Structural Determinants and Their Role in Cyanobacterial Morphogenesis
Abstract
:1. Introduction
2. How Do Cyanobacteria Modify Their Cell Shape?
2.1. Morphology and Environmental Cues
2.2. Morphological Plasticity in Cyanobacteria
2.3. Different Modes of Cell Shape Regulation in Cyanobacteria
3. The Cyanobacterial Cell Division Complex—Function and Regulation
3.1. Polymerization Properties of Cyanobacterial FtsZ
3.2. FtsZ is Essential in Cyanobacteria
3.3. Cellular Localization of FtsZ in Cyanobacteria
3.4. Transcriptional and Posttranslational Control of Cell Division in Cyanobacteria
3.5. FtsZ-Associated Regulators Control Cell Division in Cyanobacteria
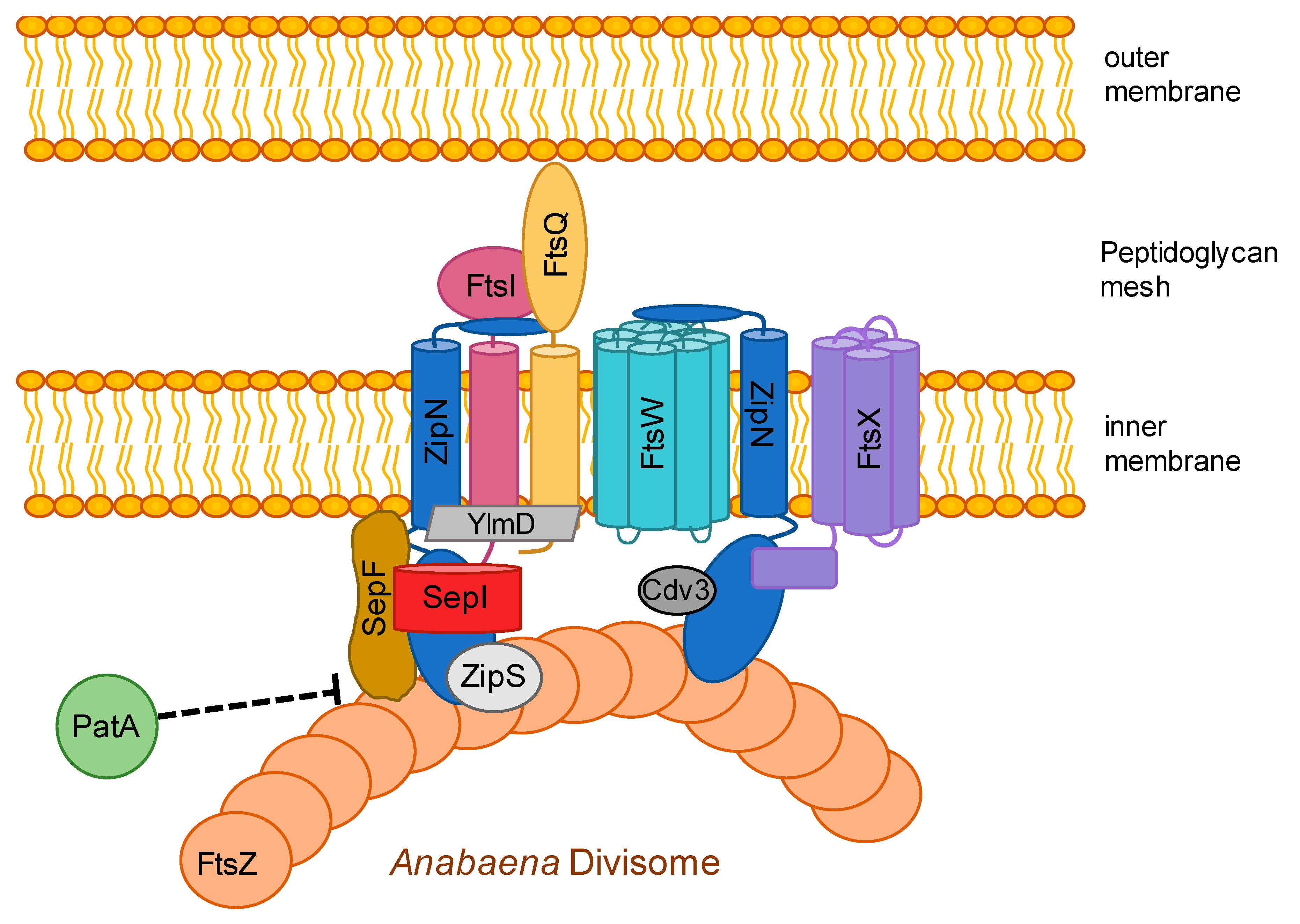
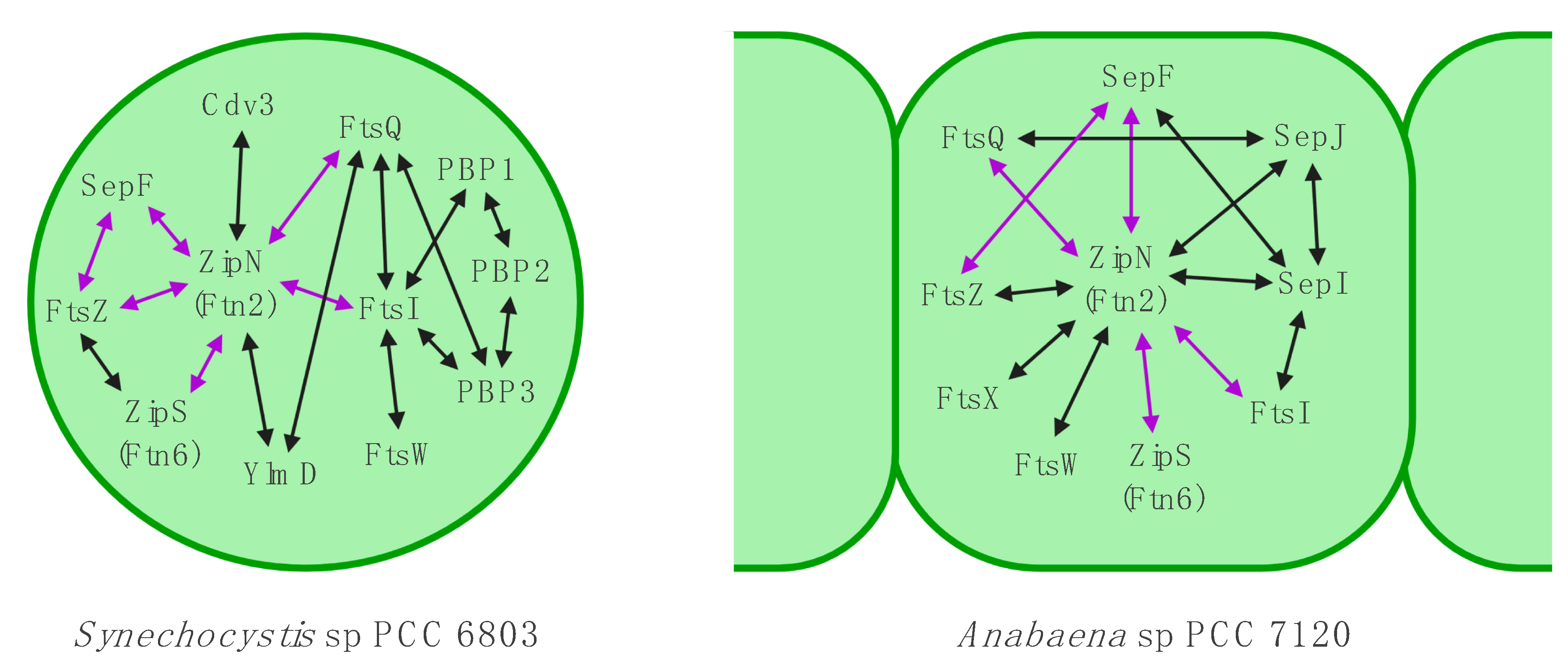
3.6. The Divisome is Linked to the Sites of Cell-cell Connections in ANABAENA
3.7. Z-Ring—All in One Place
4. Coiled-Coil-Rich Proteins in Cyanobacteria
5. Undescribed Filamentous Systems in Cyanobacteria
6. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
Data Availability
References
- Moseley, J.B. An expanded view of the eukaryotic cytoskeleton. Mol. Biol. Cell 2013, 24, 1615–1618. [Google Scholar] [CrossRef] [PubMed]
- Alberts, B.; Johnson, A.; Lewis, J.; Morgan, D.; Raff, M.; Roberts, K.; Walter, P. Molecular Biology of the Cell, 6th ed.; Garland Science: New York, NY, USA, 2014. [Google Scholar]
- Huber, F.; Boire, A.; López, M.P.; Koenderink, G.H. Cytoskeletal crosstalk: When three different personalities team up. Curr. Opin. Cell Biol. 2015, 32, 39–47. [Google Scholar] [CrossRef]
- Bi, E.; Lutkenhaus, J. FtsZ ring structure associated with division in Escherichia coli. Nature 1991, 354, 161–164. [Google Scholar] [CrossRef] [PubMed]
- De Boer, P.; Crossley, R.; Rothfield, L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature 1992, 359, 254–256. [Google Scholar] [CrossRef] [PubMed]
- Löwe, J.; Amos, L.A. Crystal structure of the bacterial cell division protein FtsZ. Nature 1998, 391, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Bork, P.; Sander, C.; Valencia, A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 1992, 89, 7290–7294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van den Ent, F.; Amos, L.A.; Löwe, J. Prokrayotic origin of the actin cytoskeleton. Nature 2001, 413, 39–44. [Google Scholar] [CrossRef]
- Ausmees, N.; Kuhn, J.R.; Jacobs-Wagner, C. The bacterial cytoskeleton: An intermediate filament-like function in cell shape. Cell 2003, 115, 705–713. [Google Scholar] [CrossRef] [Green Version]
- Kühn, J.; Briegel, A.; Mörschel, E.; Kahnt, J.; Leser, K.; Wick, S.; Jensen, G.J.; Thanbichler, M. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 2010, 29, 327–339. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Thanbichler, M. Nucleotide-independent cytoskeletal scaffolds in bacteria. Cytoskeleton 2013, 70, 409–423. [Google Scholar] [CrossRef]
- Wagstaff, J.; Löwe, J. Prokaryotic cytoskeletons: Protein filaments organizing small cells. Nat. Rev. Microbiol. 2018, 16, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Videau, P.; Rivers, O.S.; Ushijima, B.; Oshiro, R.T.; Kim, M.J.; Philmus, B.; Cozy, L.M. Mutation of the murC and murB genes impairs heterocyst differentiation in Anabaena sp. strain PCC 7120. J. Bacteriol. 2016, 198, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoiczyk, E.; Baumeister, W. Envelope structure of four gliding filamentous cyanobacteria. J. Bacteriol. 1995, 177, 2387–2395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gumbart, J.C.; Beeby, M.; Jensen, G.J.; Roux, B. Escherichia coli peptidoglycan structure and mechanics as predicted by atomic-scale simulations. PLoS Comput. Biol. 2014, 10, e1003475. [Google Scholar] [CrossRef] [Green Version]
- Hoiczyk, E.; Hansel, A. Cyanobacterial cell walls: News from an unusual prokaryotic envelope. J. Bacteriol. 2000, 182, 1191–1199. [Google Scholar] [CrossRef] [Green Version]
- Cassier-Chauvat, C.; Chauvat, F. Cell division in cyanobacteria. In The Cell Biology of Cyanobacteria; Flores, E., Herrero, A., Eds.; Caister Academic Press: Rover, UK, 2014. [Google Scholar]
- Schirrmeister, B.E.; Antonelli, A.; Bagheri, H.C. The origin of multicellularity in cyanobacteria. BMC Evol. Biol. 2011, 11, 45. [Google Scholar] [CrossRef] [Green Version]
- Rippka, R.; Stanier, R.Y.; Deruelles, J.; Herdman, M.; Waterbury, J.B. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 1979, 111, 1–61. [Google Scholar] [CrossRef] [Green Version]
- Dagan, T.; Roettger, M.; Stucken, K.; Landan, G.; Koch, R.; Major, P.; Gould, S.B.; Goremykin, V.V.; Rippka, R.; De Marsac, N.T.; et al. Genomes of stigonematalean cyanobacteria (subsection V) and the evolution of oxygenic photosynthesis from prokaryotes to plastids. Genome Biol. Evol. 2013, 5, 31–44. [Google Scholar] [CrossRef] [Green Version]
- Shih, P.M.; Wu, D.; Latifi, A.; Axen, S.D.; Fewer, D.P.; Talla, E.; Calteau, A.; Cai, F.; Tandeau de Marsac, N.; Rippka, R.; et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 1053–1058. [Google Scholar] [CrossRef] [Green Version]
- Koch, R.; Kupczok, A.; Stucken, K.; Ilhan, J.; Hammerschmidt, K.; Dagan, T. Plasticity first: Molecular signatures of a complex morphological trait in filamentous cyanobacteria. BMC Evol. Biol. 2017, 17, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Weiss, G.L.; Kieninger, A.-K.; Maldener, I.; Forchhammer, K.; Pilhofer, M. Structure and function of a bacterial gap junction analog. Cell 2019, 178, 374–384.e15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nürnberg, D.J.; Mariscal, V.; Bornikoel, J.; Nieves-Morión, M.; Krauß, N.; Herrero, A.; Maldener, I.; Flores, E.; Mullineaux, C.W. Intercellular diffusion of a fluorescent sucrose analog via the septal junctions in a filamentous cyanobacterium. MBio 2015, 6, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieninger, A.K.; Forchhammer, K.; Maldener, I. A nanopore array in the septal peptidoglycan hosts gated septal junctions for cell-cell communication in multicellular cyanobacteria. Int. J. Med. Microbiol. 2019, 309, 151303. [Google Scholar] [CrossRef] [PubMed]
- Lehner, J.; Zhang, Y.; Berendt, S.; Rasse, T.M.; Forchhammer, K.; Maldener, I. The morphogene AmiC2 is pivotal for multicellular development in the cyanobacterium Nostoc punctiforme. Mol. Microbiol. 2011, 79, 1655–1669. [Google Scholar] [CrossRef] [PubMed]
- Nayar, A.S.; Yamaura, H.; Rajagopalan, R.; Risser, D.D.; Callahan, S.M. FraG is necessary for filament integrity and heterocyst maturation in the cyanobacterium Anabaena sp. strain PCC 7120. Microbiology 2007, 153, 601–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrero, A.; Stavans, J.; Flores, E. The multicellular nature of filamentous heterocyst-forming cyanobacteria. FEMS Microbiol. Rev. 2016, 40, 831–854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flores, E.; Nieves-Morión, M.; Mullineaux, C. Cyanobacterial septal junctions: Properties and regulation. Life 2018, 9, 1. [Google Scholar] [CrossRef] [Green Version]
- Flores, E.; Herrero, A. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol. 2010, 8, 39–50. [Google Scholar] [CrossRef]
- Gaysina, L.A.; Saraf, A.; Singh, P. Cyanobacteria in diverse habitats. In Cyanobacteria—From Basic Science to Applications; Mishra, A.K., Tiwari, D.N., Rai, A.N.B.T.-C., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 1–28. [Google Scholar]
- Pospíšil, P. Production of reactive oxygen species by photosystem II as a response to light and temperature stress. Front. Plant Sci. 2016, 7, 1950. [Google Scholar] [CrossRef]
- Larkum, A.W.D.; Ritchie, R.J.; Raven, J.A. Living off the sun: Chlorophylls, bacteriochlorophylls and rhodopsins. Photosynthetica 2018, 56, 11–43. [Google Scholar] [CrossRef]
- Wiltbank, L.B.; Kehoe, D.M. Diverse light responses of cyanobacteria mediated by phytochrome superfamily photoreceptors. Nat. Rev. Microbiol. 2019, 17, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Yerrapragada, S.; Shukla, A.; Hallsworth-Pepin, K.; Choi, K.; Wollam, A.; Clifton, S.; Qin, X.; Muzny, D.; Raghuraman, S.; Ashki, H.; et al. Extreme sensory complexity encoded in the 10-megabase draft genome Sequence of the chromatically acclimating cyanobacterium tolypothrix sp. PCC 7601. Genome Announc. 2015, 3, e00355-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bennett, A.; Bogorad, L. Complementary chromatic adaptation in a filamentous blue-green alga. J. Cell Biol. 1973, 58, 419. [Google Scholar] [CrossRef] [PubMed]
- Sanfilippo, J.E.; Garczarek, L.; Partensky, F.; Kehoe, D.M. Chromatic acclimation in cyanobacteria: A diverse and widespread process for optimizing photosynthesis. Annu. Rev. Microbiol. 2019, 73, 407–433. [Google Scholar] [CrossRef] [PubMed]
- Kehoe, D.M.; Grossman, A.R. Similarity of a chromatic adaptation sensor to phytochrome and ethylene receptors. Science 1996, 273, 1409–1412. [Google Scholar] [CrossRef]
- Terauchi, K.; Montgomery, B.L.; Grossman, A.R.; Lagarias, J.C.; Kehoe, D.M. RcaE is a complementary chromatic adaptation photoreceptor required for green and red light responsiveness. Mol. Microbiol. 2004, 51, 567–577. [Google Scholar] [CrossRef]
- Bordowitz, J.R.; Montgomery, B.L. Photoregulation of cellular morphology during complementary chromatic adaptation requires sensor-kinase-class protein RcaE in Fremyella diplosiphon. J. Bacteriol. 2008, 190, 4069–4074. [Google Scholar] [CrossRef] [Green Version]
- Singh, S.P.; Montgomery, B.L. Morphogenes bolA and mreB mediate the photoregulation of cellular morphology during complementary chromatic acclimation in Fremyella diplosiphon. Mol. Microbiol. 2014, 93, 167–182. [Google Scholar] [CrossRef]
- Wilde, A.; Mullineaux, C.W. Light-controlled motility in prokaryotes and the problem of directional light perception. FEMS Microbiol. Rev. 2017, 41, 900–922. [Google Scholar] [CrossRef] [Green Version]
- Robinson, B.L.; Miller, J.H. Photomorphogenesis in the blue-green alga Nostoc commune 584. Physiol. Plant. 1970, 23, 461–472. [Google Scholar] [CrossRef]
- Meeks, J.C.; Campbell, E.L.; Summers, M.L.; Wong, F.C. Cellular differentiation in the cyanobacterium Nostoc punctiforme. Arch. Microbiol. 2002, 178, 395–403. [Google Scholar] [CrossRef]
- Damerval, T.; Guglielmi, G.; Houmard, J.; De Marsac, N.T. Hormogonium differentiation in the cyanobacterium Calothrix: A photoregulated developmental process. Plant Cell 1991, 3, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Zepu, Z.; Yuhuan, W.; Jie, X.; Lijie, F.; Dingji, S. Differentiation of hormogonia and photosynthetic characterization of Nostoc flagelliforme. Acta Bot. Sin. 2000, 42, 570–575. [Google Scholar]
- Campbell, D.; Houmard, J.; De Marsac, N.T. Electron transport regulates cellular differentiation in the filamentous cyanobacterium Calothrix. Plant Cell 1993, 5, 451–463. [Google Scholar] [CrossRef] [PubMed]
- Khamar, H.J.; Breathwaite, E.K.; Prasse, C.E.; Fraley, E.R.; Secor, C.R.; Chibane, F.L.; Elhai, J.; Chiu, W.-L. Multiple roles of soluble sugars in the establishment of Gunnera-Nostoc endosymbiosis. Plant Physiol. 2010, 154, 1381–1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meeks, J.C.; Elhai, J. Regulation of cellular differentiation in filamentous cyanobacteria in free-living and plant-associated symbiotic growth states. Microbiol. Mol. Biol. Rev. 2002, 66, 94–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liaimer, A.; Helfrich, E.J.N.; Hinrichs, K.; Guljamow, A.; Ishida, K.; Hertweck, C.; Dittmann, E. Nostopeptolide plays a governing role during cellular differentiation of the symbiotic cyanobacterium Nostoc punctiforme. Proc. Natl. Acad. Sci. USA 2015, 112, 1862–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashidoko, Y.; Nishizuka, H.; Tanaka, M.; Murata, K.; Murai, Y.; Hashimoto, M. Isolation and characterization of 1-palmitoyl-2-linoleoyl-sn-glycerol as a hormogonium-inducing factor (HIF) from the coralloid roots of Cycas revoluta (Cycadaceae). Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Nürnberg, D.J.; Mariscal, V.; Parker, J.; Mastroianni, G.; Flores, E.; Mullineaux, C.W. Branching and intercellular communication in the Section V cyanobacterium Mastigocladus laminosus, a complex multicellular prokaryote. Mol. Microbiol. 2014, 91, 935–949. [Google Scholar] [CrossRef] [Green Version]
- Campbell, E.L.; Meeks, J.C. Characteristics of hormogonia formation by symbiotic Nostoc spp. in response to the presence of anthoceros punctatus or its extracellular products. Appl. Environ. Microbiol. 1989, 55, 125–131. [Google Scholar] [CrossRef] [Green Version]
- Adams, D.G.; Bergman, B.; Nierzwicki-Bauer, S.A.; Duggan, P.S.; Rai, A.N.; Schüßler, A. Cyanobacterial-plant symbioses. In The Prokaryotes: Prokaryotic Biology and Symbiotic Associations; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 359–400. [Google Scholar]
- Decelle, J.; Colin, S.; Foster, R.A. Photosymbiosis in marine planktonic protists. In Marine Protists: Diversity and Dynamics; Ohtsuka, S., Suzaki, T., Horiguchi, T., Suzuki, N., Not, F., Eds.; Springer: Tokyo, Japan, 2015; pp. 465–500. [Google Scholar]
- Peters, G.A.; Perkins, S.K. The Azolla–Anabaena symbiosis: Endophyte continuity in the Azolla life-cycle is facilitated by epidermal trichomes. New Phytol. 1993, 123, 65–75. [Google Scholar] [CrossRef]
- Caputo, A.; Nylander, J.A.A.; Foster, R.A. The genetic diversity and evolution of diatom-diazotroph associations highlights traits favoring symbiont integration. FEMS Microbiol. Lett. 2019, 366, fny297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velázquez-Suárez, C.; Luque, I.; Herrero, A. The inorganic nutrient regime and the mre genes regulate cell and filament size and morphology in the phototrophic multicellular bacterium Anabaena. MSphere 2020, 5, e00747-20. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Montgomery, B.L. Determining cell shape: Adaptive regulation of cyanobacterial cellular differentiation and morphology. Trends Microbiol. 2011, 19, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Claessen, D.; Rozen, D.E.; Kuipers, O.P.; Søgaard-Andersen, L.; Van Wezel, G.P. Bacterial solutions to multicellularity: A tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 2014, 12, 115–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; Piao, S.; Myneni, R.B.; Huang, M.; Zeng, Z.; Canadell, J.G.; Ciais, P.; Sitch, S.; Friedlingstein, P.; Arneth, A.; et al. Greening of the Earth and its drivers. Nat. Clim. Chang. 2016, 6, 791–795. [Google Scholar] [CrossRef]
- Caccamo, P.D.; Brun, Y.V. The molecular basis of noncanonical bacterial morphology. Trends Microbiol. 2018, 26, 191–208. [Google Scholar] [CrossRef]
- Lange, R.; Hengge-Aronis, R. Growth phase-regulated expression of bolA and morphology of stationary-phase Escherichia coli cells are controlled by the novel sigma factor σ(S). J. Bacteriol. 1991, 173, 4474–4481. [Google Scholar] [CrossRef] [Green Version]
- Young, K.D. The selective value of bacterial shape. Microbiol. Mol. Biol. Rev. 2006, 70, 660–703. [Google Scholar] [CrossRef] [Green Version]
- Typas, A.; Banzhaf, M.; Gross, C.A.; Vollmer, W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat. Rev. Microbiol. 2012, 10, 123. [Google Scholar] [CrossRef] [Green Version]
- Egan, A.J.F.; Errington, J.; Vollmer, W. Regulation of peptidoglycan synthesis and remodelling. Nat. Rev. Microbiol. 2020, 18, 446–460. [Google Scholar] [CrossRef] [PubMed]
- Pinho, M.G.; Kjos, M.; Veening, J.W. How to get (a)round: Mechanisms controlling growth and division of coccoid bacteria. Nat. Rev. Microbiol. 2013, 11, 601–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Errington, J.; Wu, L.J. Cell cycle machinery in bacillus subtilis. In Prokaryotic Cytoskeletons: Filamentous Protein Polymers Active in the Cytoplasm of Bacterial and Archaeal Cells; Löwe, J., Amos, L.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 67–101. [Google Scholar]
- Den Blaauwen, T.; De Pedro, M.A.; Nguyen-Disteche, M.; Ayala, J.A. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 2008, 32, 321–344. [Google Scholar] [CrossRef] [Green Version]
- Jones, L.J.F.; Carballido-López, R.; Errington, J. Control of cell shape in bacteria: Helical, actin-like filaments in Bacillus subtilis. Cell 2001, 104, 913–922. [Google Scholar] [CrossRef]
- Kruse, T.; Bork-Jensen, J.; Gerdes, K. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol. Microbiol. 2005, 55, 78–89. [Google Scholar] [CrossRef]
- Domínguez-Escobar, J.; Chastanet, A.; Crevenna, A.H.; Fromion, V.; Wedlich-Söldner, R.; Carballido-López, R. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 2011, 333, 225–228. [Google Scholar] [CrossRef] [Green Version]
- Garner, E.C.; Bernard, R.; Wang, W.; Zhuang, X.; Rudner, D.Z.; Mitchison, T. Circumferential motions of the cell wall synthesis machinery drive cytoskeletal dynamics in B. subtilis. Science 2011, 333, 222–225. [Google Scholar] [CrossRef] [Green Version]
- Van Teeffelen, S.; Wang, S.; Furchtgott, L.; Huang, K.C.; Wingreen, N.S.; Shaevitz, J.W.; Gitai, Z. The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc. Natl. Acad. Sci. USA 2011, 108, 15822-7. [Google Scholar] [CrossRef] [Green Version]
- Siefert, J.L.; Fox, G.E. Phylogenetic mapping of bacterial morphology. Microbiology 1998, 144, 2803–2808. [Google Scholar] [CrossRef] [Green Version]
- Yulo, P.R.J.; Hendrickson, H.L. The evolution of spherical cell shape; progress and perspective. Biochem. Soc. Trans. 2019, 47, 1621–1634. [Google Scholar] [CrossRef]
- Springstein, B.L.; Weissenbach, J.; Koch, R.; Stücker, F.; Stucken, K. The role of the cytoskeletal proteins MreB and FtsZ in multicellular cyanobacteria. FEBS Open Biol. 2020, 10, 2510–2531. [Google Scholar] [CrossRef] [PubMed]
- Waidner, B.; Specht, M.; Dempwolff, F.; Haeberer, K.; Schaetzle, S.; Speth, V.; Kist, M.; Graumann, P.L. A novel system of cytoskeletal elements in the human pathogen Helicobacter pylori. PLoS Pathog. 2009, 5, e1000669. [Google Scholar] [CrossRef] [PubMed]
- Sycuro, L.K.; Pincus, Z.; Gutierrez, K.D.; Biboy, J.; Stern, C.A.; Vollmer, W.; Salama, N.R. Peptidoglycan crosslinking relaxation promotes helicobacter pylori’s helical shape and stomach colonization. Cell 2010, 141, 822–833. [Google Scholar] [CrossRef] [Green Version]
- Zhi, P.W.; Zhao, Y. Morphological reversion of Spirulina (Arthrospira) platensis (Cyanophyta): From linear to helical. J. Phycol. 2005, 41, 622–628. [Google Scholar]
- Savage, D.F.; Afonso, B.; Chen, A.H.; Silver, P.A. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science 2010, 327, 1258–1261. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.A.; Tay, J.W.; Cameron, J.C. Multi-generational analysis and manipulation of chromosomes in a polyploid cyanobacterium. BioRxiv 2019, 661256. [Google Scholar] [CrossRef]
- Watanabe, S.; Noda, A.; Ohbayashi, R.; Uchioke, K.; Kurihara, A.; Nakatake, S.; Morioka, S.; Kanesaki, Y.; Chibazakura, T.; Yoshikawa, H. ParA-like protein influences the distribution of multi-copy chromosomes in cyanobacterium Synechococcus elongatus PCC 7942. Microbiology 2018, 164, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Yang, G.; Zhao, W.; Zhang, Y.; Zhao, J. MreB is important for cell shape but not for chromosome segregation of the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 2007, 63, 1640–1652. [Google Scholar] [CrossRef]
- Watanabe, S. Cyanobacterial multi-copy chromosomes and their replication. Biosci. Biotechnol. Biochem. 2020, 84, 1309–1321. [Google Scholar] [CrossRef]
- Miyagishima, S.Y.; Wolk, P.P.; Osteryoung, K.W. Identification of cyanobacterial cell division genes by comparative and mutational analyses. Mol. Microbiol. 2005, 56, 126–143. [Google Scholar] [CrossRef]
- Ramos-León, F.; Mariscal, V.; Frías, J.E.; Flores, E.; Herrero, A. Divisome-dependent subcellular localization of cell-cell joining protein SepJ in the filamentous cyanobacterium Anabaena. Mol. Microbiol. 2015, 96, 566–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazouni, K.; Domain, F.; Cassier-Chauvat, C.; Chauvat, F. Molecular analysis of the key cytokinetic components of cyanobacteria: FtsZ, ZipN and MinCDE. Mol. Microbiol. 2004, 52, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Koksharova, O.A.; Wolk, C.P. A novel gene that bears a DnaJ motif influences cyanobacterial cell division. J. Bacteriol. 2002, 184, 5524–5528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marbouty, M.; Saguez, C.; Cassier-Chauvat, C.; Chauvat, F. Characterization of the FtsZ-interacting septal proteins SepF and Ftn6 in the spherical-celled cyanobacterium Synechocystis strain PCC 6803. J. Bacteriol. 2009, 191, 6178–6185. [Google Scholar] [CrossRef] [Green Version]
- MacCready, J.S.; Schossau, J.; Osteryoung, K.W.; Ducat, D.C. Robust min-system oscillation in the presence of internal photosynthetic membranes in cyanobacteria. Mol. Microbiol. 2017, 103, 483–503. [Google Scholar] [CrossRef]
- Marbouty, M.; Saguez, C.; Cassier-Chauvat, C.; Chauvat, F. ZipN, an FtsA-like orchestrator of divisome assembly in the model cyanobacterium Synechocystis PCC6803. Mol. Microbiol. 2009, 74, 409–420. [Google Scholar] [CrossRef]
- Stucken, K.; Ilhan, J.; Roettger, M.; Dagan, T.; Martin, W.F. Transformation and conjugal transfer of foreign genes into the filamentous multicellular cyanobacteria (subsection V) Fischerella and Chlorogloeopsis. Curr. Microbiol. 2012, 65, 552–560. [Google Scholar] [CrossRef]
- Liu, P.; Zheng, H.; Meng, Q.; Terahara, N.; Gu, W.; Wang, S.; Zhao, G.; Nakane, D.; Wang, W.; Miyata, M. Chemotaxis without conventional two-component system, based on cell polarity and aerobic conditions in helicity-switching swimming of Spiroplasma eriocheiris. Front. Microbiol. 2017, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kürner, J.; Frangakis, A.S.; Baumeister, W. Cryo-electron tomography reveals the cytoskeletal structure of Spiroplasma melliferum. Science 2005, 307, 436–438. [Google Scholar] [CrossRef]
- Ehlers, K.; Oster, G. On the mysterious propulsion of synechococcus. PLoS ONE 2012, 7, e36081. [Google Scholar] [CrossRef] [Green Version]
- Campbell, E.L.; Summers, M.L.; Christman, H.; Martin, M.E.; Meeks, J.C. Global gene expression patterns of Nostoc punctiforme in steady-state dinitrogen-grown heterocyst-containing cultures and at single time points during the differentiation of akinetes and hormogonia. J. Bacteriol. 2007, 189, 5247–5256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, E.L.; Christman, H.; Meeks, J.C. DNA microarray comparisons of plant factor- and nitrogen deprivation-induced hormogonia reveal decision-making transcriptional regulation patterns in Nostoc punctiforme. J. Bacteriol. 2008, 190, 7382LP–7391LP. [Google Scholar] [CrossRef] [Green Version]
- Kabeya, Y.; Nakanishi, H.; Suzuki, K.; Ichikawa, T.; Kondou, Y.; Matsui, M.; Miyagishima, S.-Y. The YlmG protein has a conserved function related to the distribution of nucleoids in chloroplasts and cyanobacteria. BMC Plant Biol. 2010, 10, 57. [Google Scholar] [CrossRef] [Green Version]
- Zheng, X.; O’Shea, E.K. Cyanobacteria maintain constant protein concentration despite genome copy-number variation. Cell Rep. 2017, 19, 497–504. [Google Scholar] [CrossRef] [Green Version]
- Burnat, M.; Schleiff, E.; Flores, E. Cell envelope components influencing filament length in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 2014, 196, 4026–4035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, L.M.; Makarova, K.S.; Koonin, E.V.; Aravind, L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: Implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004, 32, 5260–5279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mandakovic, D.; Trigo, C.; Andrade, D.; Riquelme, B.; Gómez-Lillo, G.; Soto-Liebe, K.; Díez, B.; Vásquez, M. CyDiv, a conserved and novel filamentous cyanobacterial cell division protein involved in septum localization. Front. Microbiol. 2016, 7, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leganés, F.; Blanco-Rivero, A.; Fernández-Piñas, F.; Redondo, M.; Fernández-Valiente, E.; Fan, Q.; Lechno-Yossef, S.; Wolk, C.P. Wide variation in the cyanobacterial complement of presumptive penicillin-binding proteins. Arch. Microbiol. 2005, 184, 234–248. [Google Scholar] [CrossRef]
- Berendt, S.; Lehner, J.; Zhang, Y.V.; Rasse, T.M.; Forchhammer, K.; Maldener, I. Cell wall amidase amic1 is required for cellular communication and heterocyst development in the cyanobacterium Anabaena PCC 7120 but not for filament integrity. J. Bacteriol. 2012, 194, 5218–5227. [Google Scholar] [CrossRef] [Green Version]
- Rexroth, S.; Mullineaux, C.W.; Ellinger, D.; Sendtko, E.; Rögner, M.; Koenig, F. The plasma membrane of the cyanobacterium Gloeobacter violaceus contains segregated bioenergetic domains. Plant Cell 2011, 23, 2379LP–2390. [Google Scholar] [CrossRef] [Green Version]
- Mitschke, J.; Georg, J.; Scholz, I.; Sharma, C.M.; Dienst, D.; Bantscheff, J.; Voß, B.; Steglich, C.; Wilde, A.; Vogel, J.; et al. An experimentally anchored map of transcriptional start sites in the model cyanobacterium Synechocystis sp. PCC6803. Proc. Natl. Acad. Sci. USA 2011, 108, 2124LP–2129LP. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitschke, J.; Vioque, A.; Haas, F.; Hess, W.R.; Muro-Pastor, A.M. Dynamics of transcriptional start site selection during nitrogen stress-induced cell differentiation in Anabaena sp. PCC7120. Proc. Natl. Acad. Sci. USA 2011, 108, 20130–20135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flaherty, B.L.; Van Nieuwerburgh, F.; Head, S.R.; Golden, J.W. Directional RNA deep sequencing sheds new light on the transcriptional response of Anabaena sp. strain PCC 7120 to combined-nitrogen deprivation. BMC Genom. 2011, 12, 332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stöckel, J.; Elvitigala, T.R.; Liberton, M.; Pakrasi, H.B. Carbon availability affects diurnally controlled processes and cell morphology of cyanothece 51142. PLoS ONE 2013, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Khayatan, B.; Meeks, J.C.; Risser, D.D. Evidence that a modified type IV pilus-like system powers gliding motility and polysaccharide secretion in filamentous cyanobacteria. Mol. Microbiol. 2015, 98, 1021–1036. [Google Scholar] [CrossRef]
- Marbouty, M.; Mazouni, K.; Saguez, C.; Cassier-Chauvat, C.; Chauvat, F. Characterization of the Synechocystis strain PCC 6803 penicillin-binding proteins and cytokinetic proteins FtsQ and FtsW and their network of interactions with ZipN. J. Bacteriol. 2009, 191, 5123–5133. [Google Scholar] [CrossRef] [Green Version]
- Bornikoel, J.; Staiger, J.; Madlung, J.; Forchhammer, K.; Maldener, I. LytM factor Alr3353 affects filament morphology and cell–cell communication in the multicellular cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 2018, 108, 187–203. [Google Scholar] [CrossRef] [Green Version]
- Flores, E.; Pernil, R.; Muro-Pastor, A.M.; Mariscal, V.; Maldener, I.; Lechno-Yossef, S.; Fan, Q.; Wolk, C.P.; Herrero, A. Septum-localized protein required for filament integrity and diazotrophy in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 2007, 189, 3884–3890. [Google Scholar] [CrossRef] [Green Version]
- Merino-Puerto, V.; Mariscal, V.; Mullineaux, C.W.; Herrero, A.; Flores, E. Fra proteins influencing filament integrity, diazotrophy and localization of septal protein SepJ in the heterocyst-forming cyanobacterium Anabaena sp. Mol. Microbiol. 2010, 75, 1159–1170. [Google Scholar] [CrossRef]
- Sakr, S.; Thyssen, M.; Denis, M.; Zhang, C.C. Relationship among several key cell cycle events in the developmental cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 2006, 188, 5958–5965. [Google Scholar] [CrossRef] [Green Version]
- Koksharova, O.A.; Klint, J.; Rasmussen, U. Comparative proteomics of cell division mutants and wild-type of Synechococcus sp. strain PCC 7942. Microbiology 2007, 153, 2505–2517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández-Muñiz, W.; Stevens, S.E. Characterization of the motile hormogonia of Mastigocladus laminosus. J. Bacteriol. 1987, 169, 218LP–223LP. [Google Scholar] [CrossRef] [Green Version]
- González, A.; Fillat, M.F.; Bes, M.-T.; Peleato, M.-L.; Sevilla, E. The challenge of Iron stress in Cyanobacteria. In Cyanobacteria; IntechOpen: London, UK, 2018. [Google Scholar]
- González, A.; Bes, M.T.; Valladares, A.; Peleato, M.L.; Fillat, M.F. FurA is the master regulator of iron homeostasis and modulates the expression of tetrapyrrole biosynthesis genes in A nabaena sp. PCC 7120. Environ. Microbiol. 2012, 14, 3175–3187. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Bes, M.T.; Barja, F.; Peleato, M.L.; Fillat, M.F. Overexpression of FurA in Anabaena sp. PCC 7120 reveals new targets for this regulator involved in photosynthesis, iron uptake and cellular morphology. Plant Cell Physiol. 2010, 51, 1900–1914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, Y.-P.; Rittichier, J.; Kuru, E.; Yablonowski, J.; Pasciak, E.; Tekkam, S.; Hall, E.; Murphy, B.; Lee, T.K.; Garner, E.C.; et al. Full color palette of fluorescent d-amino acids for in situ labeling of bacterial cell walls. Chem. Sci. 2017, 8, 6313–6321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.Y.; Lin, G.M.; Xing, W.Y.; Zhang, C.C. Diversity of growth patterns probed in live cyanobacterial cells using a fluorescent analog of a peptidoglycan precursor. Front. Microbiol. 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Laddomada, F.; Miyachiro, M.M.; Dessen, A. Structural insights into protein-protein interactions involved in bacterial cell wall biogenesis. Antibiotics 2016, 5, 14. [Google Scholar] [CrossRef] [Green Version]
- Lázaro, S.; Fernández-Piñas, F.; Fernández-Valiente, E.; Blanco-Rivero, A.; Leganés, F. PbpB, a gene coding for a putative penicillin-binding protein, is required for aerobic nitrogen fixation in the cyanobacterium Anabaena sp. strain PCC7120. J. Bacteriol. 2001, 183, 628–636. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Jäger, K.; Black, T.; Zarka, K.; Koksharova, O.; Wolk, C.P. HcwA, an autolysin, is required for heterocyst maturation in Anabaena sp. strain PCC 7120. J. Bacteriol. 2001, 183, 6841LP–6851LP. [Google Scholar] [CrossRef] [Green Version]
- Fenton, A.K.; Gerdes, K. Direct interaction of FtsZ and MreB is required for septum synthesis and cell division in Escherichia coli. EMBO J. 2013, 32, 1953–1965. [Google Scholar] [CrossRef]
- Osawa, M.; Anderson, D.E.; Erickson, H.P. Reconstitution of contractile FtsZ rings in liposomes. Science 2008, 320, 792–794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osawa, M.; Anderson, D.E.; Erickson, H.P. Curved FtsZ protofilaments generate bending forces on liposome membranes. EMBO J. 2009, 28, 3476–3484. [Google Scholar] [CrossRef] [PubMed]
- Lutkenhaus, J.; Du, S.E. coli Cell Cycle Machinery. In Prokaryotic Cytoskeletons: Filamentous Protein Polymers Active in the Cytoplasm of Bacterial and Archaeal Cells; Löwe, J., Amos, L.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 27–65. [Google Scholar]
- Den Blaauwen, T.; Hamoen, L.W.; Levin, P.A. The divisome at 25: The road ahead. Curr. Opin. Microbiol. 2017, 36, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Camargo, S.; Picossi, S.; Corrales-Guerrero, L.; Valladares, A.; Arévalo, S.; Herrero, A. ZipN is an essential FtsZ membrane tether and contributes to the septal localization of SepJ in the filamentous cyanobacterium Anabaena. Sci. Rep. 2019, 9, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valladares, A.; Velázquez-Suárez, C.; Herrero, A. Interactions of PatA with the divisome during heterocyst differentiation in Anabaena. mSphere 2020, 5, e00188-20. [Google Scholar] [CrossRef] [PubMed]
- Gorelova, O.A.; Baulina, O.I.; Rasmussen, U.; Koksharova, O.A. The pleiotropic effects of ftn2 and ftn6 mutations in cyanobacterium Synechococcus sp. PCC 7942: An ultrastructural study. Protoplasma 2013, 250, 931–942. [Google Scholar] [CrossRef]
- Chen, C.; MacCready, J.S.; Ducat, D.C.; Osteryoung, K.W. The molecular machinery of chloroplast division. Plant Physiol. 2018, 176, 138–151. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, Y.; Mogi, Y.; TerBush, A.D.; Osteryoung, K.W. Chloroplast FtsZ assembles into a contractible ring via tubulin-like heteropolymerization. Nat. Plants 2016, 2, 16095. [Google Scholar] [CrossRef]
- Miyagishima, S.; Nakamura, M.; Uzuka, A.; Era, A. FtsZ-less prokaryotic cell division as well as FtsZ- and dynamin-less chloroplast and non-photosynthetic plastid division. Front. Plant Sci. 2014, 5, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.-C.C.; Hugenin, S.; Friry, A.; Huguenin, S.; Friry, A. Analysis of genes encoding the cell division protein FtsZ and a glutathione synthetase homologue in the cyanobacterium Anabaena sp. PCC 7120. Res. Microbiol. 1995, 146, 445–455. [Google Scholar] [CrossRef]
- Wang, N.; Bian, L.; Ma, X.; Meng, Y.; Chen, C.S.; Ur Rahman, M.; Zhang, T.; Li, Z.; Wang, P.; Chen, Y. Assembly properties of the bacterial tubulin homolog FtsZ from the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 2019, 294, 16309–16319. [Google Scholar] [CrossRef]
- Corrales-Guerrero, L.; Camargo, S.; Valladares, A.; Picossi, S.; Luque, I.; Ochoa De Alda, J.A.G.; Herrero, A. FtsZ of filamentous, heterocyst-forming cyanobacteria has a conserved N-terminal peptide required for normal FtsZ polymerization and cell division. Front. Microbiol. 2018, 9, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Duman, R.; Ishikawa, S.; Celik, I.; Strahl, H.; Ogasawara, N.; Troc, P.; Löwe, J.; Hamoen, L.W. Structural and genetic analyses reveal the protein SepF as a new membrane anchor for the Z ring. Proc. Natl. Acad. Sci. USA 2013, 110, E4601–E4610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erickson, H.P.; Anderson, D.E.; Osawa, M. FtsZ in bacterial cytokinesis: Cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 2010, 74, 504–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, R.; Liu, Y.; Liu, S.; Wang, Y.; Li, K.; Li, N.; Xu, D.; Zeng, Q. Three-dimensional superresolution imaging of the FtsZ ring during cell division of the cyanobacterium prochlorococcus. MBio 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abed, R.M.; Garcia-Pichel, F.; Hernández-Mariné, M. Polyphasic characterization of benthic, moderately halophilic, moderately thermophilic cyanobacteria with very thin trichomes and the proposal of Halomicronema excentricum gen. nov., sp. nov. Arch. Microbiol. 2002, 177, 361–370. [Google Scholar] [CrossRef]
- Sarcina, M.; Mullineaux, C.W. Effects of tubulin assembly inhibitors on cell division in prokaryotes in vivo. FEMS Microbiol. Lett. 2000, 191, 25–29. [Google Scholar] [CrossRef]
- Billi, D. Loss of topological relationships in a Pleurocapsalean cyanobacterium (Chroococcidiopsis sp.) with partially inactivatedftsZ. Ann. Microbiol. 2009, 59, 235. [Google Scholar] [CrossRef]
- Mori, T.; Johnson, C.H. Independence of circadian timing from cell division in cyanobacteria. J. Bacteriol. 2001, 183, 2439–2444. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, T.; Maki, M.; Okamoto, H.; Hiroishi, S. Coordination of DNA replication and cell division in Cyanobacteria Microcystis aeruginosa. FEMS Microbiol. Lett. 2005, 251, 149–154. [Google Scholar] [CrossRef] [Green Version]
- Esteves-Ferreira, A.A.; Inaba, M.; Obata, T.; Fort, A.; Fleming, G.T.A.; Araújo, W.L.; Fernie, A.R.; Sulpice, R. A novel mechanism, linked to cell density, largely controls cell division in synechocystis. Plant Physiol. 2017, 174, 2166–2182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, G.; Yang, Q.; Wang, Q.; Kim, Y.-I.; Wood, T.L.; Osteryoung, K.W.; van Oudenaarden, A.; Golden, S.S. Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell 2010, 140, 529–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandh, G.; El-Shehawy, R.; Díez, B.; Bergman, B. Temporal separation of cell division and diazotrophy in the marine diazotrophic cyanobacterium Trichodesmium erythraeum IMS101. FEMS Microbiol. Lett. 2009, 295, 281–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakr, S.; Jeanjean, R.; Zhang, C.-C.; Arcondeguy, T. Inhibition of cell division suppresses heterocyst development in Anabaena sp. strain PCC 7120. J. Bacteriol. 2006, 188, 1396–1404. [Google Scholar] [CrossRef] [Green Version]
- Klint, J.; Rasmussen, U.; Bergman, B. FtsZ may have dual roles in the filamentous cyanobacterium Nostoc/Anabaena sp. strain PCC 7120. J. Plant Physiol. 2007, 164, 11–18. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, X. Regulation by hetC of genes required for heterocyst differentiation and cell division in Anabaena sp. strain PCC 7120. J. Bacteriol. 2005, 187, 8489–8493. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, I.; Peng, L.; Bedu, S. Developmental regulation of the cell division protein FtsZ in Anabaena sp. strain PCC 7120, a cyanobacterium capable of terminal differentiation. J. Bacteriol. 2000, 182, 4640–4643. [Google Scholar] [CrossRef] [Green Version]
- Golubić, S.; Hernández-Mariné, M.; Hoffmann, L. Developmental aspects of branching in filamentous Cyanophyta/cyanobacteria. Algol. Stud. Für Hydrobiol. Suppl. Vol. 1996, 83, 303–329. [Google Scholar] [CrossRef]
- Lopes Pinto, F.; Erasmie, S.; Blikstad, C.; Lindblad, P.; Oliveira, P. FtsZ degradation in the cyanobacterium Anabaena sp. strain PCC 7120. J. Plant Physiol. 2011, 168, 1934–1942. [Google Scholar] [CrossRef]
- Goclaw-Binder, H.; Sendersky, E.; Shimoni, E.; Kiss, V.; Reich, Z.; Perelman, A.; Schwarz, R. Nutrient-associated elongation and asymmetric division of the cyanobacterium Synechococcus PCC 7942. Environ. Microbiol. 2012, 14, 680–690. [Google Scholar] [CrossRef]
- Yamauchi, Y.; Kaniya, Y.; Kaneko, Y.; Hihara, Y. Physiological roles of the cyAbrB transcriptional regulator pair Sll0822 and Sll0359 in synechocystis sp. strain PCC 6803. J. Bacteriol. 2011, 193, 3702–3709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, D.; Xu, X. CalA, a cyAbrB protein, binds to the upstream region of ftsZ and is down-regulated in heterocysts in Anabaena sp. PCC 7120. Arch. Microbiol. 2010, 192, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Jain, I.H.; Vijayan, V.; O’Shea, E.K. Spatial ordering of chromosomes enhances the fidelity of chromosome partitioning in cyanobacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 13638–13643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, A.H.; Afonso, B.; Silver, P.A.; Savage, D.F. Spatial and temporal organization of chromosome duplication and segregation in the cyanobacterium synechococcus elongatus PCC 7942. PLoS ONE 2012, 7, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szwedziak, P.; Wang, Q.; Freund, S.M.; Löwe, J. FtsA forms actin-like protofilaments. EMBO J. 2012, 31, 2249–2260. [Google Scholar] [CrossRef] [Green Version]
- Loose, M.; Mitchison, T.J. The bacterial cell division proteins ftsA and ftsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol. 2014, 16, 38–46. [Google Scholar] [CrossRef] [Green Version]
- Du, S.; Lutkenhaus, J. Assembly and activation of the Escherichia coli divisome. Mol. Microbiol. 2017, 105, 177–187. [Google Scholar] [CrossRef] [Green Version]
- Pichoff, S.; Lutkenhaus, J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002, 21, 685–693. [Google Scholar] [CrossRef] [Green Version]
- Jensen, S.O.; Thompson, L.S.; Harry, E.J. Cell division in Bacillus subtilis: FtsZ and FtsA association is Z-ring independent, and FtsA is required for efficient midcell Z-ring assembly. J. Bacteriol. 2005, 187, 6536LP–6544LP. [Google Scholar] [CrossRef] [Green Version]
- Hamoen, L.W.; Meile, J.-C.; De Jong, W.; Noirot, P.; Errington, J. SepF, a novel FtsZ-interacting protein required for a late step in cell division. Mol. Microbiol. 2006, 59, 989–999. [Google Scholar] [CrossRef]
- Vitha, S.; Froehlich, J.E.; Koksharova, O.; Pyke, K.A.; van Erp, H.; Osteryoung, K.W. ARC6 is a J-domain plastid division protein and an evolutionary descendant of the cyanobacterial cell division protein Ftn2. Plant Cell 2003, 15, 1918–1933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, J.K.; Makde, R.D.; Kumar, V.; Panda, D. SepF increases the assembly and bundling of FtsZ polymers and stabilizes FtsZ protofilaments by binding along its length. J. Biol. Chem. 2008, 283, 31116–31124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacCready, J.S.; Hakim, P.; Young, E.J.; Hu, L.; Liu, J.; Osteryoung, K.W.; Vecchiarelli, A.G.; Ducat, D.C. Protein gradients on the nucleoid position the carbon-fixing organelles of cyanobacteria. elife 2018, 7, 1–33. [Google Scholar] [CrossRef] [PubMed]
- Springstein, B.L.; Arévalo, S.; Helbig, A.O.; Herrero, A.; Stucken, K.; Flores, E.; Dagan, T. A novel septal protein of multicellular heterocystous cyanobacteria is associated with the divisome. Mol. Microbiol. 2020, 113, 1140–1154. [Google Scholar] [CrossRef] [PubMed]
- Marbouty, M.; Saguez, C.; Chauvat, F. The cyanobacterial cell division factor Ftn6 contains an N-terminal DnaD-like domain. BMC Struct. Biol. 2009, 9, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Rudolf, M.; Tetik, N.; Ramos-León, F.; Flinner, N.; Ngo, G.; Stevanovic, M.; Burnat, M.; Pernil, R.; Flores, E.; Schleiff, E. The peptidoglycan-binding protein SjcF1 influences septal junction function and channel formation in the filamentous cyanobacterium Anabaena. MBio 2015, 6, e00376–15. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Omairi-Nasser, A.; Li, X.; Dong, C.; Lin, Y.; Haselkorn, R.; Zhao, J. An amidase is required for proper intercellular communication in the filamentous cyanobacterium Anabaena sp. PCC 7120. Proc. Natl. Acad. Sci. USA 2017, 114, E1405–E1412. [Google Scholar] [CrossRef] [Green Version]
- D’Ulisse, V.; Fagioli, M.; Ghelardini, P.; Paolozzi, L. Three functional subdomains of the Escherichia coli FtsQ protein are involved in its interaction with the other division proteins. Microbiology 2007, 153, 124–138. [Google Scholar] [CrossRef] [Green Version]
- Mariscal, V.; Nürnberg, D.J.; Herrero, A.; Mullineaux, C.W.; Flores, E. Overexpression of SepJ alters septal morphology and heterocyst pattern regulated by diffusible signals in Anabaena. Mol. Microbiol. 2016, 101, 968–981. [Google Scholar] [CrossRef]
- Karimova, G.; Dautin, N.; Ladant, D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 2005, 187, 2233LP–2243LP. [Google Scholar] [CrossRef] [Green Version]
- Cho, H. The role of cytoskeletal elements in shaping bacterial cells. J. Microbiol. Biotechnol. 2015, 25, 307–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szwedziak, P.; Ghosal, D. FtsZ-ring architecture and its control by MinCD. In Prokaryotic Cytoskeletons: Filamentous Protein Polymers Active in the Cytoplasm of Bacterial and Archaeal Cells; Löwe, J., Amos, L.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 213–244. [Google Scholar]
- Mukherjee, A.; Cao, C.; Lutkenhaus, J. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proc. Natl. Acad. Sci. USA 1998, 95, 2885LP–2890LP. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, E.; Lutkenhaus, J. Cell division inhibitors SulA and MinCD prevent formation of the FtsZ ring. J. Bacteriol. 1993, 175, 1118LP–1125LP. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raynaud, C.; Cassier-Chauvat, C.; Perennes, C.; Bergounioux, C. An Arabidopsis homolog of the bacterial cell division inhibitor SulA is involved in plastid division. Plant Cell 2004, 16, 1801–1811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukherjee, A.; Lutkenhaus, J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998, 17, 462–469.e6. [Google Scholar] [CrossRef]
- Murray, S.M.; Howard, M. Center finding in E. coli and the role of mathematical modeling: Past, present and future. J. Mol. Biol. 2019, 431, 928–938. [Google Scholar] [CrossRef]
- Jordan, A.; Chandler, J.; MacCready, J.S.; Huang, J.; Osteryoung, K.W.; Ducat, D.C. Engineering cyanobacterial cell morphology for enhanced recovery and processing of biomass. Appl. Environ. Microbiol. 2017, 83, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Rust, M.J. The min oscillator defines sites of asymmetric cell division in cyanobacteria during stress recovery. Cell Syst. 2018, 7, 471–481.e6. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, E.; Weber, K. Intermediate Filaments: Structure, dynamics, function and disease. Annu. Rev. Biochem. 1994, 63, 345–382. [Google Scholar] [CrossRef]
- Herrmann, H.; Aebi, U. Intermediate filaments: Structure and assembly. Cold Spring Harb. Perspect. Biol. 2016, 8, a018242. [Google Scholar] [CrossRef]
- Sundararajan, K.; Goley, E.D. Cytoskeletal proteins in caulobacter crescentus: Spatial orchestrators of cell cycle progression, development, and cell shape. Subcell. Biochem. 2017, 84, 103–137. [Google Scholar] [PubMed] [Green Version]
- Van Teeseling, M.C.F.; de Pedro, M.A.; Cava, F. Determinants of bacterial morphology: From fundamentals to possibilities for antimicrobial targeting. Front. Microbiol. 2017, 8, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurme, R.; Namork, E.; Nurmiaho-Lassila, E.L.; Rhen, M. Intermediate filament-like network formed in vitro by a bacterial coiled coil protein. J. Biol. Chem. 1994, 269, 10675–10682. [Google Scholar]
- Kelemen, G.H. Intermediate filaments supporting cell shape and growth in bacteria. In Prokaryotic Cytoskeletons: Filamentous Protein Polymers Active in the Cytoplasm of Bacterial and Archaeal Cells; Löwe, J., Amos, L.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 161–211. [Google Scholar]
- Erickson, H.P. Evolution of the cytoskeleton. Bioessays 2007, 29, 668–677. [Google Scholar] [CrossRef] [Green Version]
- Wickstead, B.; Gull, K. The evolution of the cytoskeleton. J. Cell Biol. 2011, 194, 513–525. [Google Scholar] [CrossRef]
- Fiuza, M.; Letek, M.; Leiba, J.; Villadangos, A.F.; Vaquera, J.; Zanella-Cléon, I.; Mateos, L.M.; Molle, V.; Gil, J.A. Phosphorylation of a novel cytoskeletal protein (RsmP) regulates rod-shaped morphology in Corynebacterium glutamicum. J. Biol. Chem. 2010, 285, 29387–29397. [Google Scholar] [CrossRef] [Green Version]
- Bagchi, S.; Tomenius, H.; Belova, L.M.; Ausmees, N. Intermediate filament-like proteins in bacteria and a cytoskeletal function in Streptomyces. Mol. Microbiol. 2008, 70, 1037–1050. [Google Scholar] [PubMed] [Green Version]
- Holmes, N.A.; Walshaw, J.; Leggett, R.M.; Thibessard, A.; Dalton, K.A.; Gillespie, M.D.; Hemmings, A.M.; Gust, B.; Kelemen, G.H. Coiled-coil protein Scy is a key component of a multiprotein assembly controlling polarized growth in Streptomyces. Proc. Natl. Acad. Sci. USA 2013, 110, E397–E406. [Google Scholar] [CrossRef] [Green Version]
- Fröjd, M.J.; Flärdh, K. Apical assemblies of intermediate filament-like protein FilP are highly dynamic and affect polar growth determinant DivIVA in Streptomyces venezuelae. Mol. Microbiol. 2019, 112, 47–61. [Google Scholar] [CrossRef]
- England, P.; Bourhy, P.; Picardeau, M.; Saint Girons, I.; Mazouni, K.; Pehau-Arnaudet, G. The scc spirochetal coiled-coil protein forms helix-like filaments and binds to nucleic acids generating nucleoprotein structures. J. Bacteriol. 2005, 188, 469–476. [Google Scholar]
- Yang, R.; Bartle, S.; Otto, R.; Rogers, M.; Plamann, L.; Hartzell, P.L.; Stassinopoulos, A. AglZ is a filament-forming coiled-coil protein required for adventurous gliding motility of Myxococcus xanthus. J. Bacteriol. 2004, 186, 6168–6178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Specht, M.; Schätzle, S.; Graumann, P.L.; Waidner, B. Helicobacter pylori possesses four coiled-coil-rich proteins that form extended filamentous structures and control cell shape and motility. J. Bacteriol. 2011, 193, 4523–4530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Köster, S.; Weitz, D.A.; Goldman, R.D.; Aebi, U.; Herrmann, H. Intermediate filament mechanics in vitro and in the cell: From coiled coils to filaments, fibers and networks. Curr. Opin. Cell Biol. 2015, 32, 82–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Springstein, B.L.; Woehle, C.; Weissenbach, J.; Helbig, A.O.; Dagan, T.; Stucken, K. Identification and characterization of novel filament-forming proteins in cyanobacteria. Sci. Rep. 2020, 10, 1894. [Google Scholar] [CrossRef]
- Scanlan, D.J.; Ostrowski, M.; Mazard, S.; Dufresne, A.; Garczarek, L.; Hess, W.R.; Post, A.F.; Hagemann, M.; Paulsen, I.; Partensky, F. Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 2009, 73, 249–299. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.W.; Gonzales, A.; Harwood, T.V.; Huynh, J.; Hwang, Y.; Park, J.S.; Trieu, A.Q.; Italia, P.; Pallipuram, V.K.; Risser, D.D. Dynamic localization of HmpF regulates type IV pilus activity and directional motility in the filamentous cyanobacterium Nostoc punctiforme. Mol. Microbiol. 2017, 106, 252–265. [Google Scholar] [CrossRef] [Green Version]
- Bhaya, D.; Takahashi, A.; Shahi, P.; Arthur, R. Novel motility mutants of synechocystis strain PCC 6803 generated by in vitro transposon mutagenesis. J. Bacteriol. 2001, 183, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, H.; Aebi, U. Intermediate filaments: Molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Annu. Rev. Biochem. 2004, 73, 749–789. [Google Scholar] [CrossRef]
- Šmarda, J.; Šmajs, D.; Komrska, J.; Krzyžánek, V. S-layers on cell walls of cyanobacteria. Micron 2002, 33, 257–277. [Google Scholar] [CrossRef]
- Cohen, S.E.; McKnight, B.M.; Golden, S.S. Roles for ClpXP in regulating the circadian clock in Synechococcus elongatus. Proc. Natl. Acad. Sci. USA 2018, 115, E7805–E7813. [Google Scholar] [CrossRef] [Green Version]
- Springstein, B.L.; Nürnberg, D.J.; Woehle, C.; Weissenbach, J.; Theune, M.L.; Helbig, A.O.; Maldener, I.; Dagan, T.; Stucken, K. Two novel heteropolymer-forming proteins maintain the multicellular shape of the cyanobacterium Anabaena sp. PCC 7120. FEBS J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lupas, A.; Van Dyke, M.; Stock, J. Predicting coiled coils from protein sequences. Science 1991, 252, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Bailey-Watts, A.E.; Bindloss, M.E. Freshwater primary production by a blue–green alga of bacterial size. Nature 1968, 220, 1344–1345. [Google Scholar] [CrossRef]
- Bisalputra, T.; Oakley, B.R.; Walker, D.C.; Shields, C.M. Microtubular complexes in blue-green algae. Protoplasma 1975, 86, 19–28. [Google Scholar] [CrossRef]
- Jensen, T.E.; Ayala, R.P. The fine structure of striated microtubules and sleeve bodies in several species of Anabaena. J. Ultrasruct. Res. 1976, 57, 185–193. [Google Scholar] [CrossRef]
- Bermudes, D.; Hinkle, G.; Margulis, L. Do prokaryotes contain microtubules? Microbiol. Rev. 1994, 58, 387–400. [Google Scholar] [CrossRef]
- Rast, A.; Schaffer, M.; Albert, S.; Wan, W.; Pfeffer, S.; Beck, F.; Plitzko, J.M.; Nickelsen, J.; Engel, B.D. Biogenic regions of cyanobacterial thylakoids form contact sites with the plasma membrane. Nat. Plants 2019, 5, 436–446. [Google Scholar] [CrossRef]
- Jensen, T.E.; Ayala, R.P. Microtubule-like inclusions in isolates of the blue-green bacteria Anabaena and Nostoc. Cytologia 1980, 45, 315–326. [Google Scholar] [CrossRef] [Green Version]
- Basler, M.; Pilhofer, M.; Henderson, G.P.; Jensen, G.J.; Mekalanos, J.J. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 2012, 483, 182–186. [Google Scholar] [CrossRef]
- Porta, D.; Rippka, R.; Hernández-Mariné, M. Unusual ultrastructural features in three strains of Cyanothece (cyanobacteria). Arch. Microbiol. 2000, 173, 154–163. [Google Scholar] [CrossRef]
- Medeiros, J.M.; Böck, D.; Weiss, G.L.; Kooger, R.; Wepf, R.A.; Pilhofer, M. Robust workflow and instrumentation for cryo-focused ion beam milling of samples for electron cryotomography. Ultramicroscopy 2018, 190, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Cyanobase Locus Tags and NCBI Accession Numbers | ||||
|---|---|---|---|---|
| Proteins | Synechocystis | Synechococcus | Anabaena | Function |
| FtsZ | Sll1633 (WP_010872126.1) | Synpcc7942_2378 (WP_011244037.1) | Alr3858 (WP_010997999.1) | Cell division |
| ZipN (Ftn2) | Sll0169 (WP_010873289.1) | Synpcc7942_1943 (WP_011244461.1) | All2707 (WP_010996860.1) | Cell division |
| ZipS (Ftn6) | Sll1939 (WP_010871735.1) | Synpcc7942_1707 (WP_011244694.1) | All1616 (BAB77982.1) | Cell division |
| Cdv1 | Sll0227 (WP_010871341.1) | Synpcc7942_0653 (WP_011243187.1) | All4287 (BAB75986.1) | Cell division |
| SepF (Cdv2) | Slr2073 (WP_010872037.1) | Synpcc7942_2059 (WP_011378295.1) | Alr0487 (WP_010994663.1) | Cell division |
| Cdv3 | Slr0848 (WP_010873766.1) | Synpcc7942_2006 (WP_011244399.1) | Alr4701 (WP_010998832.1) | Cell division |
| YlmD | Slr1573 (WP_010874196.1) a | Synpcc7942_0346 (WP_011243479.1) | All5255 (WP_010999379.1) | Cell wall synthesis |
| YlmE | Slr0556 (WP_010874100.1) | Synpcc7942_2060 (WP_011244343.1) | Alr0486 (WP_010994662.1) | Unknown |
| YlmG | Ssr2142 (WP_010871471.1) | Synpcc7942_0477 (WP_011243354.1) b | Asl2061 (WP_010996222.1) | Cell division |
| Ssl0353 (WP_010873648.1) | Synpcc7942_2017 (WP_011244388.1) | Asl0940 (WP_010995114.1) | ||
| YlmH | Sll1252 (WP_010872783.1) c | Synpcc7942_1503 (WP_011378057.1) c | Alr2890 (WP_010997041.1) c | Unknown |
| MinC | Sll0288 (WP_010873891.1) | Synpcc7942_2001 (ABB58031.1) | Alr3455 (BAB75154.1) | Cell division |
| MinD | Sll0289 (WP_010873890.1) | Synpcc7942_0896 (WP_011242956.1) d | Alr3456 (WP_010997606.1) e | Cell division |
| --- | Synpcc7942_0220 (WP_011243604.1) | All2033 (WP_010996194.1) | ||
| --- | --- | All2797 (WP_010996948.1) | ||
| MinE | Ssl0546 (WP_010873889.1) | Synpcc7942_0897 (WP_011242955.1) | Asr3457 (WP_010997607.1) | Cell division |
| SulA | Slr1223 (WP_014407090.1) | Synpcc7942_2477 (WP_011243937.1) | All2390 (WP_010996546.1) | Cell division |
| FtsE | Slr0544 (WP_010874063.1) f | Synpcc7942_1414 (WP_011242455.1) f | Alr1706 (BAB78072.1) | Cell division |
| FtsI | Sll1833 (WP_010871772.1) | Synpcc7942_0482 (WP_011243349.1) | Alr0718 (WP_010994893.1) g | Cell division |
| FtsK/SpoIIIE | Sll0284 (WP_010873902.1) h | Synpcc7942_0981 (WP_011242875.1) h | Alr3799 (WP_010997940.1) h | Cell division |
| --- | --- | All7666 (WP_010993994.1) i | ||
| FtsN | Slr0702 (WP_010873961.1) | N/A | N/A | Cell division |
| FtsQ | Sll1632 (WP_010872127.1) | Synpcc7942_2377 (WP_011378434.1) | Alr3857 (WP_010997998.1) | Cell division |
| FtsW | Slr1267 (WP_010872891.1) | Synpcc7942_0324 (WP_011377535.1) | All0154 (WP_010994331.1) | Cell division |
| FtsX | N/A | N/A | All1757 (WP_010995925.1) | Cell division |
| CyDiv | N/A | N/A | All2320 (WP_010996476.1) | Cell division |
| SepI | N/A | N/A | Alr3364 (BAB75063.1) j | Cell–cell contact |
| RodA | N/A | Synpcc7942_1104 (WP_011377865.1) | Alr0653 (WP_010994829.1) | Cell elongation |
| MreB | N/A | Synpcc7942_0300 (WP_011243524.1) | All0087 (BAB77611.1) | Cell elongation |
| MreC | N/A | Synpcc7942_0299 (WP_011243525.1) | All0086 (WP_010994263.1) | Cell elongation |
| MreD | N/A | Synpcc7942_0298 (ABB56330.1) | All0085 (BAB77609.1) | Cell elongation |
| BolA | Ssr3122 (WP_010871705.1) | Synpcc7942_1146 (ABB57176.1) | Asr0798 (WP_010994972.1) | Cell elongation |
| CikA | Slr1969 (WP_010872820.1) | Synpcc7942_0644 (WP_011243194.1) | All1688 (WP_010995857.1) | Circadian rhythm |
| PBP1 | Sll0002 (WP_010873436.1) | Synpcc7942_2000 (WP_011378270.1) | Alr5101 (WP_010999227.1) | Cell wall synthesis |
| PBP2 | Slr1710 (WP_010871874.1) | Synpcc7942_0785 (ABB56817.1) | Alr4579 (WP_010998711.1) | Cell wall synthesis |
| PBP3 | Sll1434 (WP_010872930.1) | Synpcc7942_2571 (WP_011243849.1) | All2981 (WP_010997132.1) | Cell wall synthesis |
| PBP4 | Sll1833 (WP_010871772.1) | Synpcc7942_0580 (WP_011377631.1) | Alr5326 (BAB77025.1) | Cell wall synthesis |
| PBP5 | Slr0646 (WP_010873596.1) | Synpcc7942_1934 (ABB57964.1) | Alr5324 (WP_010999448.1) | Cell wall synthesis |
| PBP6 | Sll1167 (WP_010872913.1) | Synpcc7942_0482 (WP_011243349.1) | All2981 (WP_010997132.1) | Cell wall synthesis |
| PBP7 | Slr1924 (WP_010873199.1) | N/A | Alr5045 (WP_010999171.1) | Cell wall synthesis |
| PBP8 | Slr0804 (WP_010872730.1) | N/A | Alr0718 (WP_010994893.1) g | Cell wall synthesis |
| PBP9 | N/A | N/A | Alr0153 (WP_010994330.1) | Cell wall synthesis |
| PBP10 | N/A | N/A | Alr1666 (WP_010995835.1) | Cell wall synthesis |
| PBP11 | N/A | N/A | Alr0054 (WP_010994231.1) | Cell wall synthesis |
| PBP12 | N/A | N/A | All2656 (WP_010996812.1) | Cell wall synthesis |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Springstein, B.L.; Nürnberg, D.J.; Weiss, G.L.; Pilhofer, M.; Stucken, K. Structural Determinants and Their Role in Cyanobacterial Morphogenesis. Life 2020, 10, 355. https://doi.org/10.3390/life10120355
Springstein BL, Nürnberg DJ, Weiss GL, Pilhofer M, Stucken K. Structural Determinants and Their Role in Cyanobacterial Morphogenesis. Life. 2020; 10(12):355. https://doi.org/10.3390/life10120355
Chicago/Turabian StyleSpringstein, Benjamin L., Dennis J. Nürnberg, Gregor L. Weiss, Martin Pilhofer, and Karina Stucken. 2020. "Structural Determinants and Their Role in Cyanobacterial Morphogenesis" Life 10, no. 12: 355. https://doi.org/10.3390/life10120355
APA StyleSpringstein, B. L., Nürnberg, D. J., Weiss, G. L., Pilhofer, M., & Stucken, K. (2020). Structural Determinants and Their Role in Cyanobacterial Morphogenesis. Life, 10(12), 355. https://doi.org/10.3390/life10120355





