Multinomial Logit Model Building via TreeNet and Association Rules Analysis: An Application via a Thyroid Dataset
Abstract
:1. Introduction
2. Generating Variables and Interactions with TreeNet and Association Rules Analysis for the Multinomial Logit Model
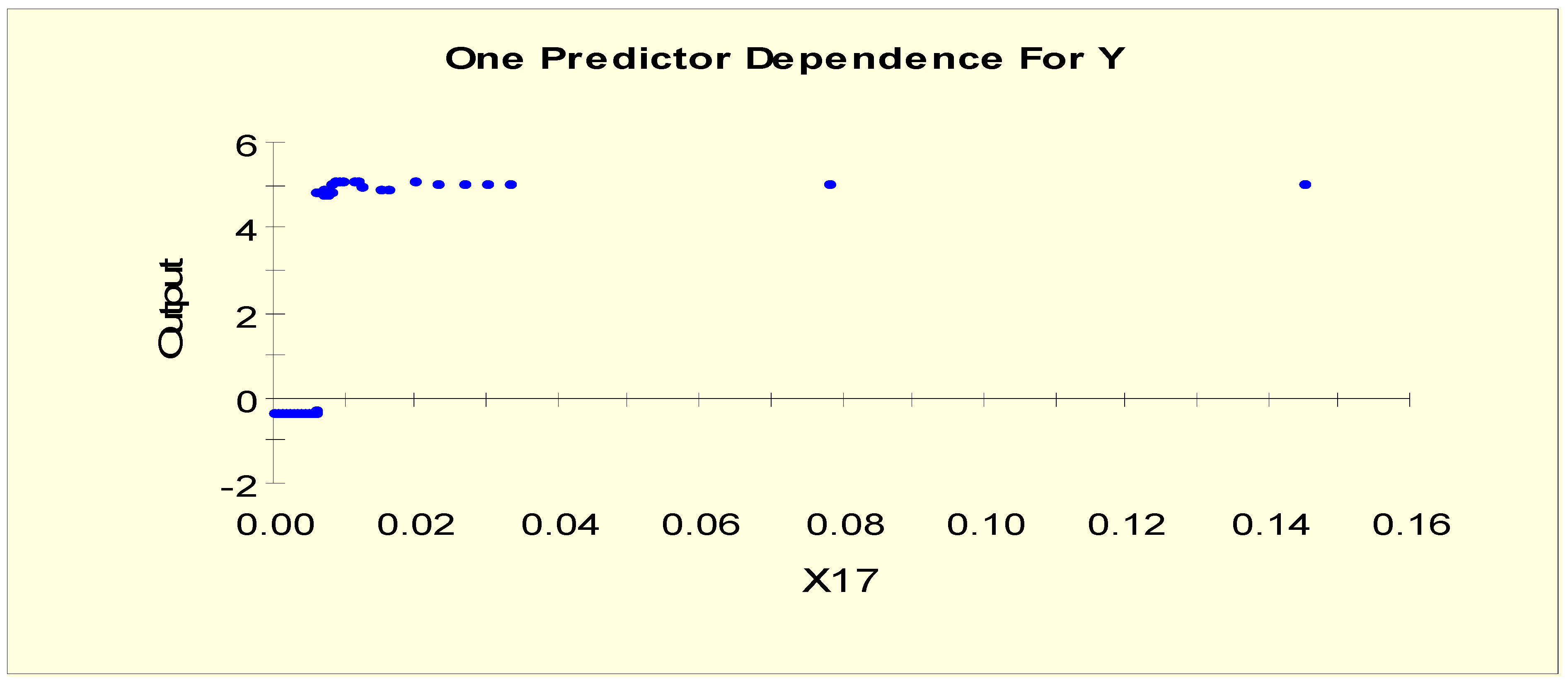
2.1. TreeNet
2.2. Association Rules Analysis (ASA)
3. Proposed Method
- Step 1:
- Discretization
- Step 2:
- Rules Generation
- Step 3:
- Rules Selection
- Step 4:
- Variable Generation
- Step 5:
- Model Selection
4. Illustrated Example: Thyroid Dataset
4.1. Results from Model 1
4.2. Results from Model 2
- Rule 6: If X20L1 = 1, X18LL1 = 0, and X21LL4 = 1, then Y = 0 with s = 8.537%, c = 100%.
- Rule 8: If X11 = 0, X19LL3 = 1, and X18LL1 = 0, then Y = 0 with s = 3.075%, c = 100%.
- Rule 22: If X8 = 0, X17L1 = 0, and X19L3 = 0, then Y = 1 with s = 1.935%, c = 97.26%.
- Rule 26: If X3 = 0, X17LL1 = 0, and X19LL2 = 1, then Y = 2 with s = 5.276%, c = 88.945%.
4.3. Performance Comparison Using the Training Set
4.4. Performance Comparison Using the Test Set
4.5. Performance Comparison with Other Methodologies
5. Discussion and Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zahid, F.M.; Tutz, G. Multinomial Logit Models with Implicit Variable Selection; Technical Report No. 89; Institute of Statistics, Ludwig-Maximilians-University: Munich, Germany, 2010. [Google Scholar]
- Cherrie, J.A. Variable Screening for Multinomial Logistic Regression on Very Large Data Sets as Applied to Direct Response Modeling. In Proceedings of the SAS Conference Proceedings, SAS Global Forum, Orlando, FL, USA, 16–19 April 2007. [Google Scholar]
- Camminatiello, I.; Lucadamo, A. Estimating multinomial logit model with multicollinearity data. Asian J. Math. Stat. 2010, 3, 93–101. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, M. Two-stage multinomial logit model. Expert Syst. Appl. 2011, 38, 6439–6446. [Google Scholar] [CrossRef]
- Changpetch, P.; Lin, D.K.J. Selection for multinomial logit models via association rules analysis. WIREs Comput. Stat. 2013, 5, 68–77. [Google Scholar] [CrossRef]
- Introducing TreeNet® Gradient Boosting Machine. Available online: https://www.minitab.com/content/dam/www/en/uploadedfiles/content/products/spm/TreeNet_Documentation.pdf (accessed on 18 February 2020).
- Changpetch, P.; Lin, D.K.J. Model selection for logistic regression via association rules analysis. J. Stat. Comput. Simul. 2013, 83, 1415–1428. [Google Scholar] [CrossRef]
- Agresti, A. Categorical Data Analysis, 2nd ed.; Wiley: Hoboken, NJ, USA, 2020. [Google Scholar]
- Yang, Y.; Webb, G.I. A Comparative Study of Discretization Methods for Naïve-Bayes Classifiers. In Proceedings of the 2002 Pacific Rim Knowledge Acquisition Workshop (PKAW’02), Tokyo, Japan, 18–19 August 2002; Yamaguchi, T., Hoffmann, A., Motoda, H., Compton, P., Eds.; Japanese Society for Artificial Intelligence: Tokyo, Japan, 2002; pp. 159–173. [Google Scholar]
- Catlett, J. On Changing Continuous Attributes into Ordered Discrete Attributes. In Proceedings of the European Working Session on Learning, European Working Session on Learning, Porto, Portugal, 6–8 March 1991; Springer: New York, NY, USA, 1991; pp. 164–178. [Google Scholar]
- Dougherty, J.; Kohavi, R.; Sahami, M. Supervised and Unsupervised Discretization of Continuous Features. In Machine Learning: Proceedings of the Twelfth International Conference on Machine Learning, Tahoe City, CA, USA, 9–12 July 1995; Morgan Kaufmann Publishers: San Francisco, CA, USA, 1995; pp. 194–202. [Google Scholar]
- Kononenko, I. Inductive and Bayesian learning in medical diagnosis. Appl. Artif. Intell. Int. J. 1993, 7, 317–337. [Google Scholar] [CrossRef]
- Kwedlo, W.; Krętowski, M. An Evolutionary Algorithm Using Multivariate Discretization for Decision Rule Induction. In Principles of Data Mining and Knowledge Discovery. PKDD 1999: Lecture Notes in Computer Science, Third European Conference, Prague, Czech Republic, 15–18 September 199; Żytkow, J.M., Rauch, J., Eds.; Springer: Heidelberg/Berlin, Germany, 1999; Volume 1704, pp. 392–397. [Google Scholar]
- Fayyad, U.M.; Irani, K.B. Multi-Interval Discretization of Continuous-Valued Attributes for Classification Learning. In Proceedings of the Thirteenth International Joint Conference on Artificial Intelligence, San Francisco, CA, USA, 28 August–3 September 1993; Morgan Kauffman: Chambéry, France, 1993; Volume 1, pp. 1022–1027. [Google Scholar]
- Pazzani, M.J. An Iterative Improvement Approach for the Discretization of Numeric Attributes in Bayesian Classifiers. In Proceedings of the First International Conference on Knowledge Discovery and Data Mining, Montreal, QC, Canada, 20–21 August 1995; Fayyad, U.M., Uthurusamy, R., Eds.; AAAI Press: Montreal, QC, Canada, 1995; pp. 228–233. [Google Scholar]
- Yang, Y.; Webb, G.I. Proportional k-interval discretization for naive-Bayes classifiers. In Machine Learning: ECML 2001: Lecture Notes in Computer Science, Freiburg, Germany, 5–7 September 2001; De Raedt, L., Flach, P., Eds.; Springer: Heidelberg/Berlin, Germany, 2001; Volume 2167, pp. 228–233. [Google Scholar]
- Hsu, C.N.; Huang, H.J.; Wong, T.T. Why Discretization Works for Naïve Bayesian Classifiers. In Proceedings of the Seventeenth International Conference on Machine Learning, Stanford, CA, USA, 29 June–2 July 2000; Langley, P., Ed.; Morgan Kaufman: San Francisco, CA, USA, 2000; pp. 309–406. [Google Scholar]
- Yang, Y.; Webb, G.I. Non-Disjoint Discretization for Naive-Bayes Classifiers. In Proceedings of the Nineteenth International Conference on Machine Learning (ICML’02), Sydney, Australia, 8–12 July 2002; Sammut, C., Hoffmann, A., Eds.; Morgan Kaufmann: San Francisco, CA, USA, 2002; pp. 666–673. [Google Scholar]
- Yang, Y.; Webb, G.I. Weighted Proportional k-Interval Discretization for Naive-Bayes Classifiers. In Advances in Knowledge Discovery and Data Mining. PAKDD 2003: Lecture Notes in Computer Science, 7th Pacific-Asia Conference, PAKDD, 2003, Seoul, Korea, 30 April 30–2 May 2003; Whang, K.Y., Jeon., J., Shim, K., Srivastava, J., Eds.; Springer: Heidelberg/Berlin, Germany, 2003; Volume 263, pp. 501–512. [Google Scholar]
- Aggarwal, M. On learning of choice models with interactive attributes. IEEE Trans. Knowl. Data Eng. 2016, 28, 2697–2708. [Google Scholar] [CrossRef]
- Berry, M.J.A.; Linoff, G. Data Mining Techniques: For Marketing, Sales, and Customer Support; John Wiley & Sons: New York, NY, USA, 1997. [Google Scholar]
- Agrawal, R.; Srikant, S. Fast Algorithms for Mining Association Rules. In Proceedings of the 20th International Conference on Very Large Data Bases, Santiago, Chile, 12–15 September 1994; Morgan Kaufmann: San Francisco, CA, USA, 1994; pp. 487–499. [Google Scholar]
- Liu, B.; Hsu, W.; Ma, Y. Integrating Classification and Association Rule Mining. In KDD-98 Proceedings of the Fourth International Conference on Knowledge Discovery and Data Mining, New York, NY, USA, 27–31 August 1998; Agrawal, R., Stolorz, P., Piatetsky-Shapiro, G., Eds.; AAAI Press: New York, NY, USA, 1998; pp. 80–86. [Google Scholar]
- Quinlan, J.R. C4.5: Programs for Machine Learning; Morgan Kaufmann: San Francisco, CA, USA, 1992. [Google Scholar]
- Changpetch, P.; Lin, D.K.J. Model selection for Poisson regression via association rules analysis. Internat. J. Stat. Prob. 2015, 4, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, G. Estimating the dimension of a model. Ann. Stat. 1978, 6, 461–464. [Google Scholar] [CrossRef]
- Schiffmann, W.; Joost, M.; Werner, R. Optimization of the Backpropagation Algorithm for Training Multilayer Perceptrons; Technical Report; University of Koblenz: Mainz, Germany, 1994. [Google Scholar]
- Keller, F.; Müller, E.; Böhm, K. HiCS: High Contrast Subspaces for Density-Based Outlier Ranking. In Proceedings of the 2012 IEEE 28th International Conference on Data Engineering (ICDE’12), Arlington, VA, USA, 1–5 April 2012; pp. 1037–1048. [Google Scholar]
- Aggarwal, C.C.; Sathe, S. Theoretical foundations and algorithms for outlier ensembles. SIGKDD Explor. Newsl. 2015, 17, 24–47. [Google Scholar] [CrossRef]
- Breunig, M.M.; Kriegel, H.; Ng, R.T.; Sander, J. LOF: Identifying Density-Based Local Outliers. In Proceedings of the 2000 ACM SIGMOD International Conference on Management of Data (SIGMOD’00), Dallas, TX, USA, 16–18 May 2000; ACM: New York, NY, USA, 2000; pp. 93–104. [Google Scholar]
- Cheng, C.; Fu, A.W.; Zhang, Y. Entropy-Based Subspace Clustering for Mining Numerical Data. In Proceedings of the Fifth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD’99), San Diego, CA, USA, 15–18 August 1999; ACM: New York, NY, USA, 1999; pp. 84–93. [Google Scholar]
- Kailing, K.; Kriegel, H.P.; Kröger, P.; Wanka, S. Ranking Interesting Subspaces for Clustering High Dimensional Data. In Knowledge Discovery in Databases: PKDD 2003: Lecture Notes in Computer Science, PKDD 2003, 7th European Conference on Principles and Practice of Knowledge Discovery in Databases, Cavtat-Dubrovnik, Croatia, 22–26 September 2003; Lavrač, N., Gamberger, D., Todorovski, L., Blockeel, H., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; Volume 2838, pp. 241–253. [Google Scholar]
- Skurichina, M.; Duin, R.P.W. Bagging, boosting and the random subspace method for linear classifiers. Pattern Anal. Appl. 2002, 5, 121–135. [Google Scholar] [CrossRef]
- Ceylan, Z. Estimation of COVID-19 prevalence in Italy, Spain, and France. Sci. Total Environ. 2020, 729, 138817. [Google Scholar] [CrossRef]
- Lukman, A.F.; Rauf, R.I.; Abiodun, O.; Oludoun, O.; Ayinde, K.; Ogundokun, R.O. COVID-19 prevalence estimation: Four most affected African countries. Infect. Dis. Model. 2020, 5, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Benvenuto, D.; Giovanetti, M.; Vassallo, L.; Angeletti, S.; Ciccozzi, M. Application of the ARIMA model on the COVID-2019 epidemic dataset. Data Brief. 2020, 29, 105340. [Google Scholar] [CrossRef] [PubMed]
- La Gatta, V.; Moscato, V.; Postiglione, M.; Sperli, G. An epidemiological neural network exploiting dynamic graph structured data applied to the covid-19 outbreak. IEEE Trans. Big Data 2020, 14. [Google Scholar] [CrossRef]
- Varotsos, C.A.; Krapivin, V.F. A new model for the spread of COVID-19 and the improvement of safety. Safety Sci. 2020, 132, 104962. [Google Scholar] [CrossRef]











| Attribute | Description | Variable |
|---|---|---|
| Age | Age in years | X1 |
| Sex | Gender | X2 = 1 if male and X2 = 0 if female |
| On Thyroxine | Patient on Thyroxine | X3 = 1 if yes and X3 = 0 if no |
| Query Thyroxine | Maybe on Thyroxine | X4 = 1 if yes and X4 = 0 if no |
| On antithyroid | On antithyroid medication | X5 = 1 if yes and X5 = 0 if no |
| Sick | Patient reports malaise | X6 = 1 if yes and X6 = 0 if no |
| Pregnant | Patient pregnant | X7 = 1 if yes and X7 = 0 if no |
| Thyroid surgery | History of thyroid surgery | X8 = 1 if yes and X8 = 0 if no |
| I131 treatment | Patient on I131 treatment | X9 = 1 if yes and X9 = 0 if no |
| Query hypothyroid | Maybe hypothyroid | X10 = 1 if yes and X10 = 0 if no |
| Query hyperthyroid | Maybe hyperthyroid | X11 = 1 if yes and X11 = 0 if no |
| Lithium | Patient on lithium | X12 = 1 if yes and X12 = 0 if no |
| Goiter | Patient has goiter | X13 = 1 if yes and X13 = 0 if no |
| Tumor | Patient has tumor | X14 = 1 if yes and X14 = 0 if no |
| Hypopituitary | Patient hypopituitary | X15 = 1 if yes and X15 = 0 if no |
| Psych | Psychological symptoms | X16 = 1 if yes and X16 = 0 if no |
| Thyroid Stimulating Hormone (TSH) | TSH value, if measured | X17 |
| Triiodothyronine (T3) | T3 value, if measured | X18 |
| Total Thyroxine (TT4) | TT4 value, if measured | X19 |
| Thyroxine Uptake (T4U) | T4U value, if measured | X20 |
| Free Thyroxine Index (FTI) | FTI—calculated from TT4 and T4U | X21 |
| Variable | Score | |
|---|---|---|
| X17 | 100.00 | |||||||||||||||||||||||||||||||||||||||||| |
| X21 | 63.88 | |||||||||||||||||||||||||| |
| X8 | 25.08 | |||||||||| |
| X3 | 17.38 | |||||| |
| X19 | 12.16 | |||| |
| X18 | 6.67 | || |
| X20 | 6.56 | || |
| X2 | 6.42 | || |
| X1 | 3.11 | |
| X10 | 2.71 | |
| X9 | 2.00 | |
| X11 | 1.93 |
| Original Variables | Generated Binary Variables |
|---|---|
| TSH (X17) | X17L1 = 1 if X17 < 0.025 and X17L1 = 0 otherwise |
| X17L2 = 1 if 0.025 ≤ X17 and X17L2 = 0 otherwise | |
| FTI (X21) | X21L1 = 1 if X21 < 0.055 and X21L1 = 0 otherwise |
| X21L2 = 1 if 0.055 ≤ X21 < 0.07 and X21L2 = 0 otherwise | |
| X21L3 = 1 if 0.07 ≤ X21 and X21L3 = 0 otherwise | |
| TT4 (X19) | X19L1 = 1 if X19 < 0.042 and X19L1 = 0 otherwise |
| X19L2 = 1 if 0.042 ≤ X19 < 0.065 and X19L2 = 0 otherwise | |
| X19L3 = 1 if 0.065 ≤ X19 and X19L3 = 0 otherwise | |
| T3 (X18) | X18L1 = 1 if X18 < 0.006 and X18L1 = 0 otherwise |
| X18L2 = 1 if 0.006 ≤ X18 and X18L2 = 0 otherwise | |
| T4U (X20) | X20L1 = 1 if X20 < 0.097 and X20L1 = 0 otherwise |
| X20L2 = 1 if 0.097 ≤ X20 and X20L2 = 0 otherwise | |
| Age (X1) | X1L1 = 1 if X1 < 0.15 and X1L1 = 0 otherwise |
| X1L2 = 1 if 0.15 ≤ X1 and X1L2 = 0 otherwise |
| Original Variables | Generated Variables with Linear Trend |
|---|---|
| FT1 (X21) | X21Q2 = X21 if 0.055 ≤ X21 < 0.07 and X21Q2 = 0 otherwise |
| TT4 (X19) | X19Q2 = X19 if 0.042 ≤ X19 < 0.065 and X19Q2 = 0 otherwise |
| Variable | Score | |
|---|---|---|
| X17 | 100.00 | |||||||||||||||||||||||||||||||||||||||||| |
| X3 | 44.88 | |||||||||||||||||| |
| X8 | 25.20 | |||||||||| |
| X19 | 22.47 | ||||||||| |
| X18 | 17.51 | ||||||| |
| X21 | 10.87 | |||| |
| X20 | 8.04 | || |
| X1 | 6.68 | || |
| X10 | 5.15 | | |
| X5 | 4.21 | | |
| X12 | 4.02 | | |
| X11 | 3.20 | |
| X2 | 0.84 |
| Original Variables | Generated Binary Variables |
|---|---|
| TSH (X17) | X17LL1 = 1 if X17 < 0.006 and X17LL1 = 0 otherwise |
| X17LL2 = 1 if 0.006 ≤ X17 and X17LL2 = 0 otherwise | |
| FTI (X21) | X21LL1 = 1 if X21 < 0.057 and X21LL1 = 0 otherwise |
| X21LL2 = 1 if 0.057 ≤ X21 < 0.071 and X21LL2 = 0 otherwise | |
| X21LL3 = 1 if 0.071 ≤ X21 < 0.115 and X21LL3 = 0 otherwise | |
| X21LL4 = 1 if 0.115 ≤ X21 < 0.217 and X21LL4 = 0 otherwise | |
| X21LL5 = 1 if 0.217 ≤ X21 and X21LL5 = 0 otherwise | |
| TT4 (X19) | X19LL1 = 1 if X19 < 0.065 and X19LL1 = 0 otherwise |
| X19LL2 = 1 if 0.065 ≤ X19 < 0.145 and X19LL2 = 0 otherwise | |
| X19LL3 = 1 if 0.145 ≤ X19 < 0.161 and X19LL3 = 0 otherwise | |
| X19LL4 = 1 if 0.161 ≤ X19 and X19LL4 = 0 otherwise | |
| T3 (X18) | X18LL1 = 1 if X18 < 0.02 and X18LL1 = 0 otherwise |
| X18LL2 = 1 if 0.02 ≤ X18 < 0.045 and X18LL2 = 0 otherwise | |
| X18LL3 = 1 if 0.045 ≤ X18 and X18LL3 = 0 otherwise | |
| T4U (X20) | X20LL1 = 1 if X20 < 0.07 and X20LL1 = 0 otherwise |
| X20LL2 = 1 if 0.07 ≤ X20 < 0.15 and X20LL2 = 0 otherwise | |
| X20LL3 = 1 if 0.15 ≤ X20 and X20LL3 = 0 otherwise | |
| Age (X1) | X1LL1 = 1 if X1 < 0.75 and X1LL1 = 0 otherwise |
| X1LL2 = 1 if 0.75 ≤ X1< 0.85 and X1LL2 = 0 otherwise | |
| X1LL3 = 1 if 0.85 ≤ X1 and X1LL3 = 0 otherwise |
| Original Variables | Generated Variables with Linear Trend |
|---|---|
| FT1 (X21) | X21QQ2 = X21 if 0.057 ≤ X21 < 0.071 and X21QQ2 = 0 otherwise |
| X21QQ4 = X21 if 0.115 ≤ X21 < 0.217 and X21QQ4 = 0 otherwise | |
| TT4 (X19) | X19QQ3 = X19 if 0.145 ≤ X19 < 0.161 and X19QQ3 = 0 otherwise |
| T3 (X18) | X18QQ2 = X18 if 0.02 ≤ X18 < 0.045 and X18QQ2 = 0 otherwise |
| Age (X1) | X1QQ2 = X1 if 0.75 ≤ X1< 0.85 and X1QQ2 = 0 otherwise |
| Model | Candidate Predictors | BIC | AIC | R2 (McFadden) | Accuracy |
|---|---|---|---|---|---|
| Model 1 | Main effects (X1–X21) | 916.05 | 841.23 | 65.58% | 97.03% |
| Model 2 | Main effects (X1–X21) All two-way interactions | 561.56 | 449.32 | 82.59% | 98.36% |
| Model 3 | Main effects (X1–X21) Generated variables from step 2 Generated interactions from step 4 | 233.41 | 121.17 | 96.41% | 99.68% |
| Actual Class | ||||
|---|---|---|---|---|
| 0 | 1 | 2 | ||
| Predict class | 0 | 3479 | 2 | 1 |
| 1 | 3 | 91 | 0 | |
| 2 | 6 | 0 | 190 | |
| Actual Class | ||||
|---|---|---|---|---|
| 0 | 1 | 2 | ||
| Predict class | 0 | 3165 | 6 | 8 |
| 1 | 4 | 67 | 0 | |
| 2 | 9 | 0 | 169 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Changpetch, P. Multinomial Logit Model Building via TreeNet and Association Rules Analysis: An Application via a Thyroid Dataset. Symmetry 2021, 13, 287. https://doi.org/10.3390/sym13020287
Changpetch P. Multinomial Logit Model Building via TreeNet and Association Rules Analysis: An Application via a Thyroid Dataset. Symmetry. 2021; 13(2):287. https://doi.org/10.3390/sym13020287
Chicago/Turabian StyleChangpetch, Pannapa. 2021. "Multinomial Logit Model Building via TreeNet and Association Rules Analysis: An Application via a Thyroid Dataset" Symmetry 13, no. 2: 287. https://doi.org/10.3390/sym13020287
APA StyleChangpetch, P. (2021). Multinomial Logit Model Building via TreeNet and Association Rules Analysis: An Application via a Thyroid Dataset. Symmetry, 13(2), 287. https://doi.org/10.3390/sym13020287





