Characterization of Resistance in Gram-Negative Urinary Isolates Using Existing and Novel Indicators of Clinical Relevance: A 10-Year Data Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design, Data Collection
2.2. Identification of Isolates during the Study Period
2.3. Susceptibility Testing of Relevant Isolates
2.4. Classification of Isolates into Resistance Categories
2.5. Statistical Analyses
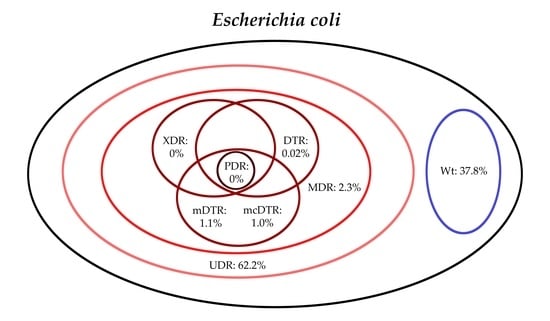
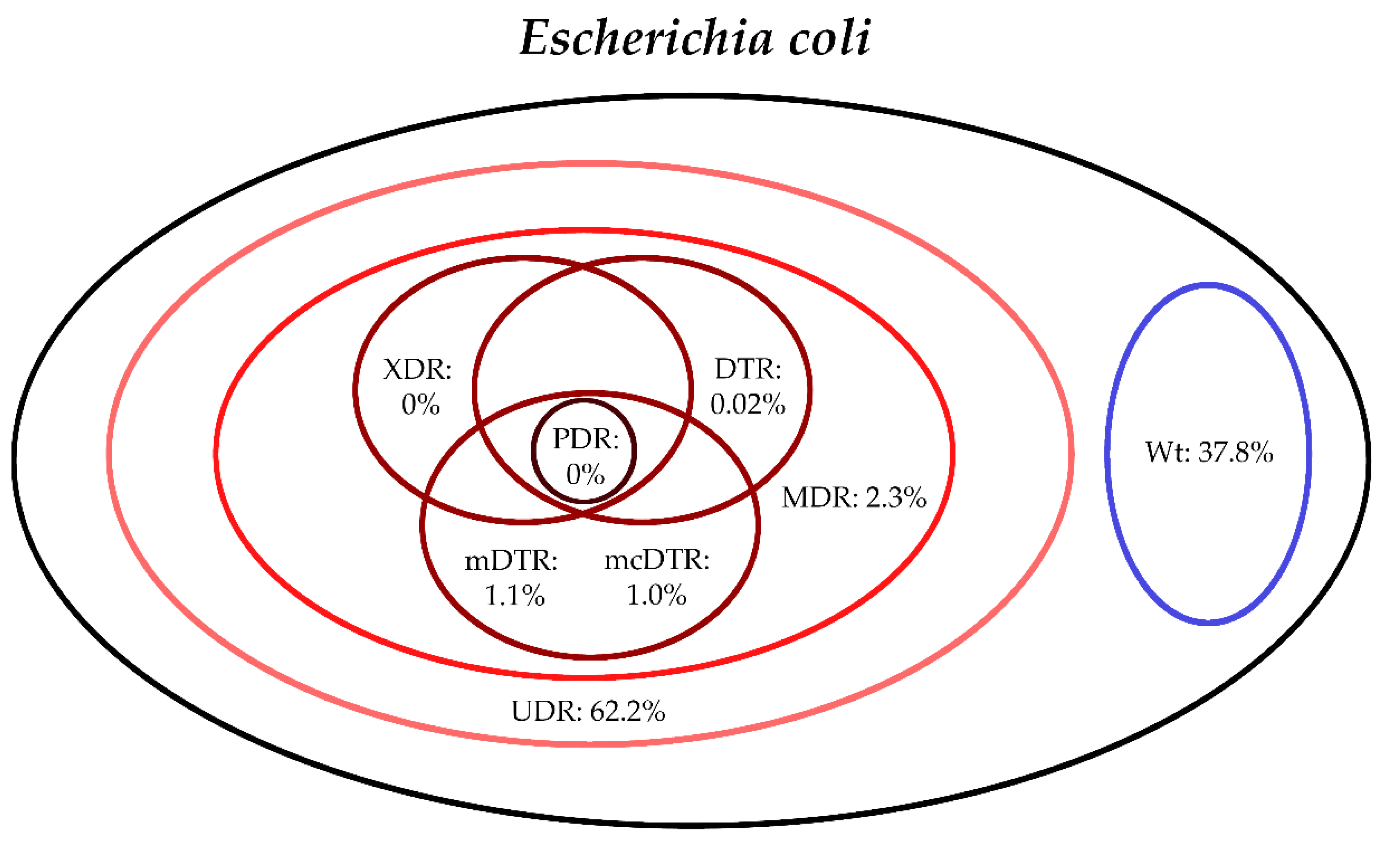
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Shallcross, L.J.; Howard, S.J.; Fowler, T.; Davies, S.C. Tackling the threat of antimicrobial resistance: From policy to sustainable action. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140082. [Google Scholar] [CrossRef] [PubMed]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.M.; Wertheim, H.F.L.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef] [Green Version]
- World Health Organisation. Antimicrobial Resistance: Global Report on Surveillance; WHO: Geneva, Switzerland, 2014; pp. 1–256. Available online: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf?ua=1 (accessed on 12 January 2020).
- Dyar, O.J.; Huttner, B.; Schouten, J.; Pulcini, C. What is antimicrobial stewardship? Clin. Microbiol. Infect. 2017, 23, 793–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gajdács, M.; Paulik, E.; Szabó, A. Knowledge, attitude and practice of community pharmacists regarding antibiotic use and infectious diseases: A cross-sectional survey in Hungary (KAPPhA-HU). Antibiotics 2020, 9, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gajdács, M. The concept of an ideal antibiotic: Implications for drug design. Molecules 2019, 24, 892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cassini, A.; Högberg, D.L.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.N.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Burden of the AMR Collaborative Group. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 55–56. [Google Scholar] [CrossRef] [Green Version]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016, 4, 481–511. [Google Scholar] [CrossRef] [Green Version]
- Darrow, J.J.; Kesselheim, A.S. Drug development and FDA approval, 1938–2013. N. Engl. J. Med. 2014, 370, e39. [Google Scholar] [CrossRef]
- Lyddiard, D.; Jones, G.L.; Greatrex, B.W. Keeping it simple: Lessons from the golden era of antibiotic discovery. FEMS Microbiol. Lett. 2016, 363, 84. [Google Scholar] [CrossRef] [Green Version]
- Infectious Diseases Society of America. The 10 × 20 Initiative: Pursuing a global commitment to develop 10 new antibacterial drugs by 2020. Clin. Infect. Dis. 2010, 50, 1081–1083. [Google Scholar] [CrossRef] [Green Version]
- Hughes, D.; Karlén, A. Discovery and preclinical development of new antibiotics. Ups. J. Med. Sci. 2014, 119, 162–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rex, J.H. ND4BB: Addressing the antimicrobial resistance crisis. Nat. Rev. Microbiol. 2014, 12, 231–232. [Google Scholar] [CrossRef]
- O’Neill, J. Antimicrobial Resistance: Tackling a Crisis for the Health and Wealth of Nations. Available online: https://amr-review.org/sites/default/files/AMRReviewPaper-Tacklingacrisisforthehealthandwealthofnations_1.pdf (accessed on 12 January 2020).
- Rice, L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: No ESKAPE. J. Infect. Dis. 2009, 197, 1079–1081. [Google Scholar] [CrossRef]
- Gajdács, M.; Albericio, F. Antibiotic resistance: From the bench to patients. Antibiotics 2019, 8, e129. [Google Scholar] [CrossRef] [Green Version]
- Santajit, S.; Indrawattana, N. Mechanisms of antimicrobial resistance in ESKAPE pathogens. Biomed. Res. Int. 2016, 2016, 2475067. [Google Scholar] [CrossRef] [Green Version]
- Medina, E.; Pieper, D.H. Tackling Threats and future problems of multidrug-resistant bacteria. Curr. Top. Microbiol. Immunol. 2016, 398, 3–33. [Google Scholar] [PubMed]
- Cui, X.; Zhang, H.; Du, H. Carbapenemases in enterobacteriaceae: Detection and antimicrobial therapy. Front Microbiol. 2019, 10, 1823. [Google Scholar] [CrossRef] [PubMed]
- Gajdács, M.; Urbán, E. Comparative epidemiology and resistance trends of proteae in urinary tract infections of inpatients and outpatients: A 10-year retrospective study. Antibiotics 2019, 8, 91. [Google Scholar] [CrossRef] [Green Version]
- Ha, D.R.; Haste, N.M.; Gluckstein, D.P. The role of antibiotic stewardship in promoting appropriate antibiotic use. Am. J. Lifestyle Med. 2017, 13, 376–383. [Google Scholar] [CrossRef] [PubMed]
- Fridkin, S.K.; Cleveland, A.A.; See, I.; Lynfield, R. Emerging infections program as surveillance for antimicrobial drug resistance. Emerg. Infect. Dis. 2015, 21, 1578–1581. [Google Scholar] [CrossRef] [Green Version]
- Magiorakos, A.P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Paterson, D.L. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadri, S.S.; Adjemian, J.; Lai, Y.L.; Spaulding, A.B.; Ricotta, E.; Prevots, D.R.; Powers, J.H., III. Difficult-to-treat resistance in gram-negative bacteremia at 173 US hospitals: Retrospective cohort analysis of prevalence, predictors, and outcome of resistance to all first-line agents. Clin. Infect. Dis. 2018, 67, 1803–1814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, M.; Daikos, G.L.; Durante-Mangoni, E.; Yahav, D.; Carmeli, Y.; Benattar, Y.D.; Zusman, O. Colistin alone versus colistin plus meropenem for treatment of severe infections caused by carbapenem-resistant Gram-negative bacteria: An open-label, randomised controlled trial. Lancet Infect. Dis. 2018, 18, 391–400. [Google Scholar] [CrossRef]
- Zak-Doron, Y.; Dishon Benattar, Y.; Pfeffer, I.; Daikos, G.L.; Skiada, A.; Antoniadou, A.; Yahav, D. The association between empirical antibiotic treatment and mortality in severe infections caused by carbapenem-resistant gram-negative bacteria: A prospective study. Clin. Infect. Dis. 2018, 67, 1815–1823. [Google Scholar] [CrossRef]
- Burnham, J.P.; Lane, M.A.; Kollef, M.H. Impact of sepsis classification and multi-drug-resistance status on outcome among patients treated with appropriate ther-apy. Crit. Care Med. 2015, 43, 1580–1586. [Google Scholar] [CrossRef]
- Al-Dulaimi, M.M.K.; Mutalib, S.A.; Ghani, M.A.; Zaini, N.A.M.; Ariffin, A.A. Multiple antibiotic resistance (MAR), plasmid profiles, and DNA polymorphisms among vibrio vulnificus isolates. Antibiotics 2019, 8, e68. [Google Scholar] [CrossRef] [Green Version]
- Gajdács, M. Epidemiology and resistance levels of enterobacteriaceaeisolates from urinary tract infections expressed as multiple antibiotic resistance (MAR) indices. J. Pharm. Res. Int. 2019, 29, 1–7. [Google Scholar] [CrossRef] [Green Version]
- McDonnell, A.; Rex, J.H.; Goosens, H.; Bonten, M.; Fowler, V.G.; Dane, A. Efficient Delivery of Investigational Antibacterial Agents via Sustainable Clinical Trial Networks. Clin. Infect. Dis. 2016, 63, S57–S59. [Google Scholar] [CrossRef]
- Rex, J.H.; Talbot, G.H.; Goldberger, M.J.; Eisenstein, B.I.; Echols, R.M.; Tomayko, J.F.; Dudley, M.N.; Dane, A. Progress in the fight against multidrug-resistant bacteria 2005–2016: Modern noninferiority trial designs enable antibiotic development in advance of epidemic bacterial resistance. Clin. Infect. Dis. 2017, 65, 141–146. [Google Scholar] [CrossRef] [Green Version]
- Hooton, T.M.; Bradley, S.F.; Cardenas, D.D.; Colgan, R.; Geerlings, S.E.; Rice, J.C.; Saint, S.; Schaeffer, A.J.; Tambayh, P.A.; Tenke, P.; et al. Diagnosis, prevention, and treatment of catheter-associated urinary tract infection in adults: 2009 international clinical practice guidelines from the infectious diseases society of America. Clin. Infect. Dis. 2010, 50, 625–663. [Google Scholar] [CrossRef]
- Wiedemann, B.; Heisig, A.; Heisig, P. Uncomplicated urinary tract infections and antibiotic resistance-epidemiological and mechanistic aspects. Antibiotics 2014, 3, 341–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simmering, J.E.; Tang, F.; Cavanaugh, J.E.; Polgreen, L.A.; Polgreen, P.M. The Increase in hospitalizations for urinary tract infections and the associated costs in the United States, 1998–2011. Open Forum. Infect. Dis. 2017, 4, ofw281. [Google Scholar] [CrossRef] [PubMed]
- Calzi, A.; Grignolo, S.; Caviglia, I.; Calevo, M.G.; Losurdo, G.; Piaggio, G.; Bandettini, R.; Castagnola, E. Resistance to oral antibiotics in 4569 Gram-negative rods isolated from urinary tract infection in children. Eur. J. Pediatr. 2016, 175, 1219–1225. [Google Scholar] [CrossRef] [PubMed]
- Stefaniuk, E.; Suchocka, U.; Bosacka, K.; Hryniewicz, W. Etiology and antibiotic susceptibility of bacterial pathogens responsible for community-acquired urinary tract infections in Poland. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1363–1369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbo, L.M.; Hooton, T.M. Antimicrobial stewardship and urinary tract infections. Antibiotics 2014, 3, 174–192. [Google Scholar] [CrossRef]
- Gajdács, M.; Urbán, E. Resistance trends and epidemiology of citrobacter-enterobacter-serratia in urinary tract infections of inpatients and outpatients (RECESUTI): A 10-year survey. Medicina 2019, 55, 285. [Google Scholar] [CrossRef] [Green Version]
- Gajdács, M.; Ábrók, M.; Lázár, A.; Burián, K. Comparative Epidemiology and resistance trends of common urinary pathogens in a tertiary-care hospital: A 10-year surveillance study. Medicina 2019, 55, 356. [Google Scholar] [CrossRef] [Green Version]
- Gajdács, M.; Burián, K.; Terhes, G. Resistance levels and epidemiology of non-fermenting gram-negative bacteria in urinary tract infections of inpatients and outpatients (RENFUTI): A 10-year epidemiological snapshot. Antibiotics 2019, 8, 143. [Google Scholar] [CrossRef] [Green Version]
- Gajdács, M.; Dóczi, I.; Ábrók, M.; Lázár, A.; Burián, K. Epidemiology of candiduria and Candida urinary tract infections in inpatients and outpatients: Results from a 10-year retrospective survey. Cent. Eur. J. Urol. 2019, 72, 209–214. [Google Scholar]
- Hospital Bed Count and Patient Turnover Report 2017. National Health Insurance Fund of Hungary. Available online: http://www.neak.gov.hu/felso_menu/szakmai_oldalak/publikus_forgalmi_adatok/gyogyito_megelozo_forgalmi_adat/fekvobeteg_szakellatas/korhazi_agyszam.html (accessed on 12 January 2020).
- Leclercq, R.; Cantón, R.; Brown, D.F.J.; Giske, C.G.; Heisig, P.; MacGowan, A.P.; Mouton, J.W.; Nordmann, P.; Rodloff, A.C.; Rossolini, G.M.; et al. EUCAST expert rules in antimicrobial susceptibility testing. Clin. Microbiol. Infect. 2013, 19, 141–160. [Google Scholar] [CrossRef] [Green Version]
- Issakhanian, L.; Behzadi, P. Antimicrobial agents and urinary tract infections. Curr. Pharm. Des. 2019, 25, 1409–1423. [Google Scholar] [CrossRef] [PubMed]
- Jahandeh, N.; Ranjbar, R.; Behzadi, P.; Behzadi, E. Uropathogenic Escherichia coli virulence genes: Invaluable approaches for designing DNA microarray probes. Cent. Eur. J. Urol. 2015, 68, 452–458. [Google Scholar]
- Gajdács, M.; Szabó, A. Physicians’ opinions towards antibiotic use and resistance in the southeastern region of Hungary. Orv. Hetil. 2019. accepted. [Google Scholar]
- Poole, K. Pseudomonas Aeruginosa: Resistance to the max. Front. Microbiol. 2011, 2, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iredell, J.; Brown, J.; Tagg, K. Antibiotic resistance in enterobacteriaceae: Mechanisms and clinical implications. BMJ 2016, 352, h6420. [Google Scholar] [CrossRef] [PubMed]
- Björn, B. Acquired resistance to colistin via chromosomal and plasmid-mediated mechanisms in klebsiella pneumoniae. Infect. Microb. Dis. 2019, 1, 10–19. [Google Scholar]
- Levison, M.E.; Levison, J.H. Pharmacokinetics and pharmacodynamics of antibacterial agents. Infect. Dis. Clin. N. Am. 2009, 23, 791–799. [Google Scholar] [CrossRef] [Green Version]
- Nation, R.L.; Garonzik, S.M.; Thamlikitkul, V.; Giamarellos-Bourboulis, E.J.; Forrest, A.; Paterson, D.L.; Li, J.; Silveira, F.P. Dosing guidance for intravenous colistin in critically Ill patients. Clin. Infect. Dis. 2017, 64, 565–571. [Google Scholar] [CrossRef]
- Corcione, S.; Lupia, T.; Maraolo, A.E.; Mornese Pinna, S.; Gentile, S.; De Rosa, F.G. Carbapenem-sparing strategy: Carbapenemase, treatment, and stewardship. Curr. Opin. Infect. Dis. 2019, 32, 663–673. [Google Scholar] [CrossRef]
- Gajdács, M.; Ábrók, M.; Lázár, A.; Burián, K. Susceptibility patterns of extended-spectrum beta-lactamase-producing (ESBL) urinary pathogens: Single-center experience. Gyógyszerészet 2019, 63, 405–411. [Google Scholar]
- Gajdács, M.; Ábrók, M.; Lázár, A.; Burián, K. Microbiology of urine samples obtained through suprapubic bladder aspiration: A 10-year epidemiological snapshot. Dev. Health Sci. 2019, 2, 76–78. [Google Scholar] [CrossRef]
- European Antimicrobial Resistance Surveillance Network (EARS-Net). Available online: https://ecdc.europa.eu/en/about-us/partnerships-and-networks/disease-and-laboratory-networks/ears-net (accessed on 12 January 2020).
- CDC Antibiotic/Antimicrobial Resistance (AR/AMR). Available online: https://www.cdc.gov/drugresistance/biggest_threats.html (accessed on 12 January 2020).
- Abat, C.; Rolain, J.M.; Dubourg, G.; Fournier, P.E.; Chaudet, H.; Raoult, D. Evaluating the clinical burden and mortality attributable to antibiotic resistance: The disparity of empirical data and simple model estimations. Clin. Infect. Dis. 2017, 65, S58–S63. [Google Scholar] [CrossRef] [PubMed]
- Abat, C.; Fournier, P.E.; Jimeno, M.T.; Rolain, J.M.; Raoult, D. Extremely and pandrug-resistant bacteria extra-deaths: Myth or reality? Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1687–1697. [Google Scholar] [CrossRef] [PubMed]
- Falagas, M.E.; Rafailidis, P.I.; Matthaiou, D.K.; Virtzili, S.; Nikita, D.; Michalopoulos, A. Pandrug-resistant Klebsiella pneumoniae, Pseudomonas aeruginosa and Acinetobacter baumannii infections: Characteristics and outcome in a series of 28 patients. Int. J. Antimicrob. Agents 2008, 32, 450–454. [Google Scholar] [CrossRef]
- Elemam, A.; Rahimian, J.; Mandell, W. Infection with panresistant Klebsiella pneumoniae: A report of 2 cases and a brief review of the literature. Clin. Infect. Dis. 2009, 49, 271–274. [Google Scholar] [CrossRef] [Green Version]











| Bacterial Isolates | Outpatients | Inpatients |
|---|---|---|
| Citrobacter-Enterobacter-Serratia | 2.6% (n = 554) | 3.0% (n = 578) |
| Acinetobacter spp. | 0.7% (n = 143) | 0.7% (n = 133) |
| Pseudomonas aeruginosa | 2.8% (n = 588) | 5.7% (n = 1096) |
| Proteus-Providencia-Morganella | 5.0% (n = 1058) | 7.2% (n = 1392) |
| Klebsiella spp. | 8.9% (n = 1895) | 13.4% (n = 2592) |
| Gram-positive cocci | 20.7% | 20.7% |
| Escherichia coli | 56.8% (n = 12002) | 42.3% (n = 8173) |
| Candida spp. | 0.4% | 6.0% |
| Other | 2.1% | 1.0% |
| Bacterial Group | Outpatients | Inpatients | Overall |
|---|---|---|---|
| Citrobacter-Enterobacter-Serratia | 37.9% | 38.2% | 38.1% |
| Acinetobacter spp. | 27.1% | 44.5% | 35.7% |
| P. aeruginosa | 18.9% | 21.3% | 20.2% |
| Proteus-Providencia-Morganella | 37.6% | 42.6% | 40.3% |
| Klebsiella spp. | 30.3% | 29.5% | 29.9% |
| E. coli | 25.3% | 27.7% | 26.1% |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gajdács, M.; Bátori, Z.; Ábrók, M.; Lázár, A.; Burián, K. Characterization of Resistance in Gram-Negative Urinary Isolates Using Existing and Novel Indicators of Clinical Relevance: A 10-Year Data Analysis. Life 2020, 10, 16. https://doi.org/10.3390/life10020016
Gajdács M, Bátori Z, Ábrók M, Lázár A, Burián K. Characterization of Resistance in Gram-Negative Urinary Isolates Using Existing and Novel Indicators of Clinical Relevance: A 10-Year Data Analysis. Life. 2020; 10(2):16. https://doi.org/10.3390/life10020016
Chicago/Turabian StyleGajdács, Márió, Zoltán Bátori, Marianna Ábrók, Andrea Lázár, and Katalin Burián. 2020. "Characterization of Resistance in Gram-Negative Urinary Isolates Using Existing and Novel Indicators of Clinical Relevance: A 10-Year Data Analysis" Life 10, no. 2: 16. https://doi.org/10.3390/life10020016
APA StyleGajdács, M., Bátori, Z., Ábrók, M., Lázár, A., & Burián, K. (2020). Characterization of Resistance in Gram-Negative Urinary Isolates Using Existing and Novel Indicators of Clinical Relevance: A 10-Year Data Analysis. Life, 10(2), 16. https://doi.org/10.3390/life10020016