Representational Difference Analysis of Transcripts Involved in Jervine Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Jervine Quantification
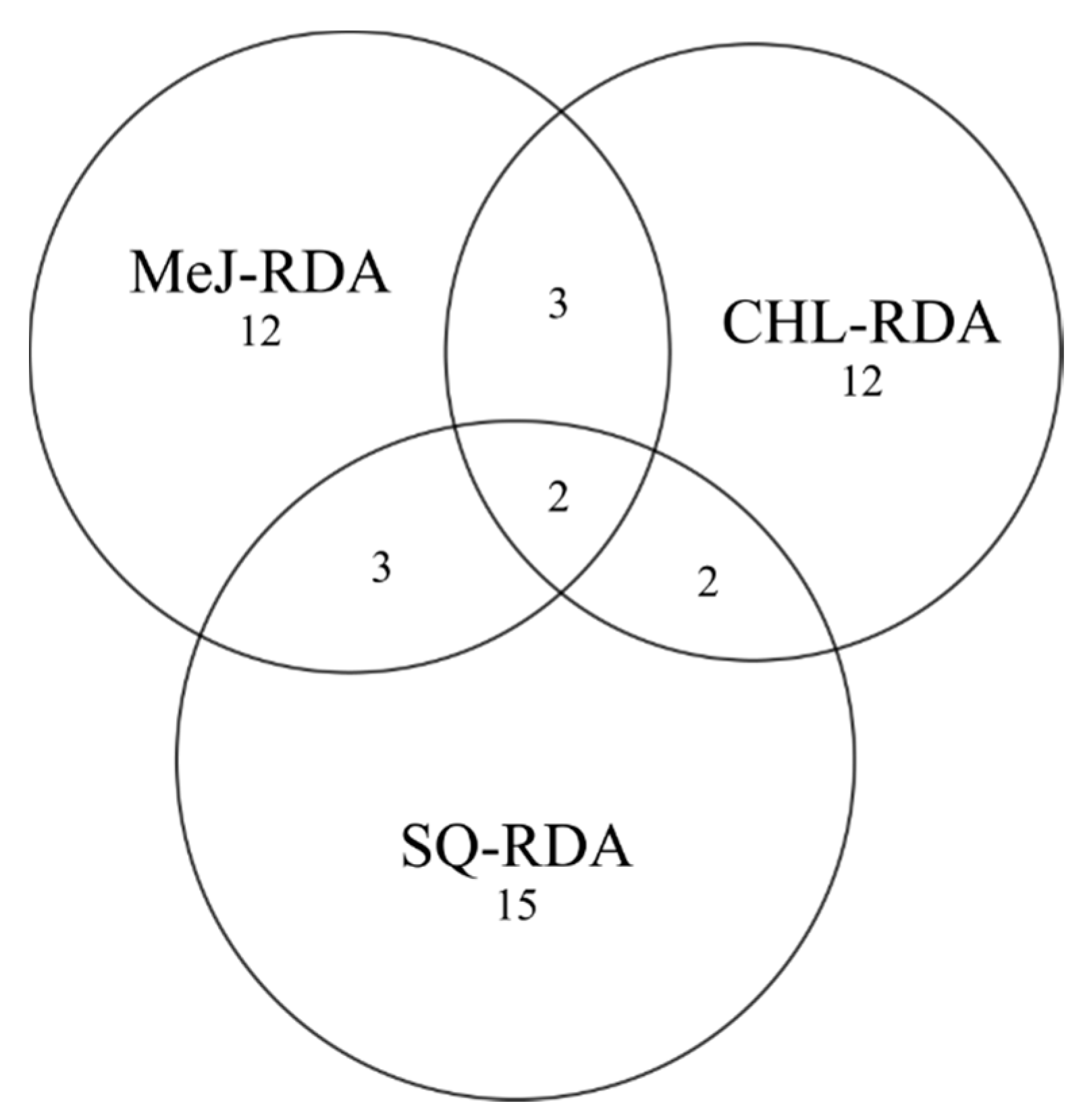
2.3. Representational Difference Analysis and Library Construction
2.4. De Novo Assembly and Functional Annotation
3. Results and Discussion
3.1. Jervine Concentration
3.2. Sequence De Novo Assembly
3.3. Functional Annotation
3.4. Identification of Genes Involved in Terpenoid Backbone and Cholesterol Biosynthesis
3.5. Identification of the Genes in Later Stages of Jervine Biosynthesis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chandler, C.M.; McDougal, O.M. Medicinal history of North American Veratrum. Phytochem. Rev. 2014, 13, 671–694. [Google Scholar] [CrossRef] [PubMed]
- Heretsch, P.; Giannis, A. The Veratrum and Solanum alkaloids. Alkaloids Chem. Biol. 2015, 74, 201–232. [Google Scholar] [PubMed]
- Schaffner, U.; Kleijn, D.; Brown, V.; Müller-Schärer, H. Veratrum album L. in montane grasslands: A model system for implementing biological control in land management practices of high biodiversity habitats. Biocontrol News Inf. 2001, 22, 19–28. [Google Scholar]
- Zomlefer, W.B.; Whitten, W.M.; Williams, N.H.; Judd, W.S. An overview of Veratrum s.l. (Liliales: Melanthiaceae) and an infrageneric phylogeny based on ITS sequence data. Syst. Bot. 2003, 28, 250–269. [Google Scholar]
- Treier, U.A.; Müller-Schärer, H. Differential effects of historical migration, glaciations and human impact on the genetic structure and diversity of the mountain pasture weed Veratrum album L. J. Biogeogr. 2011, 38, 1776–1791. [Google Scholar] [CrossRef]
- Szeliga, M.; Ciura, J.; Tyrka, M. Genetic diversity of three European Veratrum species revealed by amplified fragment length polymorphism. Biodiv. Res. Conserv. 2017, 47, 1–8. [Google Scholar] [CrossRef]
- Wu, Z.Y.; Raven, P.H. Flora of China. Vol. 24 (Flagellariaceae through Marantaceae); Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MS, USA, 2000; pp. 73–263. [Google Scholar]
- Kaźmierczakowa, R.; Bloch-Orłowska, J.; Celka, Z.; Cwener, A.; Dajdok, Z.; Michalska-Hejduk, D.; Pawlikowski, P.; Szczęśniak, E.; Ziarnek, K. Polish Red List of Pteridophytes and Flowering Plants; Instytut Ochrony Przyrody Polskiej Akademii Nauk: Kraków, Poland, 2016; p. 44. [Google Scholar]
- Barroso Mdo, S. The hellebore, the plant beloved by the Greeks: The reasons behind a myth. Vesalius 2015, 21, 30–37. [Google Scholar]
- Suladze, T.S.; Vachnadze, V.Y.; Tsakadze, D.M.; Gedevanishvili, M.D.; Tsutsunava, L.E.; Malazoniya, N.A. Alkaloid accumulation dynamics in Veratrum lobelianum growing in Georgia and biological activity of Jervine. Chem. Nat. Compd. 2006, 42, 71–74. [Google Scholar] [CrossRef]
- Cholakova, M.; Bratanov, M.; Christov, V.; Kostova, N.; Gantcheva, M.; Nikolova, E. The veratrum alkaloid, veratroylzygadenine, suppresses contact dermatitis in mice. J. Med. Plants Res. 2009, 3, 1109–1112. [Google Scholar]
- Gaillard, Y.; Pepin, G. LC-EI-MS determination of veratridine and cevadine in two fatal cases of Veratrum album poisoning. J. Anal. Toxicol. 2001, 25, 481–485. [Google Scholar] [CrossRef]
- Wang, Z.Z.; Zhao, W.J.; Zhang, X.S.; Tian, X.F.; Wang, Y.Z.; Zhang, F.; Yuan, J.C.; Han, G.Z.; Xin, K.X.; Yao, J.H. Protection of Veratrum nigrum L. var. ussuriense Nakai alkaloids against Ischemia-Reperfusion injury of the rat liver. World J. Gastroenterol. 2007, 13, 564–571. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, W.; Liu, Y. Hypotensive effect and toxicology of total alkaloids and veratramine from roots and rhizomes of Veratrum nigrum L. in spontaneously hypertensive rats. Pharmazie 2008, 63, 606–610. [Google Scholar] [PubMed]
- Li, H.J.; Jiang, Y.; Li, P. Chemistry, bioactivity and geographical diversity of steroidal alkaloids from the Liliaceae family. Nat. Prod. Rep. 2006, 23, 735–752. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, A.; Serly, J.; Christov, V.; Stamboliyska, B.; Molnar, J. Alkaloids derived from genus Veratrum and Peganum of Mongolian origin as multidrug resistance inhibitors of cancer cells. Fitoterapia 2011, 82, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Dumlu, F.A.; Aydin, T.; Odabasoglu, F.; Berktas, O.A.; Kutlu, Z.; Erol, H.E.; Halici, M.B.; Cadirci, E.; Cakir, A. Anti-Inflammatory and antioxidant properties of jervine, a sterodial alkaloid from rhizomes of Veratrum album. Phytomedicine 2019, 55, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Zhao, Y.L.; Long, C.B.; Zhu, P.F.; Liu, Y.P.; Luo, X.D. Seven new Veratramine-Type alkaloids with potent analgesic effect from Veratrum taliense. J. Ethnopharmacol. 2019, 244, 112137. [Google Scholar] [CrossRef]
- Kang, C.; Han, J.H.; Oh, J.; Kulkarni, R.; Zhou, W.; Ferreira, D.; Jang, T.S.; Myung, C.S.; Na, M. Steroidal alkaloids from Veratrum nigrum enhance glucose uptake in skeletal muscle cells. J. Nat. Prod. 2015, 78, 803–810. [Google Scholar] [CrossRef]
- Incardona, J.P.; Eaton, S. Cholesterol in signal transduction. Curr. Opin. Cell Biol. 2000, 12, 193–203. [Google Scholar] [CrossRef]
- Chen, J.K.; Taipale, J.; Cooper, M.K.; Beachy, P.A. Inhibition of hedgehog signaling by direct binding of cyclopamine to smoothened. Genes Dev. 2002, 16, 2743–2748. [Google Scholar] [CrossRef]
- Shang, Y.; Du, Q.; Liu, S.; Staadler, M.; Wang, S.; Wang, D. Antitumor activity of isosteroidal alkaloids from the plants in the genus Veratrum and Fritillaria. Curr. Protein Pept. Sci. 2018, 19, 302–310. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, Y.; Liang, Q.; Ma, Z.; Xiao, C.; Tan, H.; Gao, Y. The correlation between chemical composition, as determined by UPLC-TOF-MS, and Acute toxicity of Veratrum nigrum L. and Radix paeoniae alba. Evid. Based Complement. Altern. Med. 2014, 2014, 892797. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; Jia, W.; Chen, J.; Song, S.; Wang, J.H.; Yang, Y.H. Steroidal alkaloids from the roots and rhizomes of Vertrum nigrum L. Helv. Chim. Acta 2007, 90, 1038–1042. [Google Scholar] [CrossRef]
- Christov, V.; Mikhova, B.; Ivanova, A.; Serly, J.; Molnar, J.; Selenge, D.; Solongo, A.; Kostova, N.; Gerelt-Od, Y.; Dimitrov, D. Steroidal alkaloids of Veratrum lobelianum Bernh. and Veratrum nigrum L. Z. Nat. C J. Biosci. 2010, 65, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; Wang, J.H.; Wang, R.; Zeng, Y.M.; Liu, C.D.; Li, X. A study on the chemical constituents of Veratrum nigrum L. processed by rice vinegar. J. Asian Nat. Prod. Res. 2008, 10, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Khanfar, M.A.; El Sayed, K.A. The Veratrum alkaloids jervine, veratramine, and their analogues as prostate cancer migration and proliferation inhibitors: Biological evaluation and pharmacophore modeling. Med. Chem. Res. 2013, 22, 4775–4786. [Google Scholar] [CrossRef]
- Tang, J.; Li, H.L.; Shen, Y.H.; Jin, H.Z.; Yan, S.K.; Liu, R.H.; Zhang, W.D. Antitumor activity of extracts and compounds from the rhizomes of Veratrum dahuricum. Phytother. Res. 2008, 22, 1093–1096. [Google Scholar] [CrossRef]
- Thayer, S.P.; Di Magliano, M.P.; Heiser, P.W.; Nielsen, C.M.; Roberts, D.J.; Lauwers, G.Y.; Qi, Y.P.; Gysin, S.; Del Castillo, C.F.; Yajnik, V.; et al. Hedgehog is an early and late mediator of pancreatic cancer tumorigenesis. Nature 2003, 425, 851–856. [Google Scholar] [CrossRef]
- Heretsch, P.; Tzagkaroulaki, L.; Giannis, A. Cyclopamine and Hedgehog signaling: Chemistry, biology, medical perspectives. Angew. Chem. Int. Ed. Engl. 2010, 49, 3418–3427. [Google Scholar] [CrossRef]
- Masamune, T.; Takasugi, M.; Murai, A.; Kobayashi, K. The synthesis of Jervine and related alkaloids. J. Am. Chem. Soc. 1967, 89, 17. [Google Scholar] [CrossRef]
- Kutney, J.P. Synthetic studies in the Veratrum Alkaloid Series. The Total Synthesis of Verarine, Veratramine, Jervine, Veratrobasine, and Verticine. Bioorg. Chem. 1977, 6, 371–391. [Google Scholar] [CrossRef]
- Tang, J.; Li, H.L.; Shen, Y.H.; Jin, H.Z.; Yan, S.K.; Liu, R.H.; Zhang, W.D. Simultaneous determination of six steroidal alkaloids of Veratrum dahuricum by HPLC–ELSD and HPLC–MSn. Chromatographia 2008, 67, 15–21. [Google Scholar] [CrossRef]
- Suzuki, M.; Muranaka, T. Molecular genetics of plant sterol backbone synthesis. Lipids 2007, 42, 47–54. [Google Scholar] [CrossRef]
- Sun, C.; Sun, Y.; Song, J.; Li, C.; Li, X.; Zhang, X.; Li, Y.; Hu, S.; Luo, H.; Zhu, Y.; et al. Discovery of genes related to steroidal alkaloid biosynthesis in Fritillaria cirrhosa by generating and mining a dataset of expressed sequence tags (ESTs). J. Med. Plant Res. 2011, 5, 5307–5314. [Google Scholar]
- Augustin, M.M.; Ruzicka, D.R.; Shukla, A.K.; Augustin, J.M.; Starks, C.M.; O’Neil-Johnson, M.; McKain, M.R.; Evans, B.S.; Barrett, M.D.; Smithson, A.; et al. Elucidating steroid alkaloid biosynthesis in Veratrum californicum: Production of verazine in Sf9 cells. Plant J. 2015, 82, 991–1003. [Google Scholar] [CrossRef] [PubMed]
- Szeliga, M.; Ciura, J.; Grzesik, M.; Tyrka, M. Identification of candidate genes involved in steroidal alkaloids biosynthesis in Organ-Specific transcriptomes of Veratrum nigrum L. Gene 2019, 712, 143962. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Yu, Z.; Cai, Q.; Li, H.; Dong, Y.; Oksman-Caldentey, K.M.; Rischer, H. Agrobacterium-Mediated genetic transformation of the medicinal plant Veratrum dahuricum. Plants 2020, 9, 191. [Google Scholar] [CrossRef] [PubMed]
- Engler, C.; Kandzia, R.; Marillonnet, S.A. One pot, one step, precision cloning method with high throughput capability. PLoS ONE 2008, 3, e3647. [Google Scholar] [CrossRef] [PubMed]
- Facchini, P.J.; Bohlmann, J.; Covello, P.S.; De Luca, V.; Mahadevan, R.; Page, J.E.; Ro, D.K.; Sensen, C.W.; Storms, R.; Martin, V.J. Synthetic biosystems for the production of High-Value plant metabolites. Trends Biotechnol. 2012, 30, 127–131. [Google Scholar] [CrossRef]
- Singh, A.; Menéndez-Perdomo, I.M.; Facchini, P.J. Benzylisoquinoline alkaloid biosynthesis in opium poppy: An update. Phytochem. Rev. 2019, 18, 1457–1482. [Google Scholar] [CrossRef]
- Hubank, M.; Schatz, D.G. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 1994, 22, 5640–5648. [Google Scholar] [CrossRef]
- Sasheva, P.; Grossniklaus, U. Differentially methylated Region-Representational difference analysis (DMR-RDA): A powerful method to identify DMRs in uncharacterized genomes. Methods Mol. Biol. 2017, 1456, 113–125. [Google Scholar] [PubMed]
- Haslinger, C.; Sommergruber, W.; Voss, T.; Schreiber, M. Identification of Tumor-Specific genes. In Handbook of Immunohistochemistry and In Situ Hybridization of Human Carcinomas; Hayat, M.A., Ed.; Academic Press: Cambridge, MA, USA, 2006; Volume 4, pp. 3–21. [Google Scholar]
- Hubank, M.; Schatz, D.G. cDNA representational difference analysis: A sensitive and flexible method for identification of differentially expressed genes. Methods Enzymol. 1999, 303, 325–349. [Google Scholar] [PubMed]
- Ciura, J.; Szeliga, M.; Grzesik, M.; Tyrka, M. Next-Generation sequencing of representational difference analysis products for identification of genes involved in diosgenin biosynthesis in fenugreek (Trigonella Foenum-Graecum). Planta 2017, 245, 977–991. [Google Scholar] [CrossRef] [PubMed]
- Hagel, J.M.; Morris, J.S.; Lee, E.J.; Desgagné-Penix, I.; Bross, C.D.; Chang, L.; Chen, X.; Farrow, S.C.; Zhang, Y.; Soh, J.; et al. Transcriptome analysis of 20 taxonomically related benzylisoquinoline Alkaloid-Producing plants. BMC Plant Biol. 2015, 15, 227. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Huang, Y.; Niu, B.; Gao, Y.; Fu, L.; Li, W. CD-HIT Suite: A web server for clustering and comparing biological sequences. Bioinformatics 2010, 26, 680–682. [Google Scholar] [CrossRef]
- Wu, S.; Zhu, Z.; Fu, L.; Niu, B.; Li, W. WebMGA: A customizable web server for fast metagenomic sequence analysis. BMC Genom. 2011, 12, 444. [Google Scholar] [CrossRef]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35, W182–W185. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, W293–W297. [Google Scholar] [CrossRef]
- Tian, F.; Yang, D.C.; Meng, Y.Q.; Jin, J.P.; Gao, G. PlantRegMap: Charting functional regulatory maps in plants. Nucleic Acids Res. 2020, 48, D1104–D1113. [Google Scholar] [CrossRef] [PubMed]
- Thagun, C.; Imanishi, S.; Kudo, T.; Nakabayashi, R.; Ohyama, K.; Mori, T.; Kawamoto, K.; Nakamura, Y.; Katayama, M.; Nonaka, S.; et al. Jasmonate-Responsive ERF transcription factors regulate steroidal glycoalkaloid biosynthesis in tomato. Plant Cell Physiol. 2016, 57, 961–975. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Cheng, J.J.; Lin, C.Y.; Chen, Y.J.; Lu, M.K. Precursor-Feeding strategy for the production of solanine, solanidine and solasodine by a cell culture of Solanum lyratum. Process Biochem. 2007, 42, 899–903. [Google Scholar] [CrossRef]
- Llorca, C.M.; Potschin, M.; Zentgraf, U. bZIPs and WRKYs: Two large transcription factor families executing two different functional strategies. Front. Plant Sci. 2014, 5, 169. [Google Scholar] [CrossRef] [PubMed]
- Suttipanta, N.; Pattanaik, S.; Kulshrestha, M.; Patra, B.; Singh, S.K.; Yuan, L. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011, 157, 2081–2093. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.H.; Wang, J.W.; Wang, S.; Wang, J.Y.; Chen, X.Y. Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-δ-cadinene synthase-A. Plant Physiol. 2004, 135, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, P.; Zhang, M.; Fu, C.; Yu, L. Functional analysis of a WRKY transcription factor involved in transcriptional activation of the DBAT gene in Taxus chinensis. Plant Biol. (Stuttg.) 2013, 15, 19–26. [Google Scholar] [CrossRef]
- Ji, Y.; Xiao, J.; Shen, Y.; Ma, D.; Li, Z.; Pu, G.; Li, X.; Huang, L.; Liu, B.; Ye, H. Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol. 2014, 55, 1592–1604. [Google Scholar] [CrossRef]
- Spyropoulou, E.A.; Haring, M.A.; Schuurink, R.C. RNA Sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genom. 2014, 15, 402. [Google Scholar] [CrossRef]
- Mertens, J.; Pollier, J.; Vanden Bossche, R.; Lopez-Vidriero, I.; Franco-Zorrilla, J.M.; Goossens, A. The bHLH transcription factors TSAR1 and TSAR2 regulate triterpene saponin biosynthesis in Medicago truncatula. Plant Physiol. 2016, 170, 194–210. [Google Scholar] [CrossRef]
- Reddy, V.A.; Wang, Q.; Dhar, N.; Kumar, N.; Venkatesh, P.N.; Rajan, C.; Panicker, D.; Sridhar, V.; Mao, H.Z.; Sarojam, R. Spearmint R2R3-MYB transcription factor MsMYB negatively regulates monoterpene production and suppresses the expression of geranyl diphosphate synthase large subunit (MsGPPS.LSU). Plant Biotechnol. J. 2017, 15, 1105–1119. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, P.D.; Sonawane, P.D.; Pollier, J.; Vanden Bossche, R.; Dewangan, V.; Weithorn, E.; Tal, L.; Meir, S.; Rogachev, I.; Malitsky, S.; et al. GAME9 regulates the biosynthesis of steroidal alkaloids and upstream isoprenoids in the plant mevalonate pathway. Nat. Commun. 2016, 7, 10654. [Google Scholar] [CrossRef]
- Afrin, S.; Huang, J.J.; Luo, Z.Y. JA-Mediated transcriptional regulation of secondary metabolismin medicinal plants. Sci. Bull. 2015, 60, 1062–1072. [Google Scholar] [CrossRef]
- Schaller, H. New aspects of sterol biosynthesis in growth and development of higher plants. Plant Physiol. Biochem. 2004, 42, 465–476. [Google Scholar] [CrossRef]
- Sawai, S.; Ohyama, K.; Yasumoto, S.; Seki, H.; Sakuma, T.; Yamamoto, T.; Takebayashi, Y.; Kojima, M.; Sakakibara, H.; Aoki, T.; et al. Sterol side chain reductase 2 is a key enzyme in the biosynthesis of cholesterol, the common precursor of toxic steroidal glycoalkaloids in potato. Plant Cell 2014, 26, 3763–3774. [Google Scholar] [CrossRef]
- Diener, A.C.; Li, H.; Zhou, W.; Whoriskey, W.J.; Nes, W.D.; Fink, G.R. Sterol methyltransferase 1 controls the level of cholesterol in plants. Plant Cell 2000, 12, 853–870. [Google Scholar] [CrossRef] [PubMed]
- Nahar, N.; Westerberg, E.; Arif, U.; Huchelmann, A.; Olarte Guasca, A.; Beste, L.; Dalman, K.; Dutta, P.C.; Jonsson, L.; Sitbon, F. Transcript profiling of two potato cultivars during Glycoalkaloid-Inducing treatments shows differential expression of genes in sterol and glycoalkaloid metabolism. Sci. Rep. 2017, 7, 43268. [Google Scholar] [CrossRef] [PubMed]
- Moreau, R.A.; Nyström, L.; Whitaker, B.D.; Winkler-Moser, J.K.; Baer, D.J.; Gebauer, S.K.; Hicks, K.B. Phytosterols and their derivatives: Structural diversity, distribution, metabolism, analysis, and Health-Promoting uses. Prog. Lipid Res. 2018, 70, 35–61. [Google Scholar] [CrossRef]
- Kaneko, K.; Mitsuhashi, H.; Hirayama, K.; Ohmori, S. 11-Deoxojervine as a precursor for jervine biosynthesis in Veratrum grandiflorum. Phytochemistry 1970, 9, 2497–2501. [Google Scholar] [CrossRef]
- Choe, S.; Dilkes, B.P.; Fujioka, S.; Takatsuto, S.; Sakurai, A.; Feldmann, K.A. The DWF4 gene of arabidopsis encodes a cytochrome P450 that mediates multiple 22α-Hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 1998, 10, 231–243. [Google Scholar] [CrossRef]
- Fujita, S.; Ohnishi, T.; Watanabe, B.; Yokota, T.; Takatsuto, S.; Fujioka, S.; Yoshida, S.; Sakata, K.; Mizutani, M. Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28 and C29 sterols. Plant J. 2006, 45, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Morinaka, Y.; Ohnishi, T.; Sunohara, H.; Fujioka, S.; Ueguchi-Tanaka, M.; Mizutani, M.; Sakata, K.; Takatsuto, S.; Yoshida, S.; et al. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 2005, 24, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Ohnishi, T.; Watanabe, B.; Sakata, K.; Mizutani, M. CYP724B2 and CYP90B3 function in the early C-22 hydroxylation steps of brassinosteroid biosynthetic pathway in tomato. Biosci. Biotechnol. Biochem. 2006, 70, 2071–2080. [Google Scholar] [CrossRef]
- Itkin, M.; Heinig, U.; Tzfadia, O.; Bhide, A.J.; Shinde, B.; Cardenas, P.D.; Bocobza, S.E.; Unger, T.; Malitsky, S.; Finkers, R.; et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 2013, 341, 175–179. [Google Scholar] [CrossRef]
- Umemoto, N.; Nakayasu, M.; Ohyama, K.; Yotsu-Yamashita, M.; Mizutani, M.; Seki, H.; Saito, K.; Muranaka, T. Two cytochrome P450 Monooxygenases catalyze early hydroxylation steps in the potato steroid glycoalkaloid biosynthetic pathway. Plant Physiol. 2016, 171, 2458–2467. [Google Scholar] [CrossRef] [PubMed]
- Thornton, L.E.; Rupasinghe, S.G.; Peng, H.; Schuler, M.A.; Neff, M.M. Arabidopsis CYP72C1 is an atypical cytochrome P450 that inactivates brassinosteroids. Plant Mol. Biol. 2010, 74, 167–181. [Google Scholar] [CrossRef]
- Ohnishi, T.; Nomura, T.; Watanabe, B.; Ohta, D.; Yokota, T.; Miyagawa, H.; Sakata, K.; Mizutani, M. Tomato cytochrome P450 CYP734A7 functions in brassinosteroid catabolism. Phytochemistry 2006, 67, 1895–1906. [Google Scholar] [CrossRef]
- Sakamoto, T.; Kawabe, A.; Tokida-Segawa, A.; Shimizu, B.; Takatsuto, S.; Shimada, Y.; Fujioka, S.; Mizutani, M. Rice CYP734As function as multisubstrate and multifunctional enzymes in brassinosteroid catabolism. Plant J. 2011, 67, 1–12. [Google Scholar] [CrossRef]
- Nakayasu, M.; Umemoto, N.; Ohyama, K.; Fujimoto, Y.; Lee, H.J.; Watanabe, B.; Muranaka, T.; Saito, K.; Sugimoto, Y.; Mizutani, M. A dioxygenase catalyzes steroid 16α-Hydroxylation in steroidal glycoalkaloid biosynthesis. Plant Physiol. 2017, 75, 120–133. [Google Scholar] [CrossRef]
- Kawai, Y.; Ono, E.; Mizutani, M. Evolution and diversity of the 2-oxoglutaratedependent dioxygenase superfamily in plants. Plant J. 2014, 78, 328–343. [Google Scholar] [CrossRef]
- Miettinen, K.; Pollier, J.; Buyst, D.; Arendt, P.; Csuk, R.; Sommerwerk, S.; Moses, T.; Mertens, J.; Sonawane, P.D.; Pauwels, L.; et al. The ancient CYP716 family is a major contributor to the diversification of eudicot triterpenoid biosynthesis. Nat. Commun. 2017, 8, 14153. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Teranishi, Y.; Ueda, S.; Suzuki, H.; Kawano, N.; Yoshimatsu, K.; Saito, K.; Kawahara, N.; Muranaka, T.; Seki, H. Cytochrome P450 monooxygenase CYP716A141 is a Unique b-Amyrin C-16b oxidase involved in triterpenoid saponin biosynthesis in Platycodon grandiflorus. Plant Cell Physiol. 2017, 58, 874–884. [Google Scholar] [CrossRef] [PubMed]
- Moses, T.; Pollier, J.; Almagro, L.; Buyst, D.; Van Montagu, M.; Pedreño, M.A.; Martins, J.C.; Thevelein, J.M.; Goossens, A. Combinatorial biosynthesis of sapogenins and saponins in Saccharomyces cerevisiae using a C-16α hydroxylase from Bupleurum falcatum. Proc. Natl. Acad. Sci. USA 2014, 111, 1634–1639. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dai, L.; Yang, J.; Liu, C.; Men, Y.; Zeng, Y.; Cai, Y.; Zhu, Y.; Sun, Y. Oxidation of cucurbitadienol catalyzed by cyp87d18 in the biosynthesis of mogrosides from Siraitia grosvenorii. Plant Cell Physiol. 2016, 57, 1000–1007. [Google Scholar] [CrossRef]
- Ghosh, S. Triterpene structural diversification by plant cytochrome P450 enzymes. Front. Plant Sci. 2017, 8, 1886. [Google Scholar] [CrossRef]
- Hamberger, B.; Bak, S. Plant P450s as versatile drivers for evolution of Species-Specific chemical diversity. Philos. Trans. R. Soc. B 2013, 368, 20120426. [Google Scholar] [CrossRef]
- Seki, H.; Ohyama, K.; Sawai, S.; Mizutani, M.; Ohnishi, T.; Sudo, H.; Akashi, T.; Aoki, T.; Saito, K.; Muranaka, T. Licorice Beta-Amyrin 11-Oxidase, a cytochrome P450 with a key role in the biosynthesis of the triterpene sweetener glycyrrhizin. Proc. Natl. Acad. Sci. USA 2008, 105, 14204–14209. [Google Scholar] [CrossRef]
- Han, J.Y.; Kim, H.J.; Kwon, Y.S.; Choi, Y.E. The Cyt P450 enzyme CYP716A47 catalyzes the formation of protopanaxadiol from Dammarenediol-II during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2011, 52, 2062–2073. [Google Scholar] [CrossRef]
- Jeske, L.; Placzek, S.; Schomburg, I.; Chang, A.; Schomburg, D. BRENDA in 2019: A European ELIXIR core data resource. Nucleic Acids Res. 2019, 47, D542–D549. [Google Scholar] [CrossRef]
- Roth, S.; Kilgore, M.B.; Kutchan, T.M.; Müller, M. Exploiting the catalytic diversity of Short-Chain dehydrogenases/reductases: Versatile enzymes from plants with extended imine substrate scope. ChemBioChem 2018, 19, 1849–1852. [Google Scholar] [CrossRef]
- Persson, B.; Kallberg, Y.; Bray, J.E.; Bruford, E.; Dellaporta, S.L.; Favia, A.D.; Duarte, R.G.; Jörnvall, H.; Kavanagh, K.L.; Kedishvili, N.; et al. The SDR (Short-Chain dehydrogenase/reductase and related enzymes) nomenclature initiative. Chem. Biol. Interact. 2009, 178, 94–98. [Google Scholar] [CrossRef] [PubMed]








| CHL-RDA | MeJ-RDA | SQ-RDA | |
|---|---|---|---|
| Total number of contigs | 3547 | 3712 | 4789 |
| Total bases in contigs (bp) | 988,613 | 1,047,608 | 1,339,219 |
| Min length of contigs (bp) | 74 | 34 | 60 |
| Max length of contigs (bp) | 922 | 1003 | 886 |
| Average length of contigs (bp) | 279 | 282 | 280 |
| GC content (%) | 49.5 | 48.3 | 47.1 |
| N75 (bp) | 233 | 235 | 234 |
| N50 (bp) | 275 | 279 | 279 |
| N25 (bp) | 339 | 343 | 339 |
| Number of contigs <500 bp | 3480 | 3621 | 4708 |
| Number of contigs ≥500 bp | 67 | 91 | 81 |
| Database | RDA Library | ||
|---|---|---|---|
| CHL | MeJ | SQ | |
| NT | 3175 | 3474 | 4379 |
| NR | 3005 | 3308 | 4188 |
| KOG | 1648 | 1721 | 2193 |
| Pfam | 1801 | 1858 | 2383 |
| KEGG | 1415 | 1438 | 1845 |
| GO | 2082 | 2647 | 3308 |
| Total | 3528 | 3662 | 4701 |
| Unigenes ID | Function | Reference Gene | Blastx E-Value |
|---|---|---|---|
| CHL-RDA_contig_1929 | C-22 hydroxylation | PGA2 (Solanum tuberosum) | 2.90 × 10−12 |
| CHL-RDA_contig_1930 | C-22 hydroxylation | PGA2 (Solanum tuberosum) | 2.40 × 10−12 |
| CHL-RDA_contig_3050 | C-26 hydroxylation | CYP94N1v2 (Veratrum californicum) | 9.27 × 10−15 |
| MeJ-RDA_contig_1334 | C-26 hydroxylation | CYP734A7 (Solanum lycopersicum) | 3.00 × 10−38 |
| MeJ-RDA_contig_1251 | transamination | GABAT2 (Veratrum californicum) | 2.51 × 10−44 |
| CHL-RDA_contig_648 | C-11 hydroxylation | CYP88D6 (Glycyrrhiza glabra) | 2.59 × 10−16 |
| SQ-RDA_contig_3964 | C-16 hydroxylation | CYP86A2 (Arabidopsis thaliana) | 1.05 × 10−33 |
| MeJ-RDA_contig_2593 | C-16 hydroxylation | 16DOX (Solanum lycopersicum) | 5.60 × 10−26 |
| MeJ-RDA_contig_81 | C-16 hydroxylation | 16DOX (Solanum lycopersicum) | 5.55 × 10−12 |
| SQ-RDA_contig_2664 | reduction of heterocycles | PYCR (Medicago truncatula) | 2.00×10−55 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szeliga, M.; Ciura, J.; Tyrka, M. Representational Difference Analysis of Transcripts Involved in Jervine Biosynthesis. Life 2020, 10, 88. https://doi.org/10.3390/life10060088
Szeliga M, Ciura J, Tyrka M. Representational Difference Analysis of Transcripts Involved in Jervine Biosynthesis. Life. 2020; 10(6):88. https://doi.org/10.3390/life10060088
Chicago/Turabian StyleSzeliga, Magdalena, Joanna Ciura, and Mirosław Tyrka. 2020. "Representational Difference Analysis of Transcripts Involved in Jervine Biosynthesis" Life 10, no. 6: 88. https://doi.org/10.3390/life10060088
APA StyleSzeliga, M., Ciura, J., & Tyrka, M. (2020). Representational Difference Analysis of Transcripts Involved in Jervine Biosynthesis. Life, 10(6), 88. https://doi.org/10.3390/life10060088





