Predicting the Distribution of Ailanthus altissima Using Deep Learning-Based Analysis of Satellite Imagery
Abstract
1. Introduction
2. Materials and Methods
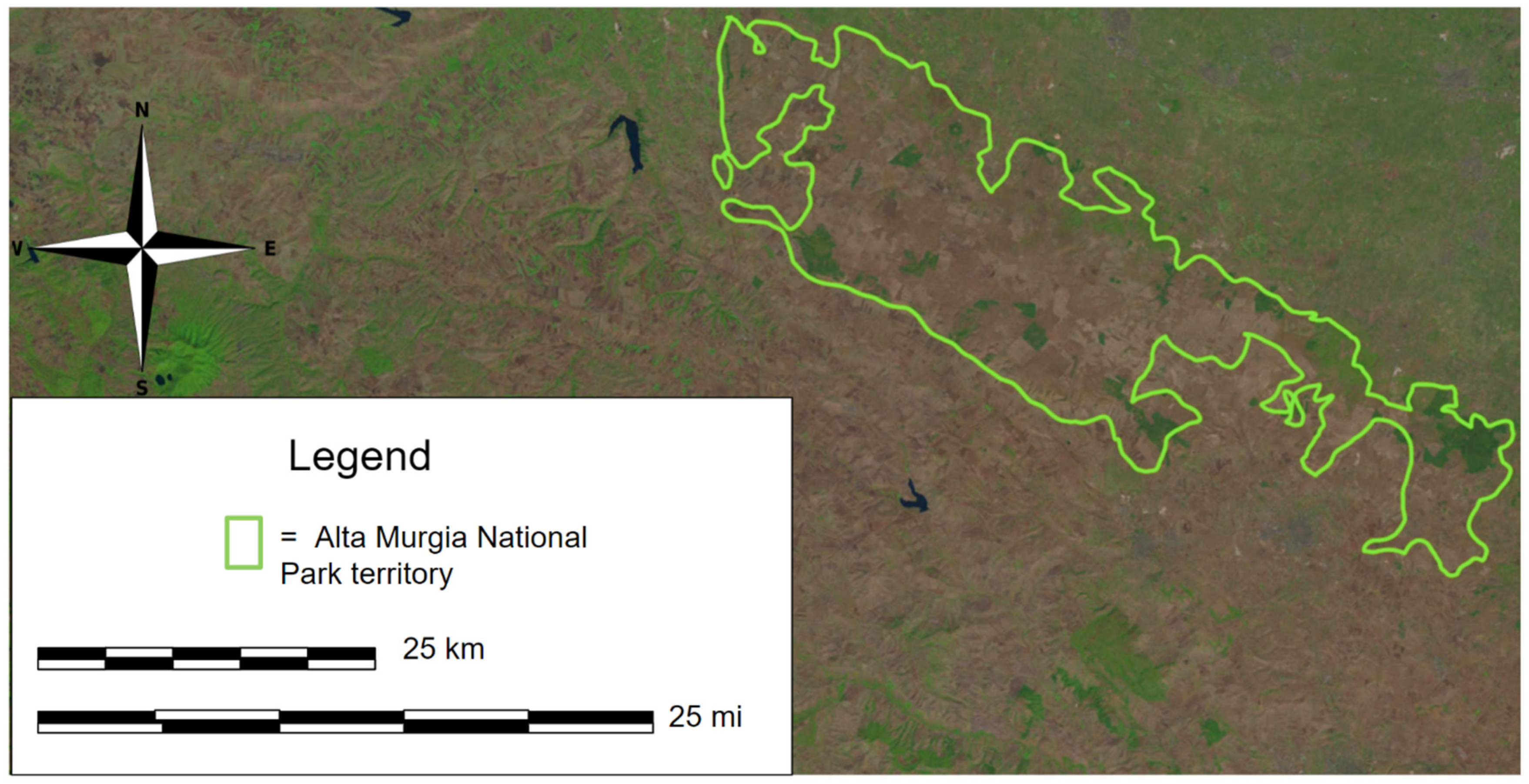
2.1. Study Area
2.2. Data Source
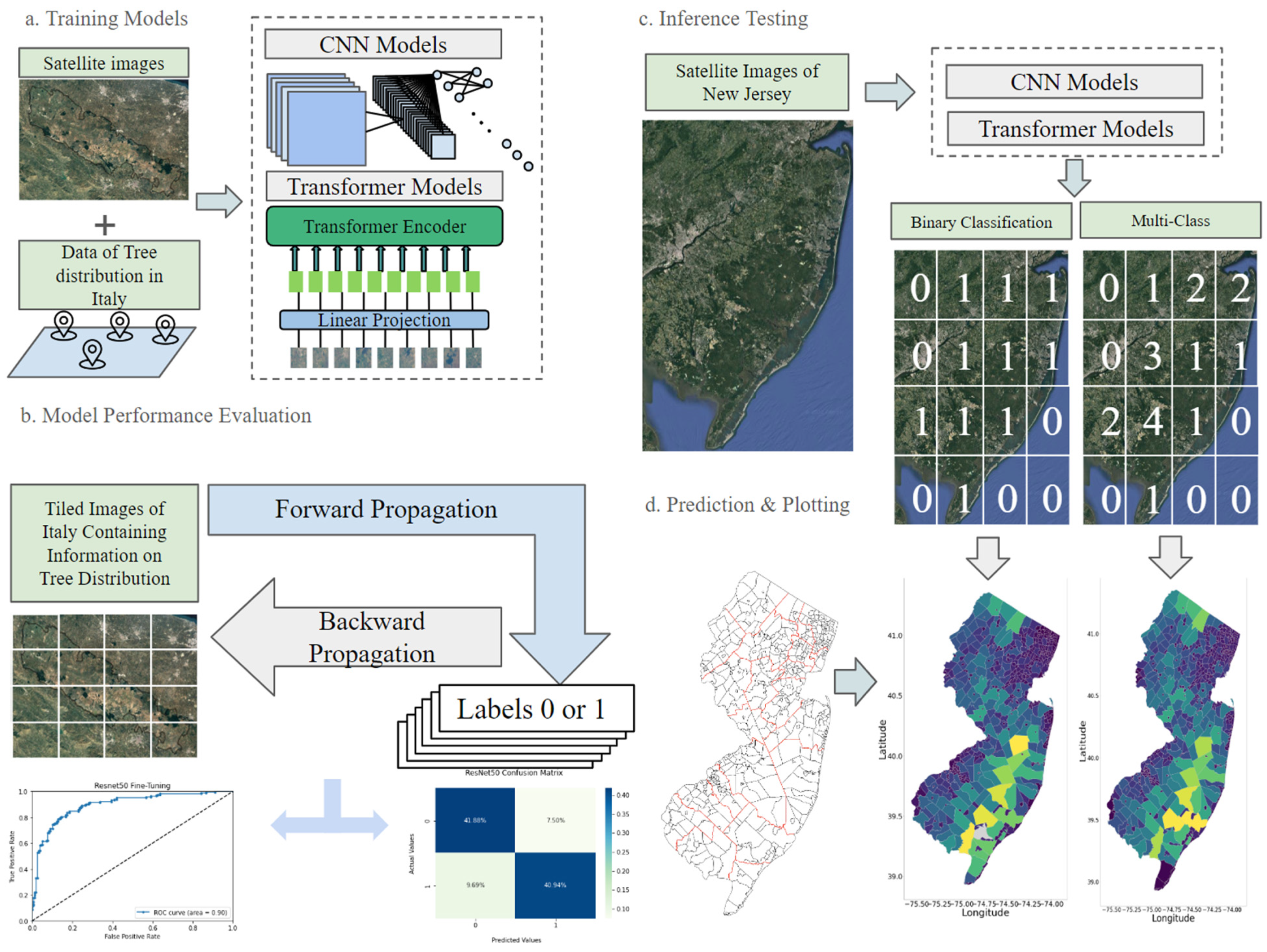
2.3. Model Frameworks
2.4. Experimental Procedures
2.4.1. Image Tilting and Augmentation
2.4.2. Ground Truth and Binary Classification
2.4.3. Inference Testing in USA
2.4.4. Multi-Class Classification Validation
2.5. Tools Utilized
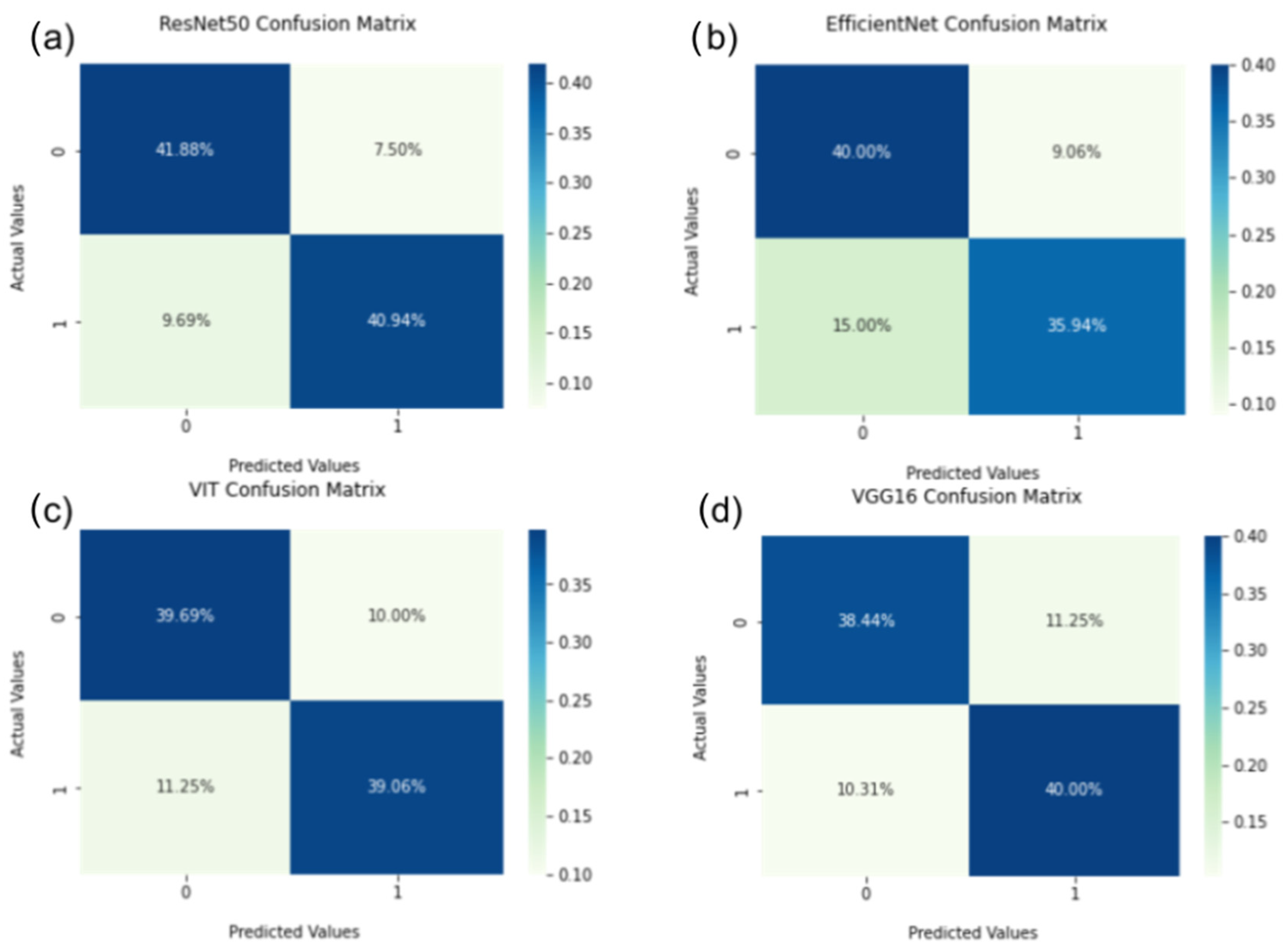
3. Results
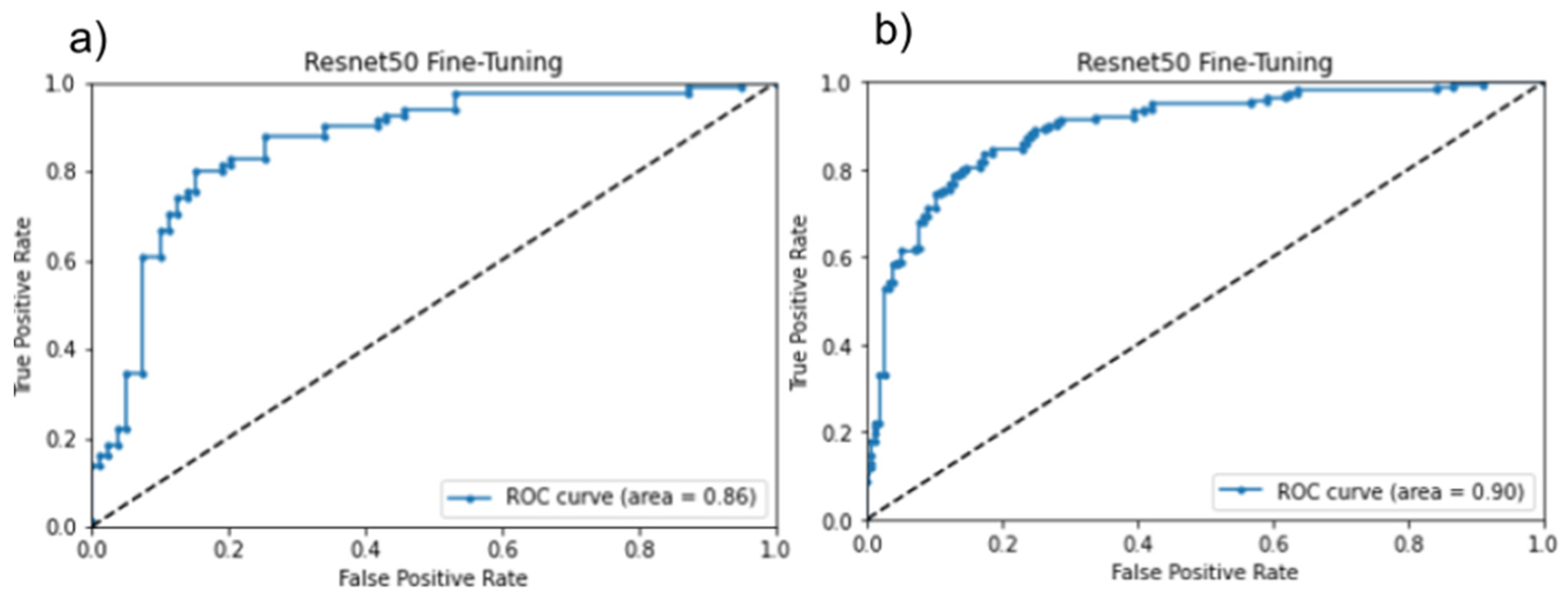
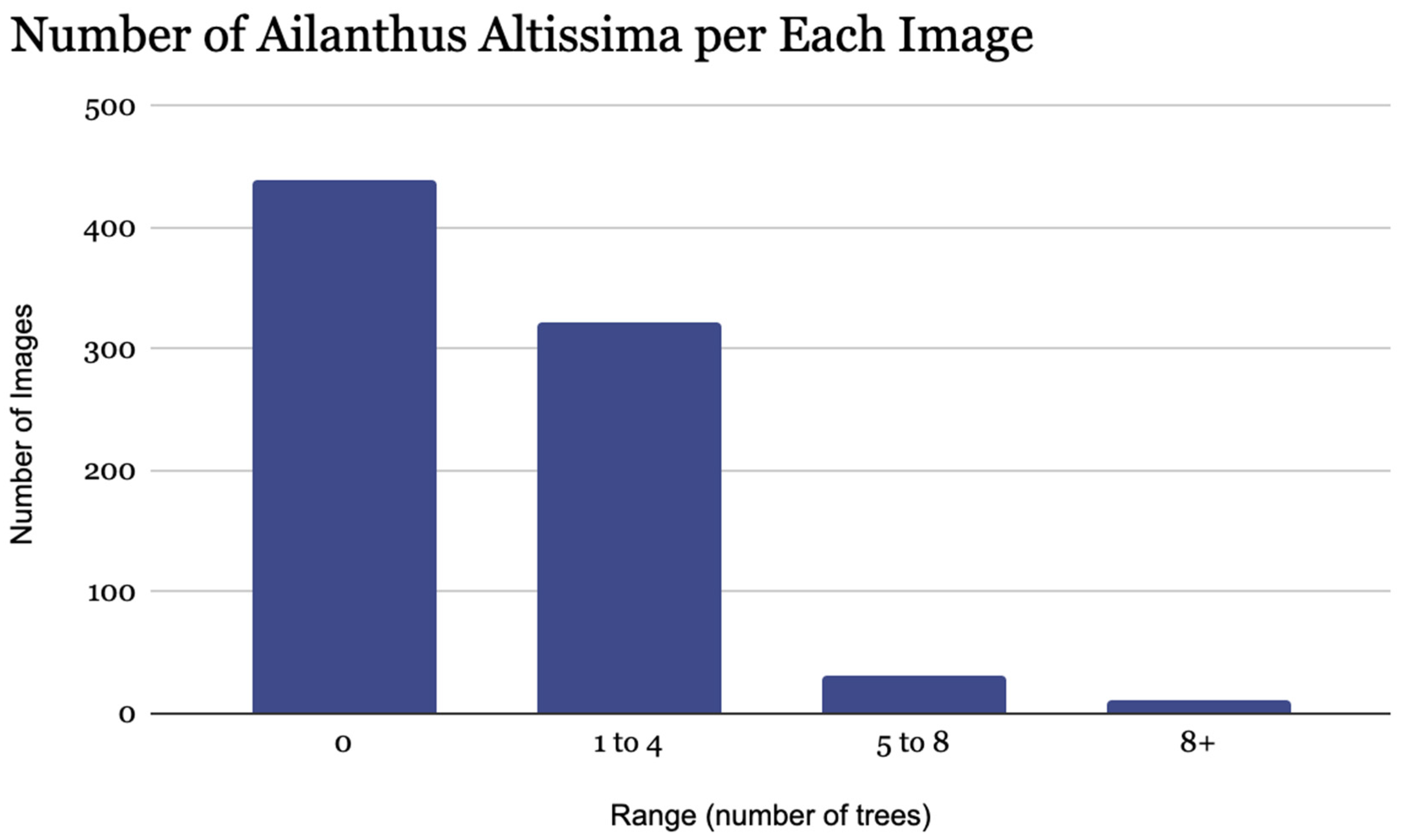
3.1. Ground Truth Result
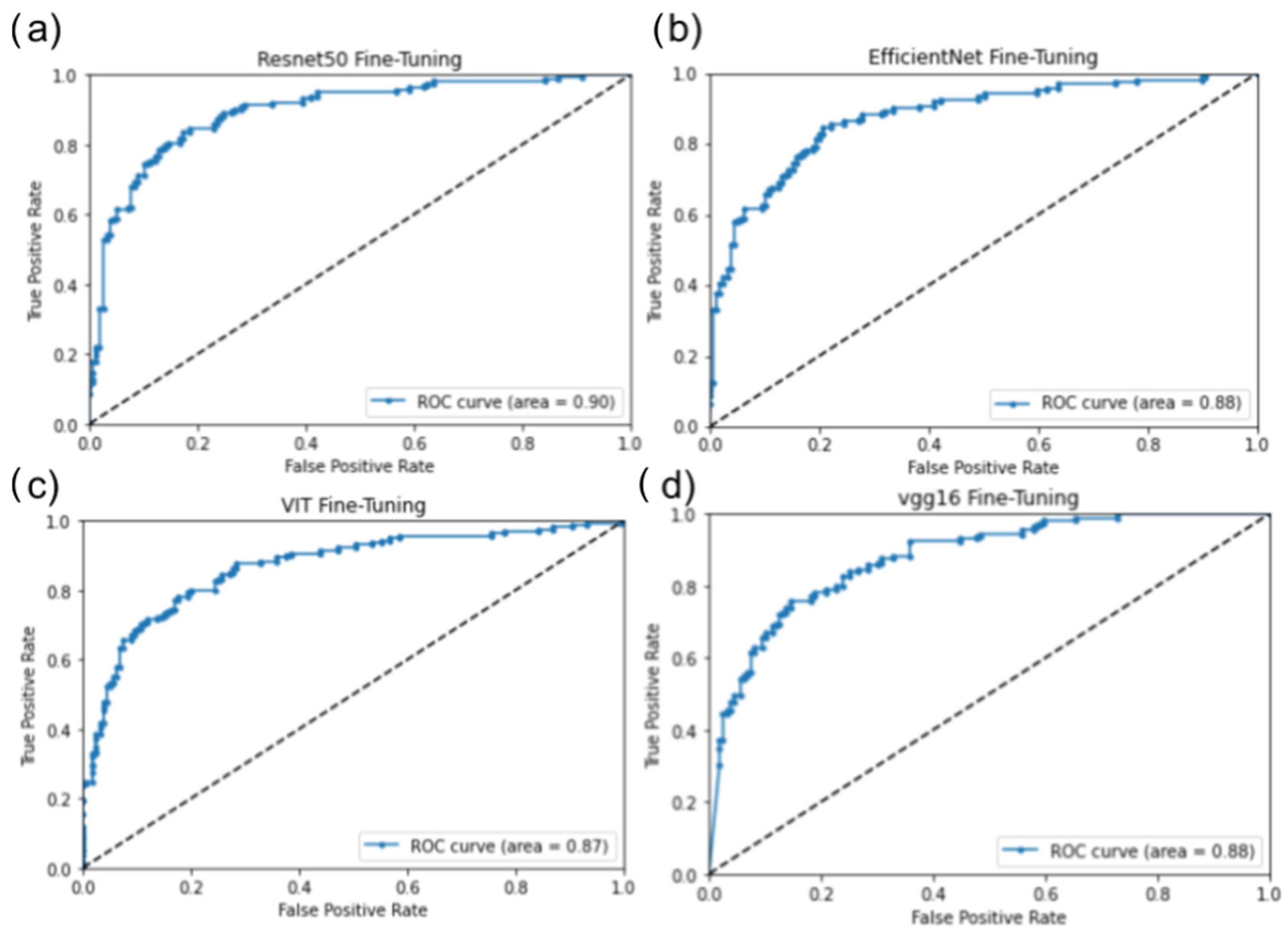
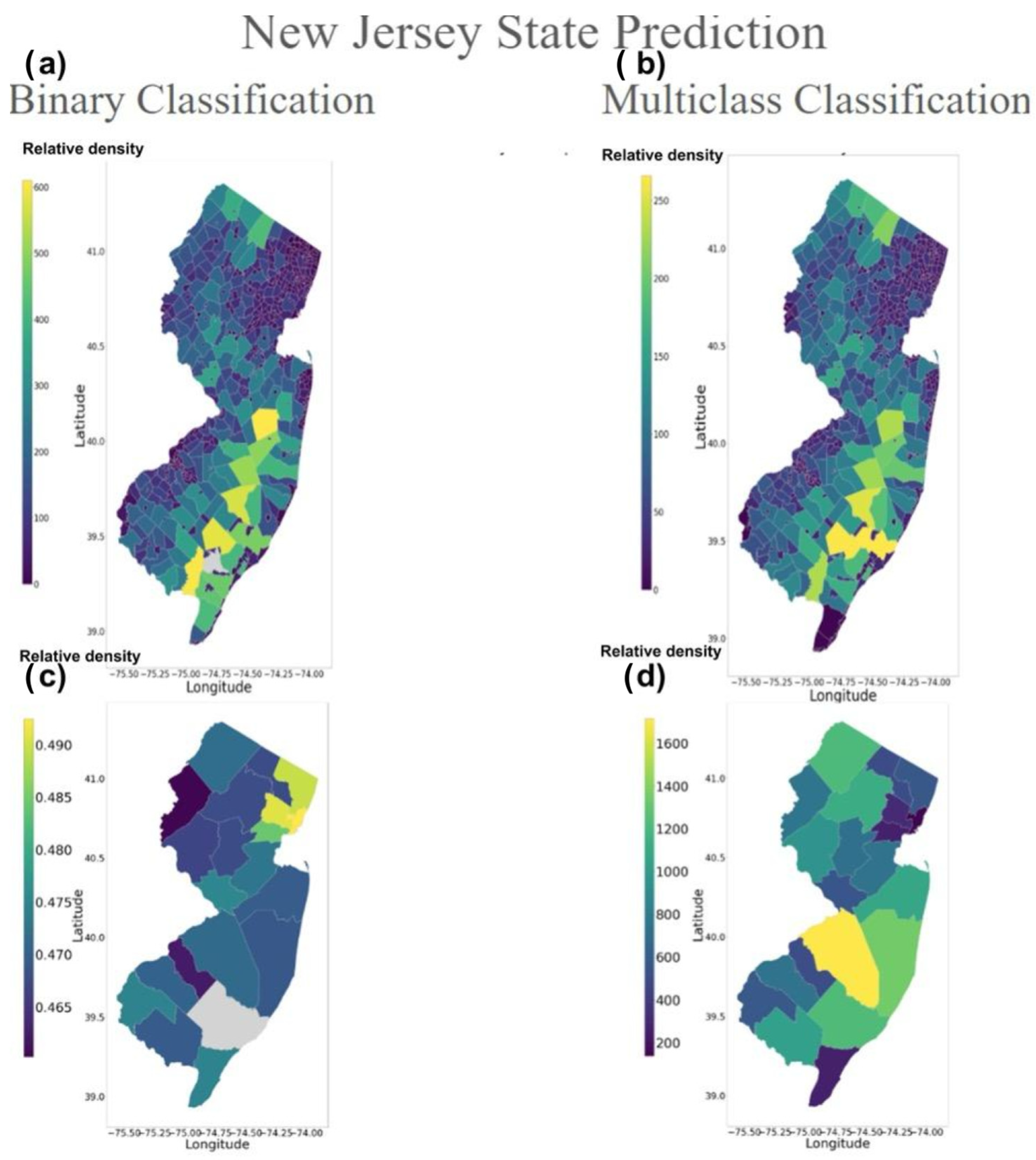
3.2. Inference Testing Result
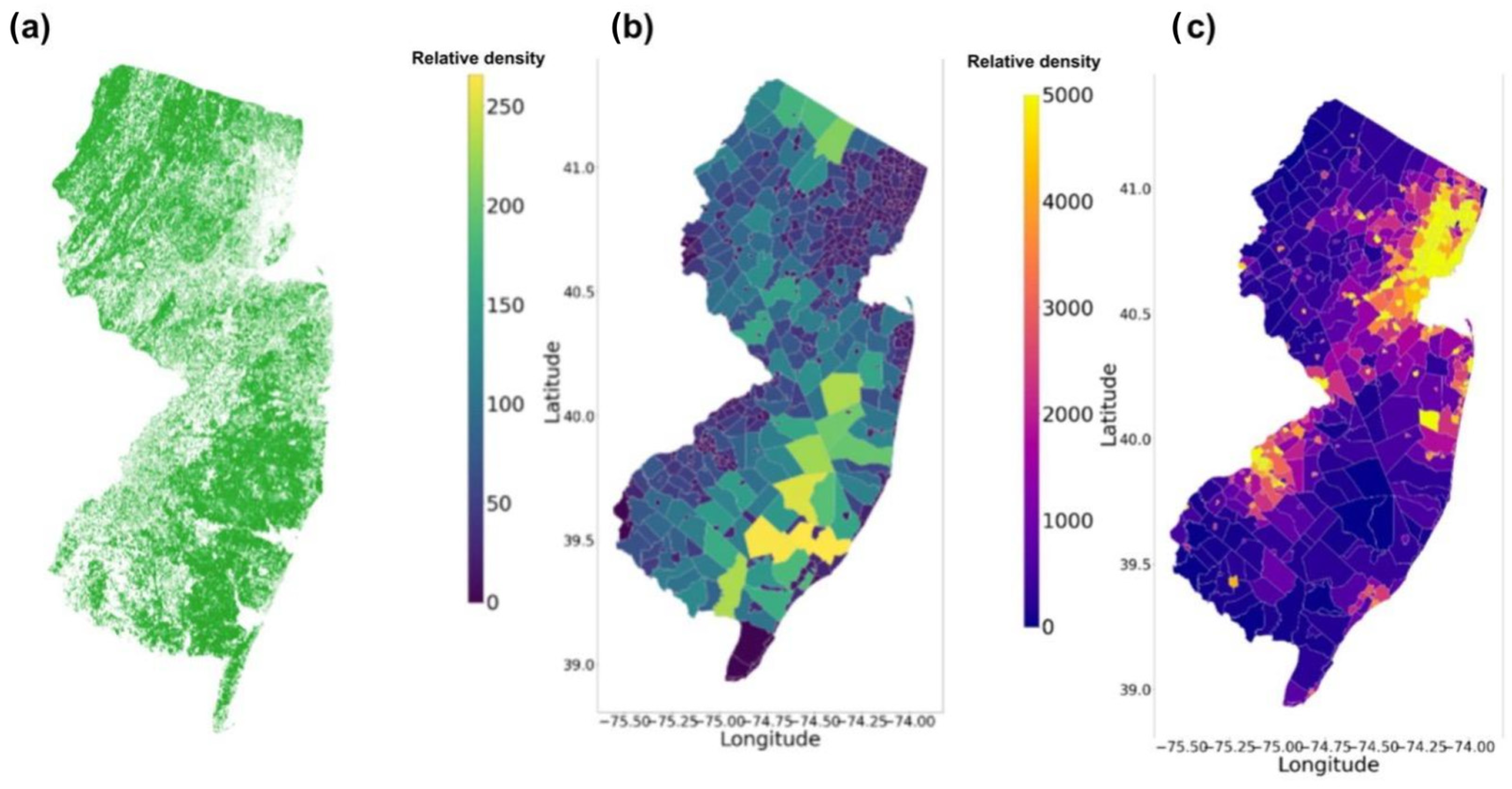
3.3. Feature Extraction Result
4. Analysis and Discussion
4.1. Validation of Inference Testing
4.2. Analysis of Tree Distribution on New Jersey Prediction Map
4.3. Analysis in Reference to Previous Studies
4.4. Machine Learning Performance Result Analysis
4.5. Limitations and Future Studies
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pyšek, P.; Richardson, D.M. Invasive Species, Environmental Change and Management, and Health. Annu. Rev. Environ. Resour. 2010, 35, 25–55. [Google Scholar] [CrossRef]
- What Is an Invasive Species and Why Are They a Problem? U.S. Geological Survey. Available online: https://www.usgs.gov/faqs/what-invasive-species-and-why-are-they-problem (accessed on 27 July 2023).
- Kumar Rai, P.; Singh, J.S. Invasive Alien Plant Species: Their Impact on Environment, Ecosystem Services and Human Health. Ecol. Indic. 2020, 111, 106020. [Google Scholar] [CrossRef] [PubMed]
- Tree of Heaven (Ailanthus altissima): Invasive Species ID. Available online: https://www.nature.org/en-us/about-us/where-we-work/united-states/indiana/stories-in-indiana/journey-with-nature--tree-of-heaven/ (accessed on 27 July 2023).
- Tree of Heaven; Invasive Species Centre: Marie, ON, Canada. Available online: https://www.invasivespeciescentre.ca/invasive-species/meet-the-species/invasive-plants/tree-of-heaven/ (accessed on 27 July 2023).
- Wolde, B.; Lal, P. Invasive-Plant-Removal Frequency—Its Impact on Species Spread and Implications for Further Integration of Forest-Management Practices. Forests 2018, 9, 502. [Google Scholar] [CrossRef]
- Sitzia, T.; Campagnaro, T.; Kowarik, I.; Trentanovi, G. Using Forest Management to Control Invasive Alien Species: Helping Implement the New European Regulation on Invasive Alien Species. Biol. Invasions 2016, 18, 1–7. [Google Scholar] [CrossRef]
- Martin, P.A.; Shackelford, G.E.; Bullock, J.M.; Gallardo, B.; Aldridge, D.C.; Sutherland, W.J. Management of UK Priority Invasive Alien Plants: A Systematic Review Protocol. Environ. Evid. 2020, 9, 1. [Google Scholar] [CrossRef]
- Warzinaick, T.; Haight, R.G.; Yemshanov, D.; Apriesnig, J.L.; Holmes, T.P.; Countryman, A.M.; Rothlisberger, J.D.; Haberland, C. Economics of Invasive Species. In Invasive Species in Forests and Rangelands of the United States; Springer: Cham, Switzerland, 2021. [Google Scholar] [CrossRef]
- Crystal-Ornelas, R.; Hudgins, E.J.; Cuthbert, R.N.; Haubrock, P.J.; Fantle-Lepczyk, J.; Angulo, E.; Kramer, A.M.; Ballesteros-Mejia, L.; Leroy, B.; Leung, B.; et al. Economic Costs of Biological Invasions within North America. NeoBiota 2021, 67, 485–510. [Google Scholar] [CrossRef]
- Zenni, R.D.; Essl, F.; García-Berthou, E.; McDermott, S.M. The Economic Costs of Biological Invasions around the World. NeoBiota 2021, 67, 1–9. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, X.; Kuang, Y.; Chen, Y.V.; Yang, B. Deep CNN-Based Methods to Evaluate Neighborhood-Scale Urban Valuation Through Street Scenes Perception. In Proceedings of the 2018 IEEE Third International Conference on Data Science in Cyberspace (DSC), Guangzhou, China, 8–21 June 2018; pp. 20–27. [Google Scholar] [CrossRef]
- Bibault, J.-E.; Bassenne, M.; Ren, H.; Xing, L. Deep Learning Prediction of Cancer Prevalence from Satellite Imagery. Cancers 2020, 12, 3844. [Google Scholar] [CrossRef] [PubMed]
- Tarantino, C.; Casella, F.; Adamo, M.; Lucas, R.; Beierkuhnlein, C.; Blonda, P. Ailanthus Altissima Mapping from Multi-Temporal Very High Resolution Satellite Images. ISPRS J. Photogramm. Remote Sens. 2019, 147, 90–103. [Google Scholar] [CrossRef]
- Niphadkar, M.; Nagendra, H.; Tarantino, C.; Adamo, M.; Blonda, P. Comparing Pixel and Object-Based Approaches to Map an Understorey Invasive Shrub in Tropical Mixed Forests. Front. Plant Sci. 2017, 8, 892. [Google Scholar] [CrossRef]
- Lake, T.A.; Briscoe Runquist, R.D.; Moeller, D.A. Deep Learning Detects Invasive Plant Species across Complex Landscapes Using Worldview-2 and Planetscope Satellite Imagery. Remote Sens. Ecol. Conserv. 2022, 8, 875–889. [Google Scholar] [CrossRef]
- Gonçalves, C.; Santana, P.; Brandão, T.; Guedes, M. Automatic Detection of Acacia Longifolia Invasive Species Based on UAV-Acquired Aerial Imagery. Inf. Process. Agric. 2022, 9, 276–287. [Google Scholar] [CrossRef]
- Rebbeck, J.; Kloss, A.; Bowden, M.; Coon, C.; Hutchinson, T.F.; Iverson, L.; Guess, G. Aerial Detection of Seed-Bearing Female Ailanthus altissima: A Cost-Effective Method to Map an Invasive Tree in Forested Landscapes. For. Sci. 2015, 61, 1068–1078. [Google Scholar] [CrossRef]
- Zhao, J.; Liu, X.; Guo, C.; Qian, Z.C.; Chen, Y.V. Phoenixmap: An Abstract Approach to Visualize 2D Spatial Distributions. IEEE Trans. Vis. Comput. Graph. 2021, 27, 2000–2014. [Google Scholar] [CrossRef] [PubMed]
- Kowarik, I.; Säumel, I. Biological Flora of Central Europe: Ailanthus altissima (Mill.) Swingle. Perspect. Plant Ecol. Evol. Syst. 2007, 8, 207–237. [Google Scholar] [CrossRef]
- Tree-of-Heaven. Available online: https://extension.psu.edu/tree-of-heaven (accessed on 4 August 2023).
- Almeida, M.T.; Mouga, T.; Barracosa, P. The Weathering Ability of Higher Plants. The Case of Ailanthus altissima (Miller) Swingle. Int. Biodeterior. Biodegrad. 1994, 33, 333–343. [Google Scholar] [CrossRef]
- MDAR Invasive Pest Dashboard. Available online: https://experience.arcgis.com/experience/a25afa4466a54313b21dd45abc34b62d/page/Page-2/?views=Spotted-Lanternfly (accessed on 27 July 2023).
- Sánchez Valdivia, A.; De Stefano, L.G.; Ferraro, G.; Gianello, D.; Ferral, A.; Dogliotti, A.I.; Reissig, M.; Gerea, M.; Queimaliños, C.; Pérez, G.L. Characterizing Chromophoric Dissolved Organic Matter Spatio-Temporal Variability in North Andean Patagonian Lakes Using Remote Sensing Information and Environmental Analysis. Remote Sens. 2024, 16, 4063. [Google Scholar] [CrossRef]
- Genzano, N.; Pergola, N.; Marchese, F. A Google Earth Engine Tool to Investigate, Map and Monitor Volcanic Thermal Anomalies at Global Scale by Means of Mid-High Spatial Resolution Satellite Data. Remote Sens. 2020, 12, 3232. [Google Scholar] [CrossRef]
- Rashidian, V.; Baise, L.G.; Koch, M.; Moaveni, B. Detecting Demolished Buildings after a Natural Hazard Using High Resolution RGB Satellite Imagery and Modified U-Net Convolutional Neural Networks. Remote Sens. 2021, 13, 2176. [Google Scholar] [CrossRef]
- Alom, M.Z.; Taha, T.M.; Yakopcic, C.; Westberg, S.; Sidike, P.; Nasrin, M.S.; Hasan, M.; Van Essen, B.C.; Awwal, A.A.S.; Asari, V.K. A State-of-the-Art Survey on Deep Learning Theory and Architectures. Electronics 2019, 8, 292. [Google Scholar] [CrossRef]
- Using Spatial Simulations of Habitat Modification for Adaptive Management of Protected Areas: Mediterranean Grassland Modification by Woody Plant Encroachment. Environmental Conservation. Cambridge Core. Available online: https://www.cambridge.org/core/journals/environmental-conservation/article/abs/using-spatial-simulations-of-habitat-modification-for-adaptive-management-of-protected-areas-mediterranean-grassland-modification-by-woody-plant-encroachment/0EDCEADF910352D9370FEE463C795B0A (accessed on 27 July 2023).
- LIFE 3.0—LIFE Project Public Page. Available online: https://webgate.ec.europa.eu/life/publicWebsite/index.cfm?fuseaction=search.dspPage&n_proj_id=4566#administrative-data (accessed on 27 July 2023).
- Tree-of-Heaven (Ailanthus altissima)—EDDMapS Distribution—EDDMapS. Available online: https://www.eddmaps.org/distribution/uscounty.cfm?sub=3003&map=distribution (accessed on 27 July 2023).
- Spotted Lanternfly (Lycorma delicatula)—EDDMapS Distribution—EDDMapS. Available online: https://www.eddmaps.org/distribution/uscounty.cfm?sub=77293 (accessed on 27 July 2023).
- Google Earth. Available online: https://earth.google.com/web/ (accessed on 27 July 2023).
- NLCD 2021 USFS Tree Canopy Cover (CONUS). NLCD 2021 USFS Tree Canopy Cover (CONUS)|Multi-Resolution Land Characteristics (MRLC) Consortium. (n.d.). Available online: https://www.mrlc.gov/data/nlcd-2021-usfs-tree-canopy-cover-conus (accessed on 27 July 2023).
- Municipal Boundaries of New Jersey, Web Mercator. Available online: https://undefined.maps.arcgis.com/sharing/rest/content/items/a1b13541f0484415b06cf9c8969bfd6c/info/metadata/metadata.xml?format=default&output=html (accessed on 30 October 2023).
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385. [Google Scholar]
- Tan, M.; Le, Q.V. EfficientNetV2: Smaller Models and Faster Training. arXiv 2021, arXiv:2104.00298. [Google Scholar]
- Sarkar, A. EfficientNetV2—Faster, Smaller, and Higher Accuracy than Vision Transformers. Available online: https://medium.com/towards-data-science/efficientnetv2-faster-smaller-and-higher-accuracy-than-vision-transformers-98e23587bf04 (accessed on 27 July 2023).
- Khan, S.; Naseer, M.; Hayat, M.; Zamir, S.W.; Khan, F.S.; Shah, M. Transformers in Vision: A Survey. ACM Comput. Surv. 2022, 54, 1–41. [Google Scholar] [CrossRef]
- Radhakrishnan, P. Why Transformers Are Slowly Replacing CNNs in Computer Vision? Available online: https://becominghuman.ai/transformers-in-vision-e2e87b739feb (accessed on 27 July 2023).
- Gao, R. Determining Critical Lung Cancer Subtypes from Gigapixel Multi-Scale Whole Slide H&E Stains Images. In Proceedings of the 2022 5th International Conference on Data Science and Information Technology (DSIT), Shanghai, China, 22–24 July 2022; pp. 1–10. [Google Scholar]
- Zhao, J.; Liu, X.; Tang, H.P.; Wang, X.Y.; Yang, S.; Liu, D.F.; Chen, Y.J.; Chen, Y.V. Mesoscopic structure graphs for interpreting uncertainty in non-linear embeddings. Comput. Biol. Med. 2024, 182, 109105. [Google Scholar] [CrossRef] [PubMed]
- Hernández-García, A.; König, P. Further Advantages of Data Augmentation on Convolutional Neural Networks. In Artificial Neural Networks and Machine Learning—ICANN 2018; Kůrková, V., Manolopoulos, Y., Hammer, B., Iliadis, L., Maglogiannis, I., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 95–103. [Google Scholar]
- Mikołajczyk, A.; Grochowski, M. Data Augmentation for Improving Deep Learning in Image Classification Problem. In Proceedings of the 2018 International Interdisciplinary PhD Workshop (IIPhDW), Swinoujscie, Poland, 9–12 May 2018; pp. 117–122. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2015, arXiv:1409.1556. [Google Scholar]
- ImageNet. Available online: https://www.image-net.org/update-mar-11-2021.php (accessed on 27 July 2023).
- (PDF) Deep Learning on Private Data. Available online: https://www.researchgate.net/publication/330842645_Deep_Learning_on_Private_Data (accessed on 27 July 2023).
- Maurício, J.; Domingues, I.; Bernardino, J. Comparing Vision Transformers and Convolutional Neural Networks for Image Classification: A Literature Review. Appl. Sci. 2023, 13, 5521. [Google Scholar] [CrossRef]
- Gupta, N. A Pre-Trained Vs Fine-Tuning Methodology in Transfer Learning. J. Phys. Conf. Ser. 2021, 1947, 012028. [Google Scholar] [CrossRef]
- USDA APHIS. Spotted Lanternfly. Available online: https://www.aphis.usda.gov/plant-pests-diseases/slf (accessed on 27 July 2023).
- Homeowner Resources. Available online: https://www.nj.gov/agriculture/divisions/pi/prog/pests-diseases/spotted-lanternfly/homeowner-resources/ (accessed on 27 July 2023).
- Raza, A.; Uddin, J.; Almuhaimeed, A.; Akbar, S.; Zou, Q.; Ahmad, A. AIPs-SnTCN: Predicting Anti-Inflammatory Peptides Using fastText and Transformer Encoder-Based Hybrid Word Embedding with Self-Normalized Temporal Convolutional Networks. J. Chem. Inf. Model. 2023, 63, 6537–6554. [Google Scholar] [CrossRef]
- Akbar, S.; Raza, A.; Zou, Q. Deepstacked-AVPs: Predicting Antiviral Peptides Using Tri-Segment Evolutionary Profile and Word Embedding Based Multi-Perspective Features with Deep Stacking Model. BMC Bioinform. 2024, 25, 102. [Google Scholar] [CrossRef] [PubMed]
- Akbar, S.; Zou, Q.; Raza, A.; Alarfaj, F.K. iAFPs-Mv-BiTCN: Predicting Antifungal Peptides Using Self-Attention Transformer Embedding and Transform Evolutionary Based Multi-View Features with Bidirectional Temporal Convolutional Networks. Artif. Intell. Med. 2024, 151, 102860. [Google Scholar] [CrossRef] [PubMed]
| Aspects | CNN | ViT |
|---|---|---|
| Architecture | Uses convolutional layers for local feature extraction. | Uses self-attention for global context and relationships. |
| Strengths | Efficient at capturing spatial hierarchies; faster training. | Handles global dependencies effectively. |
| Limitations | Struggles with global context without deep architectures. | Slower processing and higher computational demand. |
| Training | Relatively faster with optimized architectures. | Requires larger datasets and more resources. |
| Application | Satellite imagery classification (e.g., ResNet50, EfficientNet). | Alternative for classification with attention mechanisms. |
| Model Name | AUC Score | Accuracy | F1 Score |
|---|---|---|---|
| ResNet50 | 0.900 | 0.822 | 0.824 |
| EfficientNetv2 | 0.880 | 0.793 | 0.778 |
| ViT | 0.868 | 0.781 | 0.780 |
| VGG16 | 0.880 | 0.784 | 0.786 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, R.; Song, Z.; Zhao, J.; Li, Y. Predicting the Distribution of Ailanthus altissima Using Deep Learning-Based Analysis of Satellite Imagery. Symmetry 2025, 17, 324. https://doi.org/10.3390/sym17030324
Gao R, Song Z, Zhao J, Li Y. Predicting the Distribution of Ailanthus altissima Using Deep Learning-Based Analysis of Satellite Imagery. Symmetry. 2025; 17(3):324. https://doi.org/10.3390/sym17030324
Chicago/Turabian StyleGao, Ruohan, Zipeng Song, Junhan Zhao, and Yingnan Li. 2025. "Predicting the Distribution of Ailanthus altissima Using Deep Learning-Based Analysis of Satellite Imagery" Symmetry 17, no. 3: 324. https://doi.org/10.3390/sym17030324
APA StyleGao, R., Song, Z., Zhao, J., & Li, Y. (2025). Predicting the Distribution of Ailanthus altissima Using Deep Learning-Based Analysis of Satellite Imagery. Symmetry, 17(3), 324. https://doi.org/10.3390/sym17030324






