Prospects and Limitations Related to the Use of MicroRNA as a Biomarker of Epilepsy in Children: A Systematic Review
Abstract
:1. Introduction
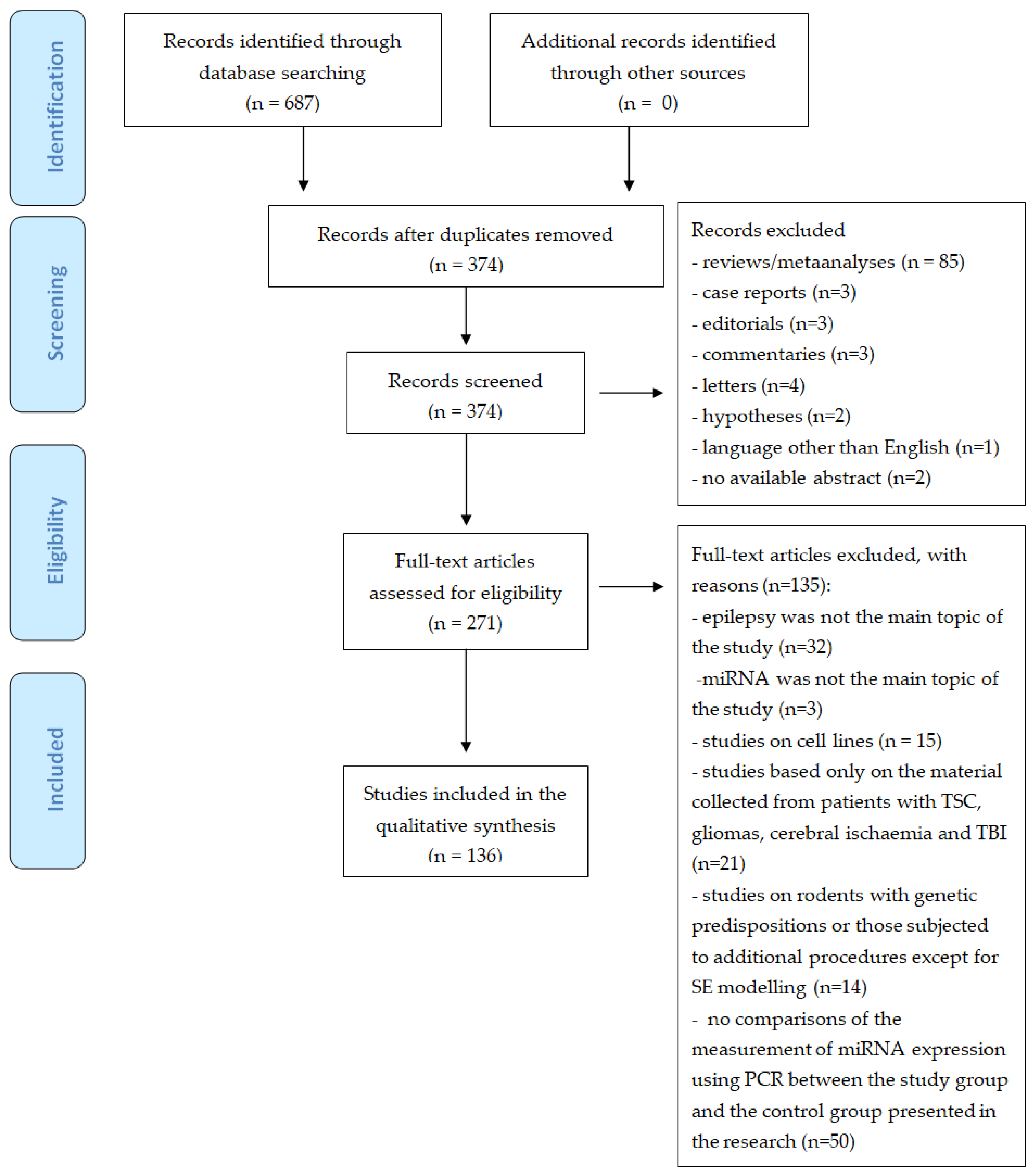
2. Materials and Methods
2.1. Search Strategy
2.2. Selection Criteria
2.3. Development of the Review
3. Results
3.1. The Level of Expression of MiRNA in Children with Epilepsy
3.2. Animal versus Human Models
3.3. Expression of MiRNAs in Blood and Other Biological Materials
3.4. Impact of Drugs on MiRNA Levels
3.5. The Most Important MiRNA in the Context of Epilepsy
4. Discussion
5. Limitations
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AP | acute phase |
| CA1 | cornu Ammonis 1 |
| CA3 | cornu Ammonis 3 |
| CP | chronic phase |
| CSF | cerebrospinal fluid |
| DG | dentate gyrus |
| FCD | Focal Cortical Dysplasia |
| GGE | Genetic Generalized Epilepsies |
| HP | hippocampus |
| KA | kainic acid |
| LP | latent phase |
| MTLE | Mesial Temporal Lobe Epilepsy |
| MTLE-HS | Mesial Temporal Lobe Epilepsy with Hippocampal Sclerosis |
| NDE | Newly Diagnosed Epilepsy |
| NS | no significance |
| PHC | parahippocampal cortex |
| PILO | pilocarpine |
| PPS | perforant pathway stimulation |
| PTZ | pentylenetetrazole |
| SE | status epilepticus |
| TLE | Temporal Lobe Epilepsy |
| TLE-HS | Temporal Lobe Epilepsy with Hippocampal Sclerosis |
References
- Nickels, K.C.; Wong-Kisiel, L.C.; Moseley, B.D.; Wirrell, E.C. Temporal lobe epilepsy in children. Epilepsy Res. Treat. 2012, 2012, 849540. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.D.; Pan, H.Y.; Huang, J.B.; Liu, X.P.; Li, J.H.; Ho, C.J.; Tsai, M.H.; Yang, J.L.; Chen, S.F.; Chen, N.C.; et al. Circulating MicroRNAs from Serum Exosomes May Serve as a Putative Biomarker in the Diagnosis and Treatment of Patients with Focal Cortical Dysplasia. Cells 2020, 9, 1867. [Google Scholar] [CrossRef] [PubMed]
- Nolan, D.; Fink, J. Genetics of epilepsy. In Handbook of Clinical Neurology; Geschwind, D.H., Paulson, H.L., Klein, C., Eds.; Elsavier: Amsterdam, The Netherlands, 2018; Volume 148, pp. 467–491. [Google Scholar]
- Josephson, C.B.; Jetté, N. Psychiatric comorbidities in epilepsy. Int. Rev. Psychiatry 2017, 29, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Lassandro, G.; Ciaccia, L.; Amoruso, A.; Palladino, V.; Palmieri, V.V.; Giordano, P. Focus on MicroRNAs as Biomarker in Pediatric Diseases. Curr. Pharm. Des. 2020, 2020. [Google Scholar] [CrossRef] [PubMed]
- Ebert, M.S.; Sharp, P.A. Roles for microRNAs in conferring robustness to biological processes. Cell 2012, 149, 515–524. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosik, K.S. The neuronal microRNA system. Nat. Rev. Neurosci. 2006, 7, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A compendium for extracellular vesicles with continuous community annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef] [Green Version]
- Gomes, A.R.; Sangani, N.B.; Fernandes, T.G.; Diogo, M.M.; Curfs, L.M.G.; Reutelingsperger, C.P. Extracellular Vesicles in CNS Developmental Disorders. Int. J. Mol. Sci. 2020, 21, 9428. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Zhang, H.; Xie, W.; Meng, F.; Zhang, K.; Jiang, Y.; Zhang, X.; Zhang, J. Altered microRNA profiles in plasma exosomes from mesial temporal lobe epilepsy with hippocampal sclerosis. Oncotarget 2017, 8, 4136–4146. [Google Scholar] [CrossRef]
- Li, N.; Pan, J.; Liu, W.; Li, Y.; Li, F.; Liu, M. MicroRNA-15a-5p serves as a potential biomarker and regulates the viability and apoptosis of hippocampus neuron in children with temporal lobe epilepsy. Diagn. Pathol. 2020, 15, 46. [Google Scholar] [CrossRef]
- Li, L.; Liu, C.Q.; Li, T.F.; Guan, Y.G.; Zhou, J.; Qi, X.L.; Yang, Y.T.; Deng, J.H.; Xu, Z.Q.; Luan, G.M. Analysis of Altered Micro RNA Expression Profiles in Focal Cortical Dysplasia IIB. J. Child Neurol. 2016, 31, 613–620. [Google Scholar] [CrossRef]
- Lee, J.Y.; Park, A.K.; Lee, E.S.; Park, W.Y.; Park, S.H.; Choi, J.W.; Phi, J.H.; Wang, K.C.; Kim, S.K. miRNA expression analysis in cortical dysplasia: Regulation of mTOR and LIS1 pathway. Epilepsy Res. 2014, 108, 433–441. [Google Scholar] [CrossRef]
- Wang, L.; Song, L.; Chen, X.; Suo, J.; Ma, Y.; Shi, J.; Liu, K.; Chen, G. microRNA-139-5p confers sensitivity to antiepileptic drugs in refractory epilepsy by inhibition of MRP1. CNS Neurosci. Ther. 2020, 26, 465–474. [Google Scholar] [CrossRef]
- Elnady, H.G.; Abdelmoneam, N.; Eissa, E.; Hamid, E.R.A.; Zeid, D.A.; Abo-Shanab, A.M.; Atta, H.; Kholoussi, N.M. MicroRNAs as Potential Biomarkers for Childhood Epilepsy. Open Access Maced. J. Med. Sci. 2019, 7, 3965–3969. [Google Scholar] [CrossRef] [Green Version]
- Ren, L.; Zhu, R.; Li, X. Silencing miR-181a produces neuroprotection against hippocampus neuron cell apoptosis post-status epilepticus in a rat model and in children with temporal lobe epilepsy. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Omran, A.; Peng, J.; Zhang, C.; Xiang, Q.L.; Xue, J.; Gan, N.; Kong, H.; Yin, F. Interleukin-1β and microRNA-146a in an immature rat model and children with mesial temporal lobe epilepsy. Epilepsia 2012, 53, 1215–1224. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Y.; Sun, Z.; Ren, S.; Yang, W.; Deng, Y.; Tian, C.; Yu, Y.; Gao, B. Molecular expression and functional analysis of genes in children with temporal lobe epilepsy. J. Integr. Neurosci. 2019, 18, 71–77. [Google Scholar] [CrossRef] [Green Version]
- Ashhab, M.U.; Omran, A.; Kong, H.; Gan, N.; He, F.; Peng, J.; Yin, F. Expressions of tumor necrosis factor alpha and microRNA-155 in immature rat model of status epilepticus and children with mesial temporal lobe epilepsy. J. Mol. Neurosci. 2013, 51, 950–958. [Google Scholar] [CrossRef]
- Peng, J.; Omran, A.; Ashhab, M.U.; Kong, H.; Gan, N.; He, F.; Yin, F. Expression patterns of miR-124, miR-134, miR-132, and miR-21 in an immature rat model and children with mesial temporal lobe epilepsy. J. Mol. Neurosci. 2013, 50, 291–297. [Google Scholar] [CrossRef]
- Brennan, G.P.; Bauer, S.; Engel, T.; Jimenez-Mateos, E.M.; Del Gallo, F.; Hill, T.D.M.; Connolly, N.M.C.; Costard, L.S.; Neubert, V.; Salvetti, B.; et al. Genome-wide microRNA profiling of plasma from three different animal models identifies biomarkers of temporal lobe epilepsy. Neurobiol. Dis. 2020, 144, 105048. [Google Scholar] [CrossRef]
- Raoof, R.; Bauer, S.; El Naggar, H.; Connolly, N.M.C.; Brennan, G.P.; Brindley, E.; Hill, T.; McArdle, H.; Spain, E.; Forster, R.J.; et al. Dual-center, dual-platform microRNA profiling identifies potential plasma biomarkers of adult temporal lobe epilepsy. EBioMedicine 2018, 38, 127–141. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Cheng, W.; Liu, L.; Tao, S.; Xia, Z.; Qi, L.; Huang, M. Identification of serum miRNAs differentially expressed in human epilepsy at seizure onset and post-seizure. Mol. Med. Rep. 2016, 14, 5318–5324. [Google Scholar] [CrossRef] [Green Version]
- Surges, R.; Kretschmann, A.; Abnaof, K.; van Rikxoort, M.; Ridder, K.; Fröhlich, H.; Danis, B.; Kaminski, R.M.; Foerch, P.; Elger, C.E.; et al. Changes in serum miRNAs following generalized convulsive seizures in human mesial temporal lobe epilepsy. Biochem. Biophys. Res. Commun. 2016, 481, 13–18. [Google Scholar] [CrossRef]
- Korotkov, A.; Broekaart, D.W.M.; Banchaewa, L.; Pustjens, B.; van Scheppingen, J.; Anink, J.J.; Baayen, J.C.; Idema, S.; Gorter, J.A.; van Vliet, E.A.; et al. microRNA-132 is overexpressed in glia in temporal lobe epilepsy and reduces the expression of pro-epileptogenic factors in human cultured astrocytes. Glia 2020, 68, 60–75. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.; Wang, H.; Wang, Q.; Chen, Y.; Chen, S. Expression of p-CREB and activity-dependent miR-132 in temporal lobe epilepsy. Int. J. Clin. Exp. Med. 2014, 7, 1297–1306. [Google Scholar]
- Reschke, C.R.; Silva, L.F.A.; Norwood, B.A.; Senthilkumar, K.; Morris, G.; Sanz-Rodriguez, A.; Conroy, R.M.; Costard, L.; Neubert, V.; Bauer, S.; et al. Potent Anti-seizure Effects of Locked Nucleic Acid Antagomirs Targeting miR-134 in Multiple Mouse and Rat Models of Epilepsy. Mol. Ther. Nucleic. Acids. 2017, 6, 45–56. [Google Scholar] [CrossRef]
- Alsharafi, W.; Xiao, B. Dynamic Expression of MicroRNAs (183, 135a, 125b, 128, 30c and 27a) in the Rat Pilocarpine Model and Temporal Lobe Epilepsy Patients. CNS Neurol. Disord. Drug Targets. 2015, 14, 1096–1102. [Google Scholar] [CrossRef]
- Alsharafi, W.A.; Xiao, B.; Li, J. MicroRNA-139-5p negatively regulates NR2A-containing NMDA receptor in the rat pilocarpine model and patients with temporal lobe epilepsy. Epilepsia 2016, 57, 1931–1940. [Google Scholar] [CrossRef]
- Korotkov, A.; Broekaart, D.W.M.; van Scheppingen, J.; Anink, J.J.; Baayen, J.C.; Idema, S.; Gorter, J.A.; Aronica, E.; van Vliet, E.A. Increased expression of matrix metalloproteinase 3 can be attenuated by inhibition of microRNA-155 in cultured human astrocytes. J. Neuroinflammation 2018, 15, 211. [Google Scholar] [CrossRef] [Green Version]
- Li, T.R.; Jia, Y.J.; Wang, Q.; Shao, X.Q.; Zhang, P.; Lv, R.J. Correlation between tumor necrosis factor alpha mRNA and microRNA-155 expression in rat models and patients with temporal lobe epilepsy. Brain. Res. 2018, 1700, 56–65. [Google Scholar] [CrossRef]
- Huang, L.G.; Zou, J.; Lu, Q.C. Silencing rno-miR-155-5p in rat temporal lobe epilepsy model reduces pathophysiological features and cell apoptosis by activating Sestrin-3. Brain. Res. 2018, 1689, 109–122. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Tang, R.; Yao, Y.; Ji, Z.; Cao, Y.; Liu, Z.; Peng, F.; Wang, W.; Can, D.; Xing, H.; et al. MiR-219 Protects Against Seizure in the Kainic Acid Model of Epilepsy. Mol. Neurobiol. 2016, 53, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Antônio, L.G.L.; Freitas-Lima, P.; Pereira-da-Silva, G.; Assirati, J.A., Jr.; Matias, C.M.; Cirino, M.L.A.; Tirapelli, L.F.; Velasco, T.R.; Sakamoto, A.C.; Carlotti, C.G., Jr.; et al. Expression of MicroRNAs miR-145, miR-181c, miR-199a and miR-1183 in the Blood and Hippocampus of Patients with Mesial Temporal Lobe Epilepsy. J. Mol. Neurosci. 2019, 69, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Cheng, Y.; Luo, H.; Rong, Z.; Li, Y.; Lu, P.; Ye, X.; Huang, W.; Qi, Z.; Li, X.; et al. Silencing MicroRNA-155 Attenuates Kainic Acid-Induced Seizure by Inhibiting Microglia Activation. Neuroimmunomodulation 2019, 26, 67–76. [Google Scholar] [CrossRef]
- Gong, G.H.; An, F.M.; Wang, Y.; Bian, M.; Wang, D.; Wei, C.X. MiR-153 regulates expression of hypoxia-inducible factor-1α in refractory epilepsy. Oncotarget 2018, 9, 8542–8547. [Google Scholar] [CrossRef] [Green Version]
- Che, N.; Zu, G.; Zhou, T.; Wang, X.; Sun, Y.; Tan, Z.; Liu, Y.; Wang, D.; Luo, X.; Zhao, Z.; et al. Aberrant Expression of miR-323a-5p in Patients with Refractory Epilepsy Caused by Focal Cortical Dysplasia. Genet. Test Mol. Biomark. 2017, 21, 3–9. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, X.; Wang, Z.; Zhang, Y.; Che, N.; Luo, X.; Tan, Z.; Sun, X.; Li, X.; Yang, K.; et al. Expression of microRNA-129-2-3p and microRNA-935 in plasma and brain tissue of human refractory epilepsy. Epilepsy Res. 2016, 127, 276–283. [Google Scholar] [CrossRef]
- Li, Y.; Huang, C.; Feng, P.; Jiang, Y.; Wang, W.; Zhou, D.; Chen, L. Aberrant expression of miR-153 is associated with overexpression of hypoxia-inducible factor-1α in refractory epilepsy. Sci. Rep. 2016, 6, 32091. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Sun, Y.; Tan, Z.; Che, N.; Ji, A.; Luo, X.; Sun, X.; Li, X.; Yang, K.; Wang, G.; et al. Serum MicroRNA-4521 is a Potential Biomarker for Focal Cortical Dysplasia with Refractory Epilepsy. Neurochem. Res. 2016, 41, 905–912. [Google Scholar] [CrossRef]
- Wang, X.; Luo, Y.; Liu, S.; Tan, L.; Wang, S.; Man, R. MicroRNA-134 plasma levels before and after treatment with valproic acid for epilepsy patients. Oncotarget 2017, 8, 72748–72754. [Google Scholar] [CrossRef] [Green Version]
- Haenisch, S.; von Rüden, E.L.; Wahmkow, H.; Rettenbeck, M.L.; Michler, C.; Russmann, V.; Bruckmueller, H.; Waetzig, V.; Cascorbi, I.; Potschka, H. miRNA-187-3p-Mediated Regulation of the KCNK10/TREK-2 Potassium Channel in a Rat Epilepsy Model. ACS Chem. Neurosci. 2016, 7, 1585–1594. [Google Scholar] [CrossRef] [PubMed]
- Organista-Juárez, D.; Jiménez, A.; Rocha, L.; Alonso-Vanegas, M.; Guevara-Guzmán, R. Differential expression of miR-34a, 451, 1260, 1275 and 1298 in the neocortex of patients with mesial temporal lobe epilepsy. Epilepsy Res. 2019, 157, 106188. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.H.; Zhang, Y.X.; Zheng, Y.; Yang, F.; Hu, Y.; Xu, S.; Yan, S.Q.; Ding, Y.; Guo, Y.; Ding, M.P. Expression of plasma microRNA-145-5p and its correlation with clinical features in patients with refractory epilepsy. Epilepsy Res. 2019, 154, 21–25. [Google Scholar] [CrossRef] [PubMed]
- An, N.; Zhao, W.; Liu, Y.; Yang, X.; Chen, P. Elevated serum miR-106b and miR-146a in patients with focal and generalized epilepsy. Epilepsy Res. 2016, 127, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Tan, L.; Tan, L.; Tian, Y.; Ma, J.; Tan, C.C.; Wang, H.F.; Liu, Y.; Tan, M.S.; Jiang, T.; et al. Circulating microRNAs are promising novel biomarkers for drug-resistant epilepsy. Sci. Rep. 2015, 5, 10201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leontariti, M.; Avgeris, M.; Katsarou, M.S.; Drakoulis, N.; Siatouni, A.; Verentzioti, A.; Alexoudi, A.; Fytraki, A.; Patrikelis, P.; Vassilacopoulou, D.; et al. Circulating miR-146a and miR-134 in predicting drug-resistant epilepsy in patients with focal impaired awareness seizures. Epilepsia 2020, 61, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Ioriatti, E.S.; Cirino, M.L.A.; Lizarte Neto, F.S.; Velasco, T.R.; Sakamoto, A.C.; Freitas-Lima, P.; Tirapelli, D.P.C.; Carlotti, C.G., Jr. Expression of circulating microRNAs as predictors of diagnosis and surgical outcome in patients with mesial temporal lobe epilepsy with hippocampal sclerosis. Epilepsy Res. 2020, 166, 106373. [Google Scholar] [CrossRef]
- Martins-Ferreira, R.; Chaves, J.; Carvalho, C.; Bettencourt, A.; Chorão, R.; Freitas, J.; Samões, R.; Boleixa, D.; Lopes, J.; Ramalheira, J.; et al. Circulating microRNAs as potential biomarkers for genetic generalized epilepsies: A three microRNA panel. Eur. J. Neurol. 2020, 27, 660–666. [Google Scholar] [CrossRef]
- Raoof, R.; Jimenez-Mateos, E.M.; Bauer, S.; Tackenberg, B.; Rosenow, F.; Lang, J.; Onugoren, M.D.; Hamer, H.; Huchtemann, T.; Körtvélyessy, P.; et al. Cerebrospinal fluid microRNAs are potential biomarkers of temporal lobe epilepsy and status epilepticus. Sci. Rep. 2017, 7, 3328. [Google Scholar] [CrossRef]
- Avansini, S.H.; de Sousa Lima, B.P.; Secolin, R.; Santos, M.L.; Coan, A.C.; Vieira, A.S.; Torres, F.R.; Carvalho, B.S.; Alvim, M.K.; Morita, M.E.; et al. MicroRNA hsa-miR-134 is a circulating biomarker for mesial temporal lobe epilepsy. PLoS ONE 2017, 12, e0173060. [Google Scholar] [CrossRef]
- Wang, J.; Yu, J.T.; Tan, L.; Tian, Y.; Ma, J.; Tan, C.C.; Wang, H.F.; Liu, Y.; Tan, M.S.; Jiang, T.; et al. Genome-wide circulating microRNA expression profiling indicates biomarkers for epilepsy. Sci. Rep. 2015, 5, 9522. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhao, Q.Y.; Edson, J.; Zhang, Z.H.; Li, X.; Wei, W.; Bredy, T.; Reutens, D.C. Genome-wide microRNA profiling in brain and blood samples in a mouse model of epileptogenesis. Epilepsy Res. 2020, 166, 106400. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Fukuda, M.; Saito, I.; Horiuchi, I.; Okazawa, T.; Ishii, E. Incidence of childhood epilepsy: A population-based study in rural Japan. Brain. Dev. 2018, 40, 904–908. [Google Scholar] [CrossRef]
- Uldall, P.; Alving, J.; Hansen, L.K.; Kibaek, M.; Buchholt, J. The misdiagnosis of epilepsy in children admitted to a tertiary epilepsy centre with paroxysmal events. Arch. Dis. Child 2006, 91, 219–221. [Google Scholar] [CrossRef] [Green Version]
- Coppola, A.; Moshé, S.L. Why is the developing brain more susceptible to status epilepticus? Epilepsia 2009, 50, 25–26. [Google Scholar] [CrossRef] [PubMed]
- Pitkänen, A.; Lukasiuk, K. Molecular and cellular basis of epileptogenesis in symptomatic epilepsy. Epilepsy Behav. 2009, 14, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.Z.; Tian, Y.; Ander, B.P.; Xu, H.; Stamova, B.S.; Zhan, X.; Turner, R.J.; Jickling, G.; Sharp, F.R. Brain and blood microRNA expression profiling of ischemic stroke, intracerebral hemorrhage, and kainate seizures. J. Cereb. Blood Flow Metab. 2010, 30, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Roncon, P.; Zucchini, S.; Ferracin, M.; Marucci, G.; Giulioni, M.; Michelucci, R.; Rubboli, G.; Simonato, M. Is autopsy tissue a valid control for epilepsy surgery tissue in microRNA studies? Epilepsia Open 2016, 2, 90–95. [Google Scholar] [CrossRef] [Green Version]
- McKiernan, R.C.; Jimenez-Mateos, E.M.; Bray, I.; Engel, T.; Brennan, G.P.; Sano, T.; Michalak, Z.; Moran, C.; Delanty, N.; Farrell, M.; et al. Reduced mature microRNA levels in association with dicer loss in human temporal lobe epilepsy with hippocampal sclerosis. PLoS ONE 2012, 7, e35921. [Google Scholar] [CrossRef] [PubMed]
- Bencurova, P.; Baloun, J.; Musilova, K.; Radova, L.; Tichy, B.; Pail, M.; Zeman, M.; Brichtova, E.; Hermanova, M.; Pospisilova, S.; et al. MicroRNA and mesial temporal lobe epilepsy with hippocampal sclerosis: Whole miRNome profiling of human hippocampus. Epilepsia 2017, 58, 1782–1793. [Google Scholar] [CrossRef] [Green Version]
- Hicks, S.D.; Ignacio, C.; Gentile, K.; Middleton, F.A. Salivary miRNA profiles identify children with autism spectrum disorder, correlate with adaptive behavior, and implicate ASD candidate genes involved in neurodevelopment. BMC Pediatr. 2016, 16, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liguori, M.; Nuzziello, N.; Licciulli, F.; Consiglio, A.; Simone, M.; Viterbo, R.G.; Creanza, T.M.; Ancona, N.; Tortorella, C.; Margari, L.; et al. Combined microRNA and mRNA expression analysis in pediatric multiple sclerosis: An integrated approach to uncover novel pathogenic mechanisms of the disease. Hum. Mol. Genet. 2018, 27, 66–79. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Mateos, E.M.; Engel, T.; Merino-Serrais, P.; McKiernan, R.C.; Tanaka, K.; Mouri, G.; Sano, T.; O’Tuathaigh, C.; Waddington, J.L.; Prenter, S.; et al. Silencing microRNA-134 produces neuroprotective and prolonged seizure-suppressive effects. Nat. Med. 2012, 18, 1087–1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jimenez-Mateos, E.M.; Engel, T.; Merino-Serrais, P.; Fernaud-Espinosa, I.; Rodriguez-Alvarez, N.; Reynolds, J.; Reschke, C.R.; Conroy, R.M.; McKiernan, R.C.; deFelipe, J.; et al. Antagomirs targeting microRNA-134 increase hippocampal pyramidal neuron spine volume in vivo and protect against pilocarpine-induced status epilepticus. Brain. Struct. Funct. 2015, 220, 2387–2399. [Google Scholar] [CrossRef]
- Zhou, X.; Chen, J.; Tao, H.; Cai, Y.; Huang, L.; Zhou, H.; Chen, Y.; Cui, L.; Zhong, W.; Li, K. Intranasal Delivery of miR-155-5p Antagomir Alleviates Acute Seizures Likely by Inhibiting Hippocampal Inflammation. Neuropsychiatr. Dis. Treat. 2020, 16, 1295–1307. [Google Scholar] [CrossRef]
- Hu, K.; Xie, Y.Y.; Zhang, C.; Ouyang, D.S.; Long, H.Y.; Sun, D.N.; Long, L.L.; Feng, L.; Li, Y.; Xiao, B. MicroRNA expression profile of the hippocampus in a rat model of temporal lobe epilepsy and miR-34a-targeted neuroprotection against hippocampal neurone cell apoptosis post-status epilepticus. BMC Neurosci. 2012, 13, 115. [Google Scholar] [CrossRef] [Green Version]
- Sano, T.; Reynolds, J.P.; Jimenez-Mateos, E.M.; Matsushima, S.; Taki, W.; Henshall, D.C. MicroRNA-34a upregulation during seizure-induced neuronal death. Cell Death Dis. 2012, 3, e287. [Google Scholar] [CrossRef] [Green Version]
- Jimenez-Mateos, E.M.; Bray, I.; Sanz-Rodriguez, A.; Engel, T.; McKiernan, R.C.; Mouri, G.; Tanaka, K.; Sano, T.; Saugstad, J.A.; Simon, R.P.; et al. miRNA Expression profile after status epilepticus and hippocampal neuroprotection by targeting miR-132. Am. J. Pathol. 2011, 179, 2519–2532. [Google Scholar] [CrossRef]
- Wang, X.; Yin, F.; Li, L.; Kong, H.; You, B.; Zhang, W.; Chen, S.; Peng, J. Intracerebroventricular injection of miR-146a relieves seizures in an immature rat model of lithium-pilocarpine induced status epilepticus. Epilepsy Res. 2018, 139, 14–19. [Google Scholar] [CrossRef]
- Zhang, H.L.; Lin, Y.H.; Qu, Y.; Chen, Q. The effect of miR-146a gene silencing on drug-resistance and expression of protein of P-gp and MRP1 in epilepsy. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2372–2379. [Google Scholar] [CrossRef]
- Iori, V.; Iyer, A.M.; Ravizza, T.; Beltrame, L.; Paracchini, L.; Marchini, S.; Cerovic, M.; Hill, C.; Ferrari, M.; Zucchetti, M.; et al. Blockade of the IL-1R1/TLR4 pathway mediates disease-modification therapeutic effects in a model of acquired epilepsy. Neurobiol. Dis. 2017, 99, 12–23. [Google Scholar] [CrossRef] [PubMed]
- Henshall, D.C. Manipulating MicroRNAs in Murine Models: Targeting the Multi-Targeting in Epilepsy. Epilepsy Curr. 2017, 17, 43–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, Y.; Liu, X.; Liao, Y.; Luo, C.; Zou, D.; Wei, X.; Huang, Q.; Wu, Y. MiR-181a influences the cognitive function of epileptic rats induced by pentylenetetrazol. Int. J. Clin. Exp. Pathol. 2015, 8, 12861–12868. [Google Scholar] [PubMed]
- Schouten, M.; Fratantoni, S.A.; Hubens, C.J.; Piersma, S.R.; Pham, T.V.; Bielefeld, P.; Voskuyl, R.A.; Lucassen, P.J.; Jimenez, C.R.; Fitzsimons, C.P. MicroRNA-124 and -137 cooperativity controls caspase-3 activity through BCL2L13 in hippocampal neural stem cells. Sci. Rep. 2015, 5, 12448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Children Diagnosed with Epilepsy | Control Children | MiRNA Expression in Children with Epilepsy | |||||||
|---|---|---|---|---|---|---|---|---|---|
| References | Patients | Number of Patients | Age (Years) | Controls | Number of Controls | Age (Years) | Samples | Up | Down |
| Li, N. et al. 2020 [11] | TLE | 63 | 9.81 ± 2.79 | healthy | 67 | 10.13 ± 2.46 | serum | miR-15a-5p | |
| Elnady, H.G. et al. 2019 [15] | epilepsy | 30 | 5–15 | Healthy | 20 | 5–15 | plasma | miR-146a | |
| miR-106b | |||||||||
| Wang, L. et al. 2020 [14] | refractory epilepsy | 26 | NDE | 35 | serum | miR-139-5p | |||
| NDE | 35 | traumatic brain injury or cerebrovascular malformation | 20 | miR-139-5p | |||||
| Wu, X. et al. 2019 [18] | TLE | 15 | 11.2 ± 2.6 | normal | 15 | 10.6 ± 3.7 | hippocampal Area CA3 | miR-135a-5p | |
| Ren, L. et al. 2016 [16] | TLE | 25 | 11 | brain tissues | miR-181a | ||||
| miR-132 | |||||||||
| miR-146a | |||||||||
| miR-34a | |||||||||
| miR-124 | |||||||||
| Li, L. et al. 2016 [12] | FCD type II B | 5 | 50–112 months | brain tissues | let-7f-1-3p | miR-6511b-5p | |||
| miR-1281 | miR-6862-5p | ||||||||
| miR-940 | |||||||||
| miR-1825 | |||||||||
| Lee, J.Y. et al. 2014 [13] | cortical dysplasia | 8 | 1–15 | deep-seated lesions | 3 | 2–13 | brain tissues | miR-21 | |
| miR-155 | |||||||||
| miR-130b | |||||||||
| miR-193b | |||||||||
| miR-199b | |||||||||
| Ashhab, M.U. et al. 2013 [19] | MTLE | 8 | 8–13 | no history of any brain disease | 8 | 6–13 | hippocampal tissues | miR-155 | |
| Peng, J. et al. 2013 [20] | MTLE | 5 | 8–12 | no history of any brain disease | 5 | 8–12 | hippocampal tissues | miR-124 | |
| miR-134 | |||||||||
| miR-132 | |||||||||
| miR-21 | |||||||||
| Omran, A. et al. 2012 [17] | MTLE | 5 | 8–12 | no history of any brain disease | 5 | 8–12 | hippocampal tissues | miR-146a | |
| References | Experimental Animals and Epilepsy Induction | Tissue | Time Points | miRNA Studied | Level of Expression in a Group with Epilepsy | Patients | Tissue | Level of Expression in a Group with Epilepsy |
|---|---|---|---|---|---|---|---|---|
| Peng, J. et al. 2013 [20] | PILO-induced SE in a rat model | HP | 2 h post SE (AP) | miR-124 | up | MTLE | HP | up |
| miR-134 | up | up | ||||||
| miR-132 | up | up | ||||||
| miR-21 | up | up | ||||||
| 3 weeks post SE (LP) | miR-124 | ns | ||||||
| miR-134 | ns | |||||||
| miR-132 | up | |||||||
| miR-21 | down | |||||||
| 8 weeks post SE (CP) | miR-124 | up | ||||||
| miR-134 | up | |||||||
| miR-132 | up | |||||||
| miR-21 | up | |||||||
| Korotkov, A. et al. 2020 [25] | tetanic stimulation-induced SE in a rat model (50 Hz) | DG | 1 day post SE (AP) | miR-132 | up | TLE-HS | HP | up |
| 1 week post SE (LP) | ns | |||||||
| 3–4 months post SE (CP) | ns | |||||||
| CA1 | 1 day post SE (AP) | ns | ||||||
| 1 week post SE (LP) | ns | |||||||
| 3–4 months post SE (CP) | ns | |||||||
| Guo, J. et al. 2014 [26] | lithium-PILO-induced epilepsy in a rat model | HP | 24 h post SE | miR-132 | up | TLE | temporal neocortex | down |
| 72 h post SE | ns | |||||||
| 7 d post SE | up | |||||||
| 14 d post SE | ns | |||||||
| 30 d post SE | ns | |||||||
| 60 d post SE | ns | |||||||
| Reschke, C.R. et al. 2017 [27] | PTZ model of generalized tonic-clonic seizures in mice | cortex | 30 min after PTZ injection | miR-134 | ns | TLE | HP | up |
| HP | up | |||||||
| PPS model of epilepsy in rats | HP | 24 h and 4 days after PPS | ns | |||||
| 14 days after PPS | ns | |||||||
| Alsharafi, W. et al. 2015 [28] | PILO-induced SE in a rat model | HP | 2 h post SE (AP) | miR-135a | up | TLE | HP | up |
| 2 months post SE (CP) | up | |||||||
| Alsharafi, W.A. et al. 2016 [29] | PILO-induced SE in a rat model | HP | 1 day after SE (AP) | miR-139-5p | down | TLE | HP | down |
| 7 days after SE (LP) | ns | |||||||
| 60 days after SE (CP) | down | |||||||
| Omran, A. et al. 2012 [17] | lithium-PILO-induced SE in a rat model | HP | 2h post SE (AP) | miR-146a | ns | MTLE | HP | up |
| 3 weeks post SE (LP) | up | |||||||
| 8 weeks post SE (CP) | up | |||||||
| Korotkov, A. et al. 2018 [30] | tetanic stimulation-induced SE in a rat model (50 Hz) | brain tissue: DG, CA1, PHC | 1 day post SE (AP) | miR-155 | up | TLE-HS | HP | up |
| 1 week post SE (LP) | up | |||||||
| 3–4 months post SE (CP) | up | |||||||
| Li, T.R. et al. 2018 [31] | KA-induced SE in a rat model | HP | 2 h after post SE (AP) | miR-155 | ns | TLE-HS | HP | up |
| 7 days post SE (LP) | up | |||||||
| 21 days post SE (LP) | up | |||||||
| 60 days post SE (CP) | up | |||||||
| Huang, L.G. et al. 2018 [32] | PILO-induced TLE in a rat model | CA1 | 0 day post-SE | miR-155 | ns | TLE | CA1, CA3 | up |
| 1 day post-SE | ns | |||||||
| 14 days post-SE | ns | |||||||
| 30 days post-SE | ns | |||||||
| 60 days post-SE | ns | |||||||
| CA3 | 0 day post-SE | ns | ||||||
| 1 day post-SE | up | |||||||
| 14 days post-SE | up | |||||||
| 30 days post-SE | up | |||||||
| 60 days post-SE | up | |||||||
| Ashhab, M.U. et al. 2013 [19] | lithium-PILO-induced SE in a rat model | HP | 2 h post SE (AP) | miR-155 | up | MTLE | HP | up |
| 3 weeks post SE (LP) | ns | |||||||
| 8 weeks post SE (CP) | up | |||||||
| Ren, L. et al. 2016 [16] | lithium-PILO-induced SE in a rat model | HP | 24 h post SE | miR-181a | up | TLE | brain tissues | up |
| 7 days post SE | up | |||||||
| 14 days post SE | up | |||||||
| 3 months post SE (TLE) | up | |||||||
| miR-132 | up | up | ||||||
| miR-146a | up | up | ||||||
| miR-34a | up | up | ||||||
| miR-124 | up | up |
| References | Patients | Number | Controls | Number | Samples | Method | miRNA Expression in Epileptic Patients | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | p-Value | Not Significant | |||||||
| Antônio, L.G.L. et al. 2019 [34] | MTLE-HS | 20 | no neurological or psychiatric medical history | 9 | hippocampal | RQ-PCR | miR-145 | p = 0.02 | miR-199a | |
| miR-1183 | ||||||||||
| miR-181c | ||||||||||
| healthy control | 10 | blood | miR-145 | p = 0.005 | ||||||
| miR-181c | p = 0.03 | |||||||||
| miR-199a | p = 0.01 | |||||||||
| miR-1183 | p = 0.001 | |||||||||
| Fu, H. et al. 2019 [35] | TLE | 12 | no history of epilepsy | 11 | brain samples | qRT-PCR | miR-155 | p < 0.05 | ||
| epilepsy | 40 | no history of epilepsy | 40 | plasma | miR-155 | p < 0.001 | ||||
| Gong, G.H. et al. 2018 [36] | MTLE | 22 | no history of epilepsy or seizures | 20 | temporal cortex | qRT-PCR | miR-153 | p < 0.01 | ||
| plasma | miR-153 | p < 0.01 | ||||||||
| Che, N. et al. 2017 [37] | FCD | 9 | hypertensive cerebral hemorrhage and no reported neurological illness | 8 | cortical samples | qRT-PCR | miR-323a-5p | p = 0.012 | ||
| 30 | healthy control | 23 | plasma | miR-323a-5p | p = 0.0320 | |||||
| Sun, Y. et al. 2016 [38] | TLE | 13 | no history of neurological diseases | 13 | cortical samples | qRT-PCR | miR-129-2-3p | p < 0.0001 | miR-935 | |
| 25 | 25 | plasma | miR-129-2-3p | p = 0.0008 | miR-935 | |||||
| Li, Y. et al. 2016 [39] | MTLE | 32 | no history of epilepsy or seizures | 18 | temporal cortex | RT-qPCR | miR-153 | p < 0.001 | miR-494 | |
| plasma | miR-153 | p < 0.001 | miR-494 | |||||||
| 56 | healthy control | 101 | plasma | miR-153 | p < 0.001 | miR-494 | ||||
| Wang, X. et al. 2016 [40] | TLE, FCD | 9 | acute intracerebral hematoma, no neurological illness associated with epilepsy | 8 | temporal cortex | RT-qPCR | hsa-miR-4521 | p = 0.001 | ||
| serum | hsa-miR-4521 | p = 0.0145 | ||||||||
| References | miRNA | Clinical Characteristics |
|---|---|---|
| Elnady, H.G. et al. 2019 [15] | miR-146a | age (p = 0.007) |
| Organista-Juárez, D. et al. 2019 [43] | miR-1260 | age (p = 0.018) |
| miR-1298 | age (p = 0.022) | |
| miR-146a | seizure frequency (p = 0.009) | |
| number of antiepileptic drugs (p = 0.03) | ||
| miR-451 | number of antiepileptic drugs (p = 0.046) | |
| Shen, C.H. et al. 2019 [44] | miR-145-5p | earlier age at epilepsy onset (p = 0.024) seizure frequency (p = 0.020) past history (head trauma, encephalitis) (p = 0.014) |
| Gong, G.H. et al. 2018 [36] | miR-153 | seizure frequency (p = 0. 018) Engel classification (p < 0. 01) |
| Huang, L.G. et al. 2018 [32] | miR-155-5p | hippocampal sclerosis (p = 4.03 × 10−5) Engel classification (p = 3.54 × 10−5) seizure frequency (p = 0.028) |
| Wang, X. et al. 2017 [41] | miR-134 | seizure severity (p = 0.016 moderate seizure group, p = 0.003 severe seizure group) |
| Yan, S. et al. 2017 [10] | miR-8071 | disease duration (p = 0.0073) seizure frequency (p = 0.0316) |
| Che, N. et al. 2017 [37] | miR-323a-5p | disease duration (p = 0.014) seizure frequency (p = 0.043) poor prognosis (p = 0.028) effectiveness of surgery (p = 0.005) |
| Surges, R. et al. 2016 [24] | miR-143-3p, miR-145-5p | total seizure duration |
| Sun, J. et al. 2016 [23] | miR-30a | seizure frequency (p < 0.01) |
| An, N. et al. 2016 [45] | miR-106b | seizure severity using the National Hospital Seizure Severity Scale (NHS3) |
| Sun, Y. et al. 2016 [38] | miR-129-2-3p | Engel classification (p = 0.005) seizure frequency (p = 0.027) |
| Wang, J. et al. 2015 [46] | miR-301a-3p | seizure severity using the National Hospital Seizure Severity Scale (NHS3) (p = 6.2 × 10−9) |
| References | miRNA | AUC |
|---|---|---|
| Leontariti, M. et al. 2020 [47] | miR-146a | 0.640 |
| miR-134 | 0.617 | |
| Shen, C.H. et al. 2019 [44] | miR-145-5p | 0.632 |
| Wang, X. et al. 2016 [40] | miR-4521 | 0.718 |
| Li, Y. et al. 2016 [39] | miR-153 | |
| Sun, Y. et al. 2016 [38] | miR-129-2-3p | 0.778 |
| Wang, J. et al. 2015 [46] | miR-301a-3p | 0.893 |
| References | miRNA | Diagnostic Marker | Sensitivity | Specificity | AUC |
|---|---|---|---|---|---|
| Ioriatti, E.S. et al. 2020 [48] | miR-328-3p | MTLE-HS | 89.30% | 90.90% | 0.935 |
| Brennan, G.P. et al. 2020 [21] | miR-93a-5p, miR-199a, miR-574-3p | TLE | 0.88–0.86 | ||
| Martins-Ferreira, R. et al. 2020 [49] | miR-146a, miR-155, miR-132 | GGE | 73% | 80% | 0.850 |
| Li, N. et al. 2020 [11] | miR-15a-5p | TLE | 82.50% | 88.10% | 0.908 |
| Shen, C.H. et al. 2019 [44] | miR-145-5p | MTLE | 0.829 | ||
| Elnady, H.G. et al. 2019 [15] | miR-106b | epilepsy | 80% | 80% | 0.885 |
| miR-146a | 73.70% | 60% | 0.763 | ||
| Raoof, R. et al. 2018 [22] | miR-27a-3p | TLE | 0.630 | ||
| GGE | 0.730 | ||||
| miR-328-3p | TLE | 0.630 | |||
| miR-654-3p | TLE | 0.870 | |||
| GGE | 0.720 | ||||
| miR-27a-3p, miR-328-3p, miR-654-3p | TLE | 0.640 | |||
| GGE | 0.740 | ||||
| Raoof, R. et al. 2017 [50] | miR-451a, mir-21-5p | TLE and SE | 0.850 | ||
| miR-19b-3p, miR-21-5p, miR-451a | 0.830 | ||||
| Avansini, S.H. et al. 2017 [51] | miR-134 | MTLE | 75% | 58% | 0.671 |
| Yan, S. et al. 2017 [10] | miR-8071 | MTLE-HS | 83.33% | 96.67% | 0.9316 |
| An, N. et al. 2016 [45] | miR-106b | epilepsy | 0.887 | ||
| miR-146a | |||||
| Wang, J. et al. 2015 [52] | miR-106b-5p | epilepsy | 80.30% | 81.20% | 0.882 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rzepka-Migut, B.; Paprocka, J. Prospects and Limitations Related to the Use of MicroRNA as a Biomarker of Epilepsy in Children: A Systematic Review. Life 2021, 11, 26. https://doi.org/10.3390/life11010026
Rzepka-Migut B, Paprocka J. Prospects and Limitations Related to the Use of MicroRNA as a Biomarker of Epilepsy in Children: A Systematic Review. Life. 2021; 11(1):26. https://doi.org/10.3390/life11010026
Chicago/Turabian StyleRzepka-Migut, Beata, and Justyna Paprocka. 2021. "Prospects and Limitations Related to the Use of MicroRNA as a Biomarker of Epilepsy in Children: A Systematic Review" Life 11, no. 1: 26. https://doi.org/10.3390/life11010026






