Comparison Analysis of Gene Expression Profiles Proximity Metrics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mutual Information Maximization Method
2.2. Modified Mutual Information Maximization Proximity Metric
2.3. A Method for Estimating the Gene Expression Profiles’ Proximity Based on Pearson’s Test
| Algorithm 1: MMIM proximity metric values calculation. |
 |
2.4. A Method for Estimating the Gene Expression Profiles Proximity Based on the Correlation Distance
3. Experiment, Results and Discussion
3.1. Evaluation of the MIM Method’s Effectiveness When Using Various Methods of Shannon Entropy Calculation
3.2. An Application and Comparison Analysis of the MMIM Proximity Metric, Pearson’s Test and Correlation Distance
3.3. The Hybrid Metric of Gene Expression Profiles Proximity Evaluation
| Algorithm 2: Hybrid metric of gene expression profiles proximity evaluation. |
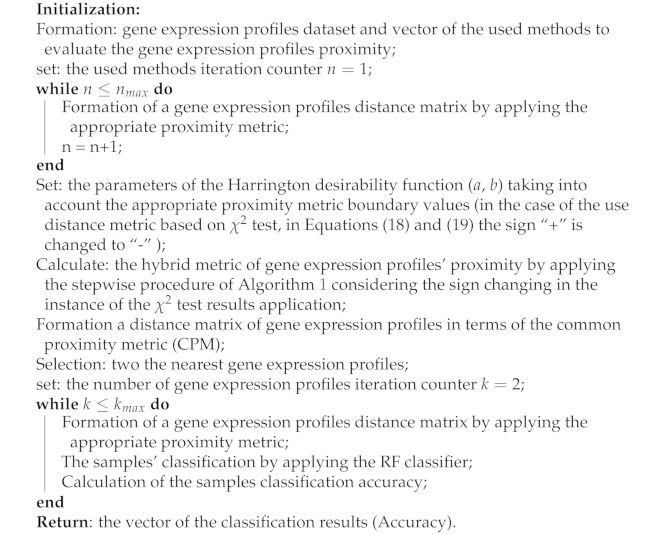 |
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| GRN | Gene Regulatory Network |
| MIM | Mutual Information Maximization |
| MMIM | Modified Mutual Information Maximization |
| RF | Random Forest |
| SOTA | Self Organizing Tree Algorithm |
| ML | Maximum Likelihood |
| JF | Jeffreys |
| LP | Laplace |
| MnM | MiniMax |
| HPM | Hybrid Proximity Metric |
References
- ArrayExpress—Functional Genomics Data. Available online: https://www.ebi.ac.uk/arrayexpress/ (accessed on 1 May 2014).
- Babichev, S.; Yasinskyi, M.; Yasinska-Damri, L.; Ratushniak, Y.; Lytvynenko, V. Current state of the problem of gene expression data processing and extraction to solve the reverse engineering tasks in the field of bioinformatics. Ceur Workshop Proc. 2021, 2853, 62–71. [Google Scholar]
- Wang, L.; Song, F.; Yin, H.; Zhu, W.; Fu, J.; Dong, Z.; Xu, P. Comparative microRNAs expression profiles analysis during embryonic development of common carp, Cyprinus carpio. Comp. Biochem. Physiol.—Part Genom. Proteom. 2021, 37, 100754. [Google Scholar] [CrossRef]
- Marchetti, M.A.; Coit, D.G.; Dusza, S.W.; Yu, A.; McLean, L.; Hu, Y.; Nanda, J.K.; Matsoukas, K.; Mancebo, S.E.; Bartlett, E.K. Performance of Gene Expression Profile Tests for Prognosis in Patients with Localized Cutaneous Melanoma: A Systematic Review and Meta-Analysis. JAMA Dermatol. 2020, 156, 953–962. [Google Scholar] [CrossRef]
- Almugren, N.; Alshamlan, H. A survey on hybrid feature selection methods in microarray gene expression data for cancer classification. IEEE Access 2019, 7, 78533–78548. [Google Scholar] [CrossRef]
- Lu, H.; Chen, J.; Yan, K.; Jin, Q.; Xue, Y.; Gao, Z. A hybrid feature selection algorithm for gene expression data classification. Neurocomputing 2017, 256, 56–62. [Google Scholar] [CrossRef]
- Vijay, S.A.A.; Kumar, P.G. Fuzzy expert system based on a novel hybrid stem cell (HSC) algorithm for classification of micro array data. J. Med. Syst. 2018, 42, 61. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.P.; Leu, Y. A novel hybrid feature selection method for microarray data analysis. Appl. Soft Comput. 2011, 11, 208–213. [Google Scholar] [CrossRef]
- Chuang, L.-Y.; Yang, C.-H.; Wu, K.-C.; Yang, C.-H. A hybrid feature selection method for DNA microarray data. Comput. Biol. Med. 2011, 41, 228–237. [Google Scholar] [CrossRef]
- Jain, I.; Jain, V.K.; Jain, R. Correlation feature selection based improved-binary particle swarm optimization for gene selection and cancer classification. Appl. Soft Comput. 2018, 62, 203–215. [Google Scholar] [CrossRef]
- Dashtban, M.; Balafar, M. Gene selection for microarray cancer classification using a new evolutionary method employing artificial intelligence concepts. Genomics 2017, 109, 91–107. [Google Scholar] [CrossRef]
- Salem, H.; Attiya, G.; El-Fishawy, N. Classification of human cancer diseases by gene expression profiles. Appl. Soft Comput. 2017, 50, 124–134. [Google Scholar] [CrossRef]
- Sharbaf, F.V.; Mosafer, S.; Moattar, M.H. A hybrid gene selection approach for microarray data classification using cellular learning automata and ant colony optimization. Genomics 2016, 107, 231–238. [Google Scholar] [CrossRef]
- Dashtban, M.; Balafar, M.; Suravajhala, P. Gene selection for tumor classification using a novel bio-inspired multi-objective approach. Genomics 2018, 110, 10–17. [Google Scholar] [CrossRef]
- Alshamlan, H.; Badr, G.; Alohali, Y. mRMR-ABC: A hybrid gene selection algorithm for cancer classification using microarray gene expression profiling. Biomed. Res. Int. 2018, 2015, 604910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alshamlan, H.M.; Badr, G.H.; Alohali, Y.A. Genetic bee colony (GBC) algorithm: A new gene selection method for microarray cancer classification. Comput. Biol. Chem. 2015, 56, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Moradi, P.; Gholampour, M. A hybrid particle swarm optimization for feature subset selection by integrating a novel local search strategy. Appl. Soft Comput. 2016, 43, 117–130. [Google Scholar] [CrossRef]
- Li, X.; Yin, M. Multiobjective binary biogeography based optimization for feature selection using gene expression data. IEEE Trans. Nanobiosci. 2013, 12, 343–353. [Google Scholar] [CrossRef] [PubMed]
- Shreem, S.S.; Abdullah, S.; Nazri, M.Z.A. Hybrid feature selection algorithm using symmetrical uncertainty and a harmony search algorithm. Int. J. Syst. Sci. 2016, 47, 1312–1329. [Google Scholar] [CrossRef]
- Izonin, I.; Tkachenko, R.; Kryvinska, N.; Zub, K.; Mishchuk, O.; Lisovych, T. Recovery of Incomplete IoT Sensed Data using High-Performance Extended-Input Neural-Like Structure. Procedia Comput. Sci. 2019, 160, 521–526. [Google Scholar] [CrossRef]
- Izonin, I.; Kryvinska, N.; Tkachenko, R.; Zub, K. An Approach towards Missing Data Recovery within IoT Smart System. Procedia Comput. Sci. 2019, 155, 11–18. [Google Scholar] [CrossRef]
- Babichev, S.; Khamula, O.; Durnyak, B.; Škvor, J. Technique of gene expression profiles selection based on SOTA clustering algorithm using statistical criteria and Shannon entropy. Adv. Intell. Syst. Comput. 2021, 1246, 23–38. [Google Scholar] [CrossRef]
- Babichev, S.; Škvor, J. Technique of Gene Expression Profiles Extraction Based on the Complex Use of Clustering and Classification Methods. Diagnostics 2020, 10, 584. [Google Scholar] [CrossRef] [PubMed]
- Babichev, S.; Barilla, J.; Fišer, J.; Škvor, J. A hybrid model of gene expression profiles reducing based on the complex use of fuzzy inference system and clustering quality criteria. In Proceedings of the 11th Conference of the European Society for Fuzzy Logic and Technology, EUSFLAT 2019, Prague, Czech Republic, 9–13 September 2020; pp. 128–133. [Google Scholar] [CrossRef] [Green Version]
- Thomas, M.C.; Joy, A.T. Elements of Information Theory, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2006; 792p. [Google Scholar]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423, 623–656. [Google Scholar] [CrossRef] [Green Version]
- Hausser, J.; Strimmer, K. Entropy inference and the James-Stein estimator, with application to nonlinear gene association networks. J. Mach. Learn. Res. 2009, 10, 1469–1484. [Google Scholar]
- Miller, G. Note on the Bias of Information Estimates. Information Theory in Psychology. 1955, pp. 95–100. Available online: https://www.scienceopen.com/document?vid=357d299f-62fa-4bda-8dd2-e4d5b5abde5d (accessed on 10 August 2021).
- Horvitz, D.G.; Thompson, D.J. A generalization of sampling without replacement from a finite universe. J. Am. Stat. Assoc. 1952, 47, 663–685. [Google Scholar] [CrossRef]
- Orlitsky, A.; Santhanam, N.P.; Zhang, J. Always Good Turing: Asymptotically optimal probability estimation. Science 2003, 302, 427–431. [Google Scholar] [CrossRef]
- Archer, E.; Park, I.M.; Pillow, J.W. Bayesian Entropy Estimation for Countable Discrete Distributions. J. Mach. Learn. Res. 2014, 15, 2833–2868. [Google Scholar]
- Harrington, J. The desirability function. Ind. Qual. Control 1965, 21, 494–498. [Google Scholar]
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Hou, J.; Aerts, J.; den Hamer, B.; van Ijcken, W.; den Bakker, M.; Riegman, P.; Van Der Leest, C.; Van Der Spek, P.; Foekens, J.A.; Hoogsteden, H.C.; et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS ONE 2010, 5, e10312. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Breiman 2001, 45, 5–32. [Google Scholar]
- Kuhn, M.; Wing, J.; Weston, S. Classification and Regression Training. Available online: https://github.com/topepo/caret/ (accessed on 18 May 2020).







| The Concentration Parameter Values | Method of the Probabilities Evaluation |
|---|---|
| 0 | Maximum Likelihood (ML) |
| 1/2 | Jeffreys (JF) |
| 1 | Laplace (LP) |
| 1/n | Schurmann–Grassberger (SG) |
| formila | Minimax (MnM) |
| Real State of the Examined Objects | Results of the Objects Classification | |
|---|---|---|
| Tumor Predicted | Healthy | |
| Tumor (1) | True positives (TP) | False negatives (FN) |
| Healthy (0) | False positives (FP) | True negatives (TN) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Babichev, S.; Yasinska-Damri, L.; Liakh, I.; Durnyak, B. Comparison Analysis of Gene Expression Profiles Proximity Metrics. Symmetry 2021, 13, 1812. https://doi.org/10.3390/sym13101812
Babichev S, Yasinska-Damri L, Liakh I, Durnyak B. Comparison Analysis of Gene Expression Profiles Proximity Metrics. Symmetry. 2021; 13(10):1812. https://doi.org/10.3390/sym13101812
Chicago/Turabian StyleBabichev, Sergii, Lyudmyla Yasinska-Damri, Igor Liakh, and Bohdan Durnyak. 2021. "Comparison Analysis of Gene Expression Profiles Proximity Metrics" Symmetry 13, no. 10: 1812. https://doi.org/10.3390/sym13101812
APA StyleBabichev, S., Yasinska-Damri, L., Liakh, I., & Durnyak, B. (2021). Comparison Analysis of Gene Expression Profiles Proximity Metrics. Symmetry, 13(10), 1812. https://doi.org/10.3390/sym13101812






