A Novel Centrosymmetric Fe(III) Complex with Anionic Bis-pyrazolyl-s-triazine Ligand; Synthesis, Structural Investigations and Antimicrobial Evaluations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Physical Measurements
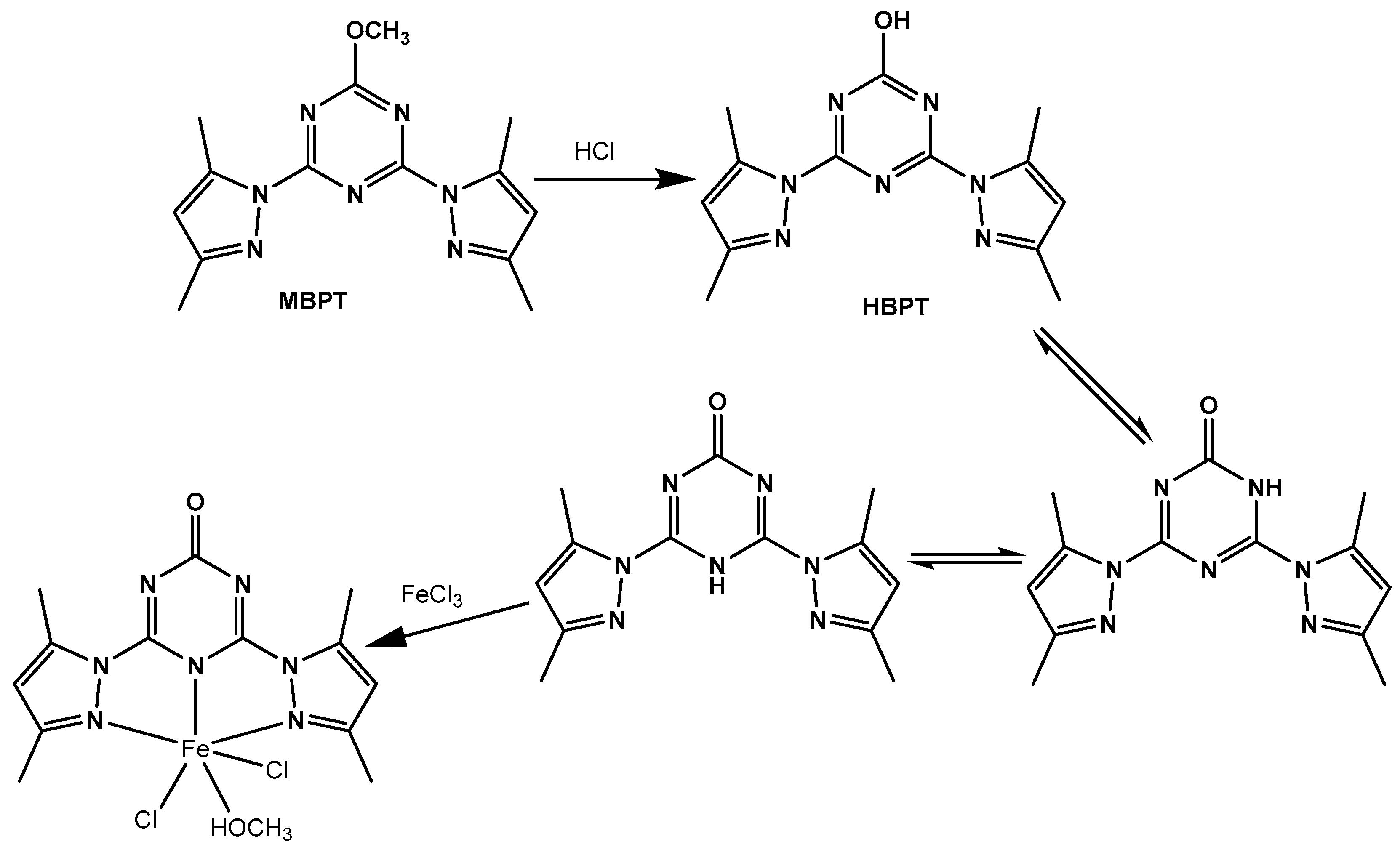
2.2. Synthesis
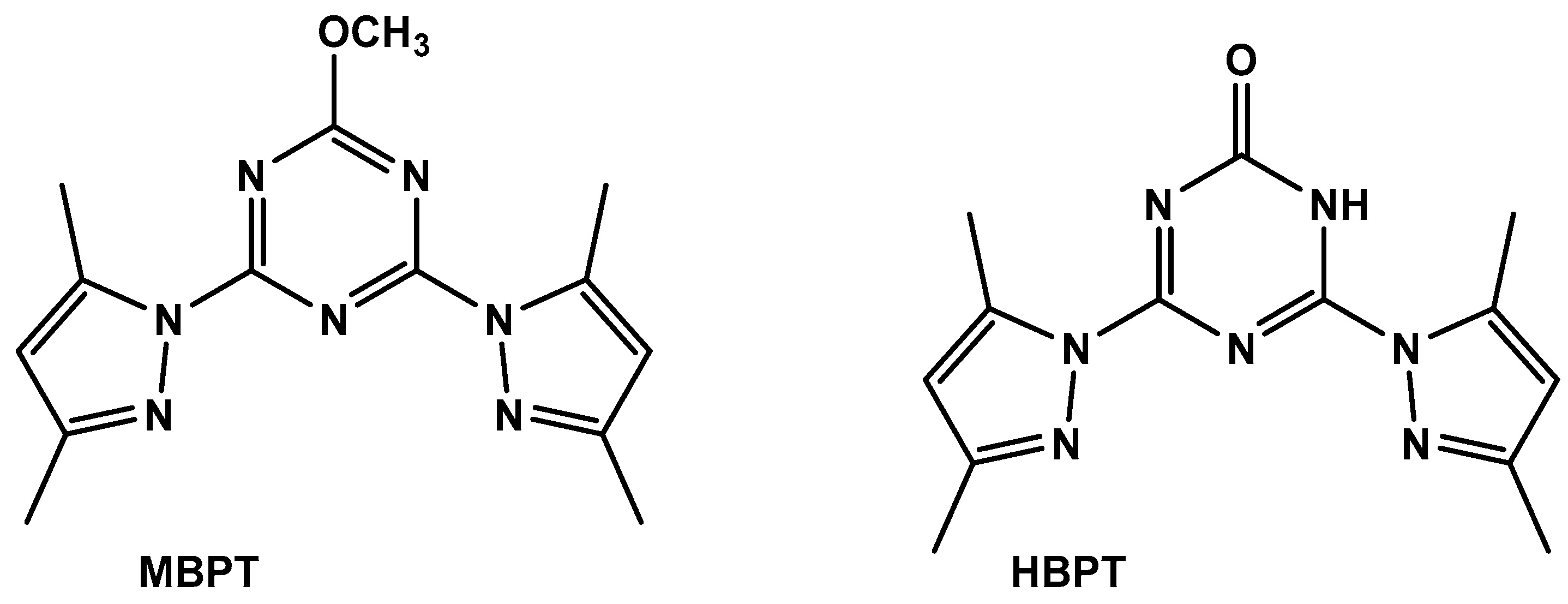
2.2.1. Synthesis of S-Triazine-Based Ligand
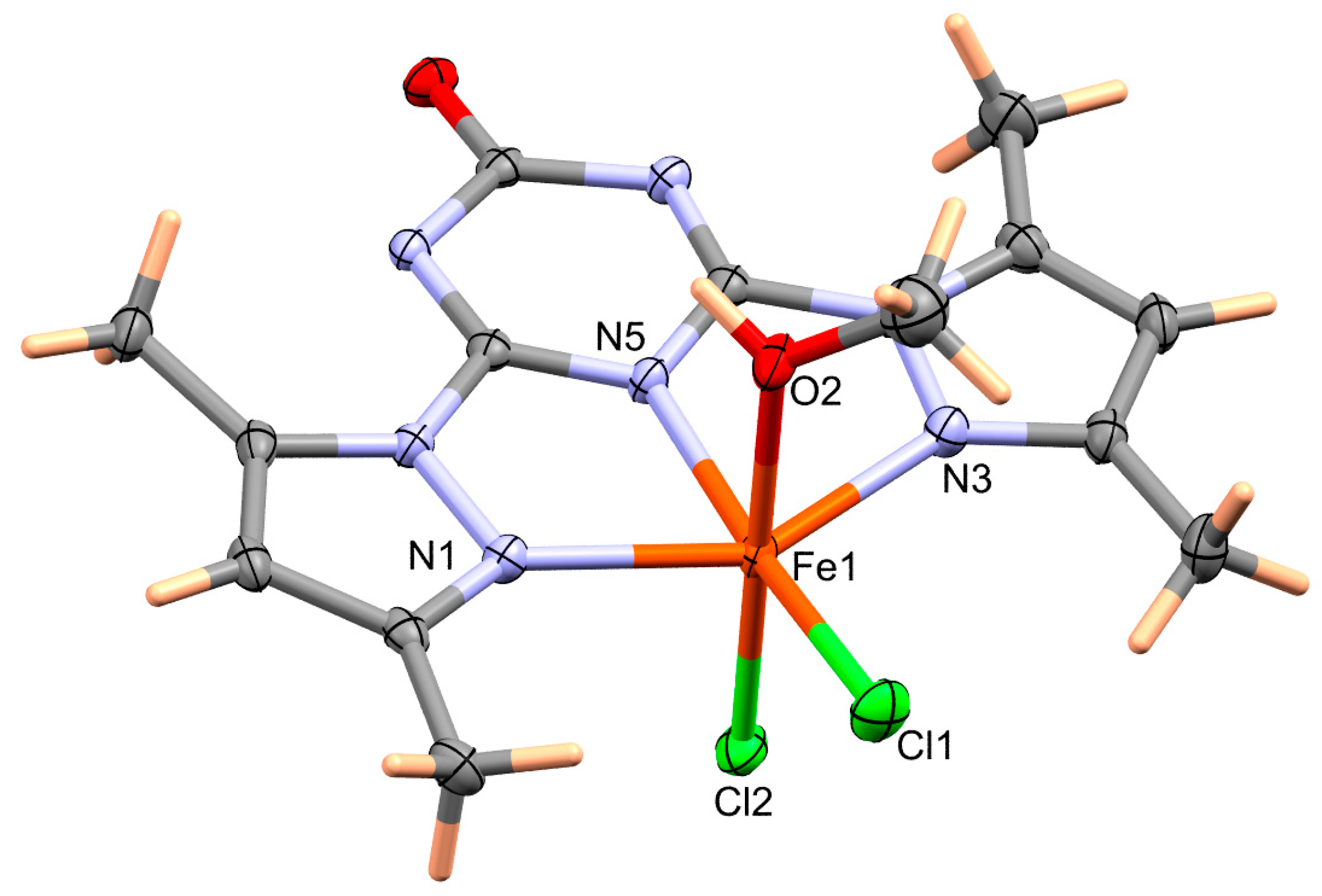
2.2.2. Synthesis of [Fe(BPT)(CH3OH)Cl2] Complex
2.3. Crystal Structure Determination
2.4. Antimicrobial Studies
2.5. Atoms in Molecules (AIM) and Natural Charge Calculations
3. Results
4. Discussion
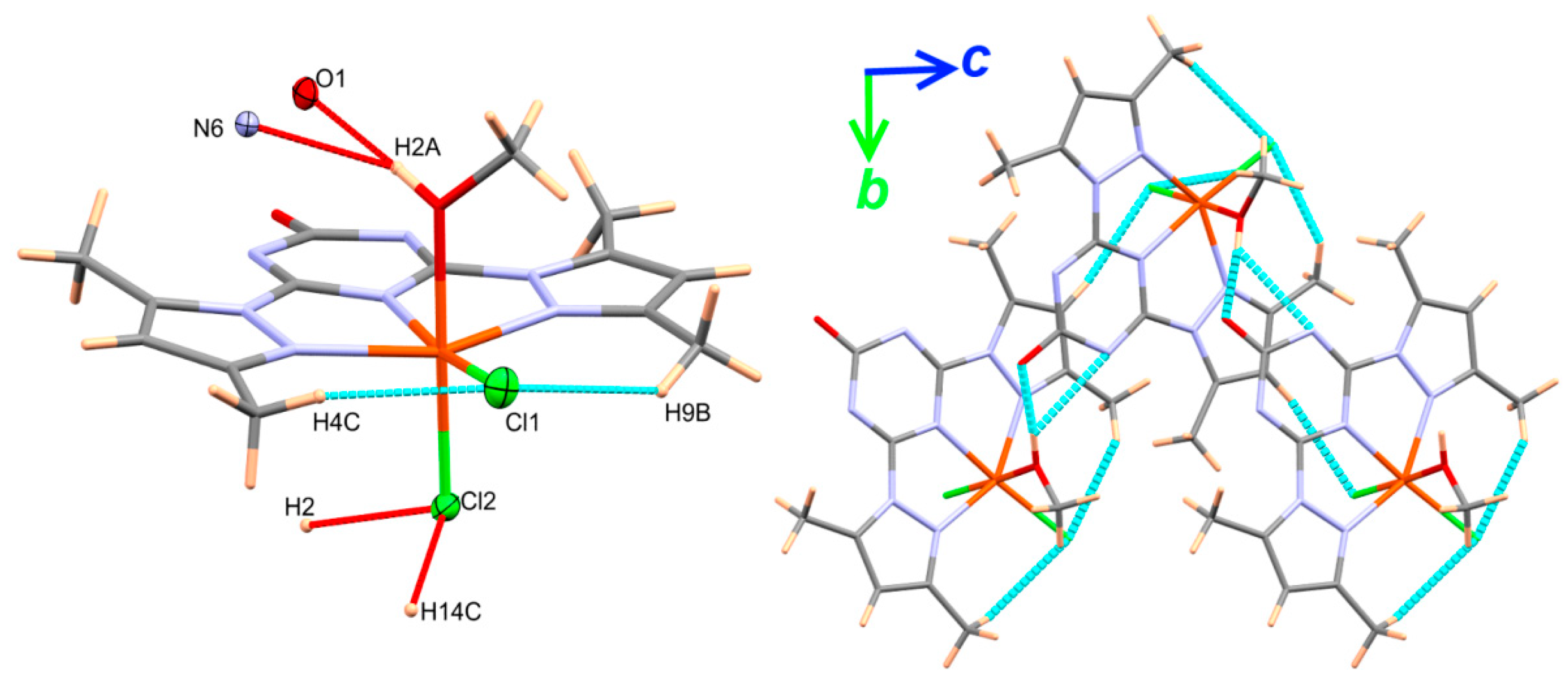
4.1. Structure Description
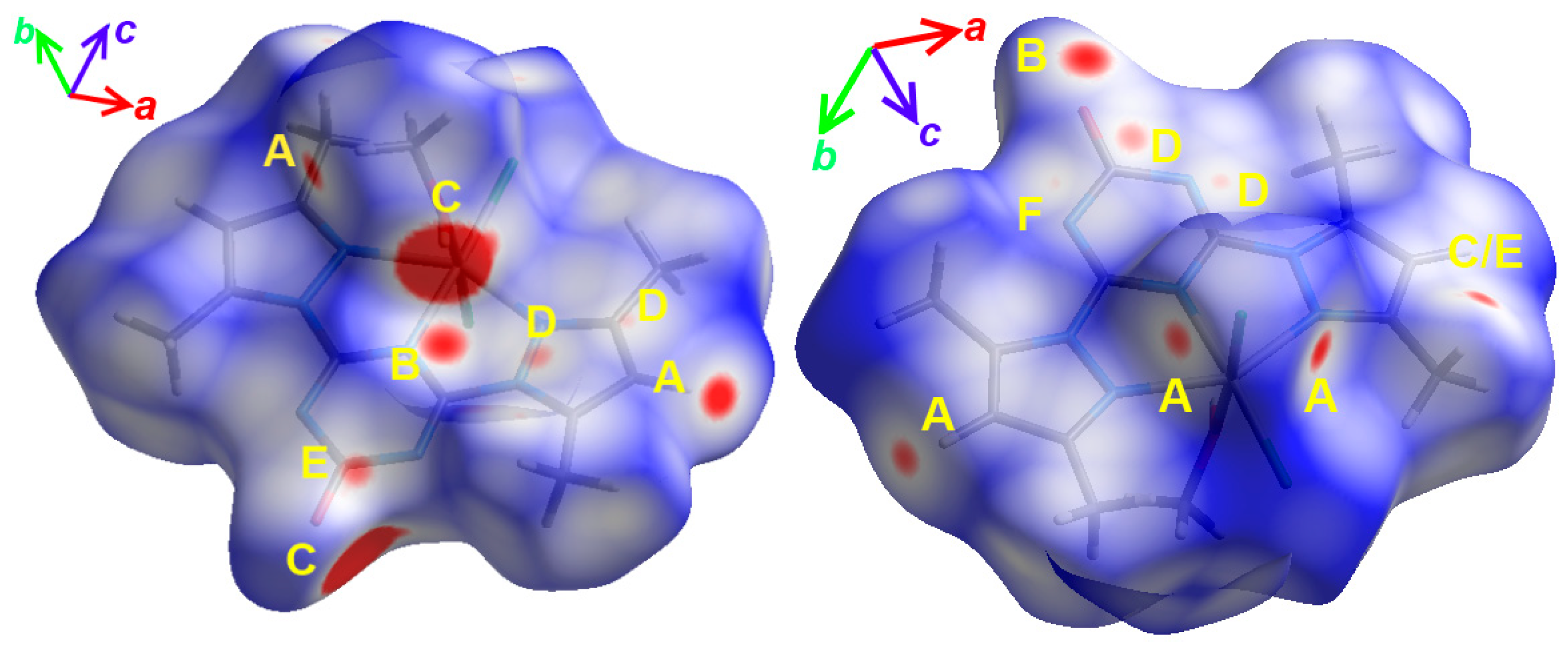
4.2. Quantitative Analysis of Molecular Packing
4.3. NBO and AIM Studies
4.4. Antimicrobial Studies
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bolm, C.; Legros, J.; Le Paih, J.; Zani, L. Iron-Catalyzed Reactions in Organic Synthesis. Chem. Rev. 2004, 104, 6217–6254. [Google Scholar] [CrossRef] [PubMed]
- Bauer, I.; Knölker, H.-J. Iron Catalysis in Organic Synthesis. Chem. Rev. 2015, 115, 3170–3387. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Darcel, C. Iron Catalysis in Reduction and Hydrometalation Reactions. Chem. Rev. 2019, 119, 2550–2610. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, J.R.; Schindler, C.S. Catalyst: Sustainable Catalysis. Chemistry 2017, 2, 313–316. [Google Scholar] [CrossRef] [Green Version]
- Egorova, K.S.; Ananikov, V.P. Toxicity of Metal Compounds: Knowledge and Myths. Organometallics 2017, 36, 4071–4090. [Google Scholar] [CrossRef] [Green Version]
- Egorova, K.S.; Ananikov, V.P. Which Metals are Green for Catalysis? Comparison of the Toxicities of Ni, Cu, Fe, Pd, Pt, Rh, and Au Salts. Angew. Chem. Int. Ed. 2016, 55, 12150–12162. [Google Scholar] [CrossRef]
- Bauer, E.B. Iron Catalysis II. Top. Organomet. Chem. 2015, 50, 1–18. [Google Scholar]
- Crisponi, G.; Remelli, M. Iron chelating agents for the treatment of iron overload. Coord. Chem. Rev. 2008, 252, 1225–1240. [Google Scholar] [CrossRef]
- Brittenham, G.M. Hematology: Basic Principles and Practice; Hoffman, R., Benz, E., Shattil, S., Furie, B., Cohen, H., Eds.; Churchill Livingstone: New York, NY, USA, 1991; p. 327. [Google Scholar]
- Graminha, A.E.; Vilhena, F.S.; Batista, A.A.; Louro, S.R.; Ziolli, R.L.; Teixeira, L.R.; Beraldo, H. 2-Pyridinoformamide-derived thiosemicarbazones and their iron(III) complexes: Potential antineoplastic activity. Polyhedron 2008, 27, 547–551. [Google Scholar] [CrossRef]
- Basha, M.T.; Bordini, J.; Richardson, D.R.; Martinez, M.; Bernhardt, P.V. Kinetico-mechanistic studies on methemoglobin generation by biologically active thiosemicarbazone iron(III) complexes. J. Inorg. Biochem. 2016, 162, 326–333. [Google Scholar] [CrossRef] [Green Version]
- Morcelli, S.R.; Kanashiro, M.M.; Candela, D.R.; Alzamora, M.; Horn, A., Jr.; Fernandes, C. Synthesis, characterization and antitumoral activity of new di-iron(III) complexes containing naphthyl groups: Effect of the isomerism on the biological activity. Inorg. Chem. Commun. 2016, 67, 22–24. [Google Scholar] [CrossRef]
- Chaston, T.B.; Lovejoy, D.B.; Watts, R.N.; Richardson, D.R. Examination of the Antiproliferative Activity of Iron Chelators: Multiple Cellular Targets and the Different Mechanism of Action of Triapine Compared with Desferrioxamine and the Potent Pyridoxal Isonicotinoyl Hydrazone Analogue 311. Clin. Cancer Res. 2003, 9, 402–414. [Google Scholar] [PubMed]
- Soliman, S.M.; El-Faham, A. Synthesis, characterization, and structural studies of two heteroleptic Mn(II) complexes with tridentate N,N,N-pincer type ligand. J. Coord. Chem. 2018, 71, 2373–2388. [Google Scholar] [CrossRef]
- Soliman, S.M.; Almarhoon, Z.; El-Faham, A. Synthesis, Molecular and Supramolecular Structures of New Cd(II) Pincer-Type Complexes with s-Triazine Core Ligand. Crystals 2019, 9, 226. [Google Scholar] [CrossRef] [Green Version]
- Soliman, S.M.; Elsilk, S.E.; El-Faham, A. Synthesis, structure and biological activity of zinc (II) pincer complexes with 2,4-bis(3,5-dimethyl-1H-pyrazol-1-yl)-6-methoxy-1,3,5-triazine. Inorg. Chim. Acta 2020, 508, 119627. [Google Scholar] [CrossRef]
- Soliman, S.M.; Elsilk, S.E.; El-Faham, A. Novel one-dimensional polymeric Cu(II) complexes via Cu(II)-assisted hydrolysis of the 2,4-bis(3,5-dimethyl-1H-pyrazol-1-yl)-6-methoxy-1,3,5-triazine pincer ligand: Synthesis, structure, and antimicrobial activities. Appl. Organomet. Chem. 2020, 34, e5941. [Google Scholar] [CrossRef]
- Lasri, J.; Haukka, M.; Al-Rasheed, H.H.; Abutaha, N.; El-Faham, A.; Soliman, S.M. Synthesis, Structure and In Vitro Anticancer Activity of Pd(II) Complex of Pyrazolyl-s-Triazine Ligand; A New Example of Metal-Mediated Hydrolysis of s-Triazine Pincer Ligand. Crystals 2021, 11, 119. [Google Scholar] [CrossRef]
- Sheldrick, G.M. Program for Empirical Absorption Correction of Area Detector Data; University of Göttingen: Göttingen, Germany, 1996. [Google Scholar]
- Sheldrick, G.M. SHELXT-Integrated space-group and crystal-structure determination. Acta Cryst. A 2015, 71, 3–8. [Google Scholar] [CrossRef] [Green Version]
- Spek, A.L. Structure validation in chemical crystallography. Acta Cryst. D 2009, 65, 148–155. [Google Scholar] [CrossRef]
- Turner, M.J.; McKinnon, J.J.; Wolff, S.K.; Grimwood, D.J.; Spackman, P.R.; Jayatilaka, D.; Spackman, M.A. Crystal Explorer 17; University of Western Australia: Perth, Australia, 2017; Available online: http://hirshfeldsurface.net (accessed on 12 June 2017).
- Spackman, M.A.; Jayatilaka, D. Hirshfeld surface analysis. CrystEngComm 2009, 11, 19–32. [Google Scholar] [CrossRef]
- Spackman, M.A.; McKinnon, J.J. Fingerprinting intermolecular interactions in molecular crystals. CrystEngComm 2002, 4, 378–392. [Google Scholar] [CrossRef]
- Bernstein, J.; Davis, R.E.; Shimoni, L.; Chang, N.-L. Patterns in hydrogen bonding: Functionality and graph set analysis in crystals. Angew. Chem. Int. Ed. Engl. 1995, 34, 1555–1573. [Google Scholar] [CrossRef]
- McKinnon, J.J.; Jayatilaka, D.; Spackman, M.A. Towards quantitative analysis of intermolecular interactions with Hirshfeld surfaces. Chem. Commun. 2007, 37, 3814–3816. [Google Scholar] [CrossRef]
- Sharma, A.; Ghabbour, H.; Khan, S.T.; de la Torre, B.G.; Albericio, F.; El-Faham, A. Novel pyrazolyl-s-triazine derivatives, molecular structure and antimicrobial activity. J. Mol. Struct. 2017, 1145, 244–253. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. GAUSSIAN 09. Revision A02; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Adamo, C.; Barone, V. Exchange functionals with improved long-range behavior and adiabatic connection methods without adjustable parameters: The mPWmPW and mPW1PWmPW1PW models. J. Chem. Phys. 1998, 108, 664–675. [Google Scholar] [CrossRef]
- Glendening, E.D.; Reed, A.E.; Carpenter, J.E.; Weinhold, F. NBO Version 3.1, CI; University of Wisconsin: Madison, WI, USA, 1998. [Google Scholar]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef]
- Lucescu, L.; Ghinet, A.; Belei, D.; Rigo, B.; Dubois, J.; Bîcu, E. Discovery of indolizines containing triazine moiety as new leads for the development of antitumoral agents targeting mitotic events. Bioorg. Med. Chem. Lett. 2015, 25, 3975–3979. [Google Scholar] [CrossRef]
- Woods, K.W.; Lai, C.; Miyashiro, J.M.; Tong, Y.; Florjancic, A.S.; Han, E.K.; Soni, N.; Shi, Y.; Lasko, L.; Leverson, J.D.; et al. Aminopyrimidinone Cdc7 Kinase Inhibitors. Bioorg. Med. Chem. Lett. 2012, 22, 1940–1943. [Google Scholar] [CrossRef]
- Ok, K.M.; Halasyamani, P.S.; Casanova, D.; Llunell, M.; Alvarez, S. Distortions in Octahedrally Coordinated d0 Transition Metal Oxides: A Continuous Symmetry Measures Approach. Chem. Mater. 2006, 18, 3176–3183. [Google Scholar] [CrossRef]
- Santiaqo, A.; David, A.; Llunell, M.; Pinsky, M. Continuous symmetry maps and shape classification. The case of six-coordinated metal compounds. New J. Chem. 2002, 26, 996–1009. [Google Scholar]
- Hagit, Z.; Shmuel, P.; David, A. Continuous symmetry measures. J. Am. Chem. Soc. 1992, 114, 7843–7851. [Google Scholar]
- Keinan, S.; Avnir, D. Quantitative Symmetry in Structure–Activity Correlations: The Near C2 Symmetry of Inhibitor/HIV Protease Complexes. J. Am. Chem. Soc. 2000, 122, 4378–4384. [Google Scholar] [CrossRef]
- Bobrov, M.F.; Popova, G.V.; Tsirelson, V.G. topological analysis of electron density and chemical bonding in cyclophosphazenes PnNnX2n (X = H, F, Cl; n = 2, 3, 4). Russ. J. Phys. Chem. 2006, 80, 584–590. [Google Scholar] [CrossRef]
- Gatti, C. Chemical bonding in crystals: New directions. Z. Kristallogr. 2005, 220, 399–457. [Google Scholar] [CrossRef]
- Gibbs, G.V.; Downs, R.T.; Cox, D.F.; Ross, N.L.; Boisen, M.B.; Rosso, K.M., Jr. Shared and closed-shell O-O interactions in silicates. J. Phys. Chem. A 2008, 112, 3693–3699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cremer, D.; Kraka, E. Chemical Bonds without Bonding Electron Density—Does the Difference Electron Density Analysis Suffice for a Description of the Chemical Bond? Angew. Chem. Int. Ed. Engl. 1984, 23, 627–628. [Google Scholar] [CrossRef]
- Soliman, S.M.; Al-Rasheed, H.H.; Albering, J.H.; El-Faham, A. Fe(III) Complexes Based on Mono- and Bis-pyrazolyl-s-triazine Ligands: Synthesis, Molecular Structure, Hirshfeld, and Antimicrobial Evaluations. Molecules 2020, 25, 5750. [Google Scholar] [CrossRef]

| Empirical formula | C14H18Cl2FeN7O2 | |
| Formula weight | 443.10 g/mol | |
| Temperature | 115(2) K | |
| Wavelength | 0.71073 Å | |
| Crystal system | monoclinic | |
| Space group | P21/c | |
| Unit cell dimensions | a = 7.309(2) Å | α = 90° |
| b = 25.461(8) Å | β = 102.646(7)° | |
| c = 9.918(3) Å | γ = 90° | |
| Volume | 1800.9(9) Å3 | |
| Z | 4 | |
| Density (calculated) | 1.634 g/cm3 | |
| Absorption coefficient | 1.160 mm−1 | |
| F(000) | 908 | |
| Crystal size, mm3 | 0.18 × 0.19 × 0.22 | |
| Theta range for data collection | 2.25 to 25.30° | |
| Index ranges | −8 ≤ h ≤ 8, −30 ≤ k ≤ 30, −11 ≤ l ≤ 11 | |
| Reflections collected | 20,165 | |
| Independent reflections | 3277 [R(int) = 0.0346] | |
| Completeness to theta | 99.90% | |
| Refinement method | Full-matrix least-squares on F2 | |
| Data/restraints/parameters | 3277/0/244 | |
| Goodness-of-fit on F2 | 1.083 | |
| Final R indices [I > 2sigma(I)] | R1 = 0.0339, wR2 = 0.0860 | |
| R indices (all data) | R1 = 0.0381, wR2 = 0.0881 | |
| Largest diff. peak and hole | 1.630 and −0.563 | |
| CCDC no. | 2090699 |
| Bond | Distance | Bond | Distance |
| Fe1–N5 | 2.079(2) | Fe1–N3 | 2.156(2) |
| Fe1–O2 | 2.112(2) | Fe1–Cl1 | 2.2550(8) |
| Fe1–N1 | 2.154(2) | Fe1–Cl2 | 2.3020(9) |
| Bond | Angle | Bond | Angle |
| N5–Fe1–O2 | 83.02(8) | N1–Fe1–Cl1 | 106.04(6) |
| N5–Fe1–N1 | 73.31(8) | N3–Fe1–Cl1 | 106.46(6) |
| O2–Fe1–N1 | 86.24(8) | N5–Fe1–Cl2 | 93.35(6) |
| N5–Fe1–N3 | 73.02(8) | O2–Fe1–Cl2 | 175.69(6) |
| O2–Fe1–N3 | 85.46(8) | N1–Fe1–Cl2 | 94.96(6) |
| N1–Fe1–N3 | 146.04(8) | N3–Fe1–Cl2 | 91.28(7) |
| N5–Fe1–Cl1 | 170.77(6) | Cl1–Fe1–Cl2 | 95.87(3) |
| O2–Fe1–Cl1 | 87.76(6) | ||
| Fe1–N5 | 2.079(2) | Fe1–N3 | 2.156(2) |
| Fe1–O2 | 2.112(2) | Fe1–Cl1 | 2.2550(8) |
| Fe1–N1 | 2.154(2) | Fe1–Cl2 | 2.3020(9) |
| Contact | Distance | Contact | Distance |
|---|---|---|---|
| O1…H2A | 1.714 | C11…O1 | 3.056 |
| O1…H4A | 2.595 | Cl1…H9C | 2.814 |
| H4A…C13 | 2.685 | Cl2…H2 | 2.656 |
| C11…C2 | 3.373 | Cl2…H7 | 2.713 |
| C13…C3 | 3.331 | Cl2…H14C | 2.685 |
| N7…H5A | 2.615 |
| Bond | ρ(r) | G(r) a | V(r) b | Eint | H(r) c | V(r)/G(r) |
|---|---|---|---|---|---|---|
| Fe1–N5 | 0.0810 | 0.1030 | −0.1250 | 39.21 | −0.0220 | 1.21 |
| Fe1–N1 | 0.0584 | 0.0728 | −0.0803 | 25.21 | −0.0075 | 1.10 |
| Fe1–N3 | 0.0580 | 0.0715 | −0.0787 | 24.70 | −0.0072 | 1.10 |
| Fe1–Cl1 | 0.0720 | 0.0730 | −0.0898 | 28.18 | −0.0168 | 1.23 |
| Fe1–Cl2 | 0.0687 | 0.0597 | −0.0762 | 23.91 | −0.0165 | 1.28 |
| Fe1–O2 | 0.0421 | 0.0795 | −0.0791 | 24.82 | 0.0004 | 0.99 |
| Microbe | [Fe(BPT)(CH3OH)Cl2] | Amoxicillin | Ampicillin |
|---|---|---|---|
| St. aureus | 6.8 | 2.7 | 2.7 |
| B. cereus | 7.9 | 2.1 | 2.1 |
| B. subtilis | 7.3 | 2.7 | 3.6 |
| E. coli | 7.9 | 2.7 | 3.6 |
| P. aeruginosa | 7.3 | 2.1 | 2.9 |
| C. albicans | 6.2 | 3.4 | 4.3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soliman, S.M.; Al-Rasheed, H.H.; Elsilk, S.E.; El-Faham, A. A Novel Centrosymmetric Fe(III) Complex with Anionic Bis-pyrazolyl-s-triazine Ligand; Synthesis, Structural Investigations and Antimicrobial Evaluations. Symmetry 2021, 13, 1247. https://doi.org/10.3390/sym13071247
Soliman SM, Al-Rasheed HH, Elsilk SE, El-Faham A. A Novel Centrosymmetric Fe(III) Complex with Anionic Bis-pyrazolyl-s-triazine Ligand; Synthesis, Structural Investigations and Antimicrobial Evaluations. Symmetry. 2021; 13(7):1247. https://doi.org/10.3390/sym13071247
Chicago/Turabian StyleSoliman, Saied M., Hessa H. Al-Rasheed, Sobhy E. Elsilk, and Ayman El-Faham. 2021. "A Novel Centrosymmetric Fe(III) Complex with Anionic Bis-pyrazolyl-s-triazine Ligand; Synthesis, Structural Investigations and Antimicrobial Evaluations" Symmetry 13, no. 7: 1247. https://doi.org/10.3390/sym13071247







