.P. Pruning filters for efficient convnets. arXiv 2016, arXiv:1608.08710. Available online: https://arxiv.org/abs/1608.08710 (accessed on 3 May 2021).
- Luo, J.-H.; Wu, J.; Lin, W. Thinet: A filter level pruning method for deep neural network compression. In Proceedings of the IEEE International Conference on Computer Vision, Venic, Italy, 27–29 October 2017; pp. 5058–5066. [Google Scholar]
- Liu, Z.; Li, J.; Shen, Z.; Huang, G.; Yan, S.; Zhang, C. Learning efficient convolutional networks through network slimming. In Proceedings of the IEEE International Conference on Computer Vision, Venic, Italy, 27–29 October 2017; pp. 2736–2744. [Google Scholar]
- Lin, M.; Ji, R.; Wang, Y.; Zhang, Y.; Zhang, B.; Tian, Y.; Shao, L. Hrank: Filter pruning using high-rank feature map. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 14–19 June 2020; pp. 1529–1538. [Google Scholar]
- Zhu, M.; Gupta, S. To prune, or not to prune: Exploring the efficacy of pruning for model compression. arXiv 2017, arXiv:1710.01878. Available online: https://arxiv.org/abs/1710.01878 (accessed on 3 May 2021).
- Hinton, G.; Vinyals, O.; Dean, J. Distilling the knowledge in a neural network. arXiv 2015, arXiv:1503.02531. Available online: https://arxiv.org/abs/1503.02531 (accessed on 3 May 2021).
- Fukuda, T.; Suzuki, M.; Kurata, G.; Thomas, S.; Cui, J.; Ramabhadran, B. Efficient Knowledge Distillation from an Ensemble of Teachers. In Proceedings of the Interspeech, Stockholm, Sweden, 20–24 August 2017; pp. 3697–3701. [Google Scholar]
- Zagoruyko, S.; Komodakis, N. Paying more attention to attention: Improving the performance of convolutional neural networks via attention transfer. arXiv 2016, arXiv:1612.03928. Available online: https://arxiv.org/abs/1612.03928 (accessed on 3 May 2021).
- Mirzadeh, S.I.; Farajtabar, M.; Li, A.; Levine, N.; Matsukawa, A.; Ghasemzadeh, H. Improved knowledge distillation via teacher assistant. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; pp. 5191–5198. [Google Scholar]
- Gong, Y.; Liu, L.; Yang, M.; Bourdev, L. Compressing deep convolutional networks using vector quantization. arXiv 2014, arXiv:1412.6115. Available online: https://arxiv.org/abs/1412.6115 (accessed on 3 May 2021).
- Vanhoucke, V.; Senior, A.; Mao, M.Z. Improving the Speed of Neural Networks on CPUs. 2011. Available online: https://www.researchgate.net/publication/319770111_Improving_the_speed_of_neural_networks_on_CPUs (accessed on 3 May 2021).
- Courbariaux, M.; Hubara, I.; Soudry, D.; El-Yaniv, R.; Bengio, Y. Binarized neural networks: Training deep neural networks with weights and activations constrained to +1 or −1. arXiv 2016, arXiv:1602.02830. Available online: https://arxiv.org/abs/1602.02830 (accessed on 3 May 2021).
- Han, S.; Mao, H.; Dally, W.J. Deep compression: Compressing deep neural networks with pruning, trained quantization and huffman coding. arXiv 2015, arXiv:1510.00149. Available online: https://arxiv.org/abs/1510.00149 (accessed on 3 May 2021).
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. Available online: https://arxiv.org/abs/1704.04861 (accessed on 3 May 2021).
- Iandola, F.N.; Han, S.; Moskewicz, M.W.; Ashraf, K.; Dally, W.J.; Keutzer, K. SqueezeNet: AlexNet-level accuracy with 50× fewer parameters and <0.5 MB model size. arXiv 2016, arXiv:1602.07360. Available online: https://arxiv.org/abs/1602.07360 (accessed on 3 May 2021).
- Ale, L.; Sheta, A.; Li, L.; Wang, Y.; Zhang, N. Deep learning based plant disease detection for smart agriculture. In Proceedings of the 2019 IEEE Globecom Workshops (GC Wkshps), Waikoloa, HI, USA, 9–13 December 2019; pp. 1–6. [Google Scholar]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Delnevo, G.; Girau, R.; Ceccarini, C.; Prandi, C. A Deep Learning and Social IoT approach for Plants Disease Prediction toward a Sustainable Agriculture. IEEE Internet Things J. 2021. Available online: https://ieeexplore.ieee.org/abstract/document/9486935 (accessed on 3 September 2021).
- Tetila, E.C.; Machado, B.B.; Menezes, G.K.; Oliveira, A.D.S.; Alvarez, M.; Amorim, W.P.; Belete, N.A.D.S.; Da Silva, G.G.; Pistori, H. Automatic recognition of soybean leaf diseases using UAV images and deep convolutional neural networks. IEEE Geosci. Remote. Sens. Lett. 2019, 17, 903–907. [Google Scholar] [CrossRef]
- Yu, H.-J.; Son, C.-H. Leaf spot attention network for apple leaf disease identification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Seattle, WA, USA, 14–19 June 2020; pp. 52–53. [Google Scholar]
- Lin, M.; Chen, Q.; Yan, S. Network in network. arXiv 2013, arXiv:1312.4400. Available online: https://arxiv.org/abs/1312.4400 (accessed on 3 September 2021).







| Class | Plant Common Name | Plant Scientific Name | Disease Common Name | Disease Scientific Name | Images (Number) |
|---|---|---|---|---|---|
| C_1 | Apple | Malus domestica | – | – | 1645 |
| C_2 | Apple | Malus domestica | Apple scab | Venturia inaequalis | 630 |
| C_3 | Apple | Malus domestica | Black rot | Botryosphaeria obtusa | 621 |
| C_4 | Apple | Malus domestica | Cedar apple rust | Gymnosporangium juniperi-virginianae | 275 |
| C_5 | Blueberry | Vaccinium spp. | – | – | 1502 |
| C_6 | Cherry (and sour) | Prunus spp. | – | – | 854 |
| C_7 | Cherry (and sour) | Prunus spp. | Powdery mildew | Podosphaera spp. | 1052 |
| C_8 | Corn (maize) | Zea mays | – | – | 1162 |
| C_9 | Corn (maize) | Zea mays | Cercospora leaf spot | Cercospora zeae-maydis | 513 |
| C_10 | Corn (maize) | Zea mays | Common rust | Puccinia sorghi | 1192 |
| C_11 | Corn (maize) | Zea mays | Northern leaf blight | Exserohilum turcicum | 987 |
| C_12 | Grape | Vitis vinifera | – | – | 423 |
| C_13 | Grape | Vitis vinifera | Black rot | Guignardia bidwellii | 1180 |
| C_14 | Grape | Vitis vinifera | Esca (Black measles) | Phaeomoniella chlamydospora | 1383 |
| C_15 | Grape | Vitis vinifera | Leaf blight | Pseudocercospora vitis | 1076 |
| C_16 | Orange | Citrus sinensis | Huanglongbing | Candidatus Liberibacter | 5507 |
| C_17 | Peach | Prunus persica | – | – | 360 |
| C_18 | Peach | Prunus persica | Bacterial spot | Xanthomonas campestris | 2297 |
| C_19 | Pepper, bell | Capsicum annuum | – | – | 1477 |
| C_20 | Pepper, bell | Capsicum annuum | Bacterial spot | Xanthomonas campestris | 997 |
| C_21 | Potato | Solanum tuberosum | – | – | 152 |
| C_22 | Potato | Solanum tuberosum | Early blight | Alternaria solani | 1000 |
| C_23 | Potato | Solanum tuberosum | Late blight | Phytophthora infestans | 1000 |
| C_24 | Raspberry | Rubus spp. | – | – | 371 |
| C_25 | Soybean | Glycine max | – | – | 5090 |
| C_26 | Squash | Cucurbita spp. | Powdery mildew | Erysiphe cichoracearum, Sphaerotheca fuliginea | 1835 |
| C_27 | Strawberry | Fragaria spp. | – | – | 456 |
| C_28 | Strawberry | Fragaria spp. | Leaf scorch | Diplocarpon earlianum | 1109 |
| C_29 | Tomato | Lycopersicum esculentum | – | – | 1591 |
| C_30 | Tomato | Lycopersicum esculentum | Bacterial spot | Xanthomonas campestris pv. Vesicatoria | 2127 |
| C_31 | Tomato | Lycopersicum esculentum | Early blight | Alternaria solani | 1000 |
| C_32 | Tomato | Lycopersicum esculentum | Late blight | Phytophthora infestans | 1909 |
| C_33 | Tomato | Lycopersicum esculentum | Leaf mold | Fulvia fulva | 952 |
| C_34 | Tomato | Lycopersicum esculentum | Septoria leaf spot | Septoria lycopersici | 1771 |
| C_35 | Tomato | Lycopersicum esculentum | Spider mites | Tetranychus urticae | 1676 |
| C_36 | Tomato | Lycopersicum esculentum | Target spot | Corynespora cassiicola | 1404 |
| C_37 | Tomato | Lycopersicum esculentum | Tomato mosaic virus | Tomato mosaic virus (ToMV) | 373 |
| C_38 | Tomato | Lycopersicum esculentum | ΤYLCV | Begomovirus (Fam. Geminiviridae) | 5357 |
| TOTAL: | 54,306 |
| VGGNet | Input Size | ||
|---|---|---|---|
| FLOPS | 15.53 G | 3.97 G | 1.06 G |
| Accuracy | 99.59% | 99.45% | 99.43% |
| (+0.0%) | (−0.14%) | (−0.16%) | |
| Model | VGGNet | VGGNet (GAP) | AlexNet | AlexNet (GAP) |
|---|---|---|---|---|
| Size | 512.79 Mb | 47.83 Mb | 218.06 Mb | 7.51 Mb |
| Accuracy | 99.43% | 99.37% | 99.18% | 98.15% |
| (+0.0%) | (−0.06%) | (+0.0%) | (−0.03%) |
| Model | Original | 85% | 90% | 95% | |
|---|---|---|---|---|---|
| Pruning | |||||
| VGGNet (GAP) | Size | 47.83 Mb | 1.18 Mb | 0.56 Mb | 0.16 Mb |
| Accuracy | 99.37% | 99.05% | 98.65% | 97.20% | |
| (+0.0%) | (−0.32%) | (−0.72%) | (−2.17%) | ||
| FLOPs | 920,230,062 | 22,974,968 | 10,764,786 | 3,240,726 | |
| AlexNet (GAP) | Size | 7.51 Mb | 0.23 Mb | 0.12 Mb | 0.04 Mb |
| Accuracy | 98.15% | 94.63% | 92.19% | 87.32% | |
| (+0.00%) | (−3.52%) | (−5.96%) | (−10.83%) | ||
| FLOPs | 21,628,408 | 1,085,788 | 666,896 | 322,200 | |
| Model | Original | 85% | 90% | 95% | |
|---|---|---|---|---|---|
| Pruning | |||||
| VGGNet (GAP) | Accuracy | 99.37% | 99.05% | 98.65% | 97.20% |
| Knowledge distillation | |||||
| Accuracy (KD) | _ | 99.06% | 98.74% | 97.26% | |
| (+0.01%) | (+0.09%) | (+0.06%) | |||
| AlexNet (GAP) | Accuracy | 98.15% | 94.63% | 92.19% | 87.32% |
| Knowledge distillation | |||||
| Accuracy (KD) | _ | 94.82% | 92.82% | 87.63% | |
| (+0.19%) | (+0.63%) | (+0.31%) | |||
| Model | Original | 85% | 90% | 95% | |
|---|---|---|---|---|---|
| Pruning | |||||
| VGGNet (GAP) | Size | 47.83 Mb | 1.18 Mb | 0.56 Mb | 0.16 Mb |
| Accuracy | 99.37% | 99.05% | 98.65% | 97.20% | |
| (+0.0%) | (−0.32%) | (−0.72%) | (−2.17%) | ||
| Fixed-point quantization with uint-8 bits. | |||||
| Size | _ | 0.29 Mb | 0.14 Mb | 0.04 Mb | |
| Accuracy | _ | 99.06% | 98.75% | 97.09% | |
| (−0.31%) | (−0.62%) | (−2.28%) | |||
| AlexNet (GAP) | Size | 7.51 Mb | 0.23 Mb | 0.12 Mb | 0.04 Mb |
| Accuracy | 98.15% | 94.63% | 92.19% | 87.32% | |
| (+0.00%) | (−3.52%) | (−5.96%) | (−10.83%) | ||
| Fixed-point quantization with uint-8 bits. | |||||
| Size | _ | 0.06 Mb | 0.03 Mb | 0.01 Mb | |
| Accuracy | _ | 94.75% | 92.78% | 87.51% | |
| (−3.40%) | (−5.37%) | (−10.64%) | |||
| Model | MobileNetV1 | MobileNetV2 |
|---|---|---|
| Size | 12.38 Mb | 8.67 Mb |
| Accuracy | 99.20% | 98.85% |
| FLOPs | 150,466,304 | 23,550,528 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Zhang, W.; Ding, J.; Xia, M.; Wang, M.; Rao, Y.; Jiang, Z. Deep Neural Network Compression for Plant Disease Recognition. Symmetry 2021, 13, 1769. https://doi.org/10.3390/sym13101769
Wang R, Zhang W, Ding J, Xia M, Wang M, Rao Y, Jiang Z. Deep Neural Network Compression for Plant Disease Recognition. Symmetry. 2021; 13(10):1769. https://doi.org/10.3390/sym13101769
Chicago/Turabian StyleWang, Ruiqing, Wu Zhang, Jiuyang Ding, Meng Xia, Mengjian Wang, Yuan Rao, and Zhaohui Jiang. 2021. "Deep Neural Network Compression for Plant Disease Recognition" Symmetry 13, no. 10: 1769. https://doi.org/10.3390/sym13101769
APA StyleWang, R., Zhang, W., Ding, J., Xia, M., Wang, M., Rao, Y., & Jiang, Z. (2021). Deep Neural Network Compression for Plant Disease Recognition. Symmetry, 13(10), 1769. https://doi.org/10.3390/sym13101769







