C1431T Variant of PPARγ Is Associated with Preeclampsia in Pregnant Women
Abstract
1. Introduction
2. Results
2.1. Overview of Maternal Clinical Features
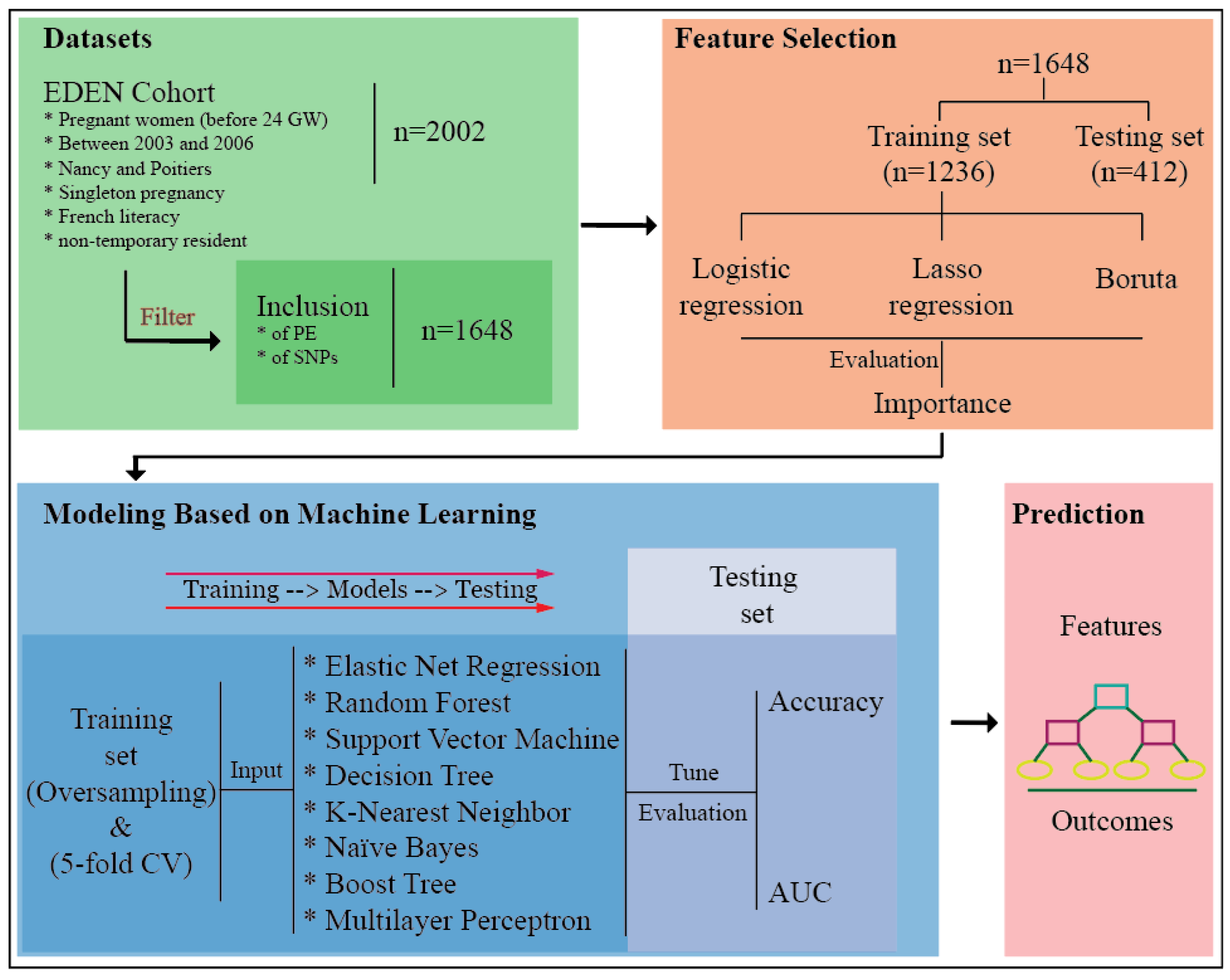
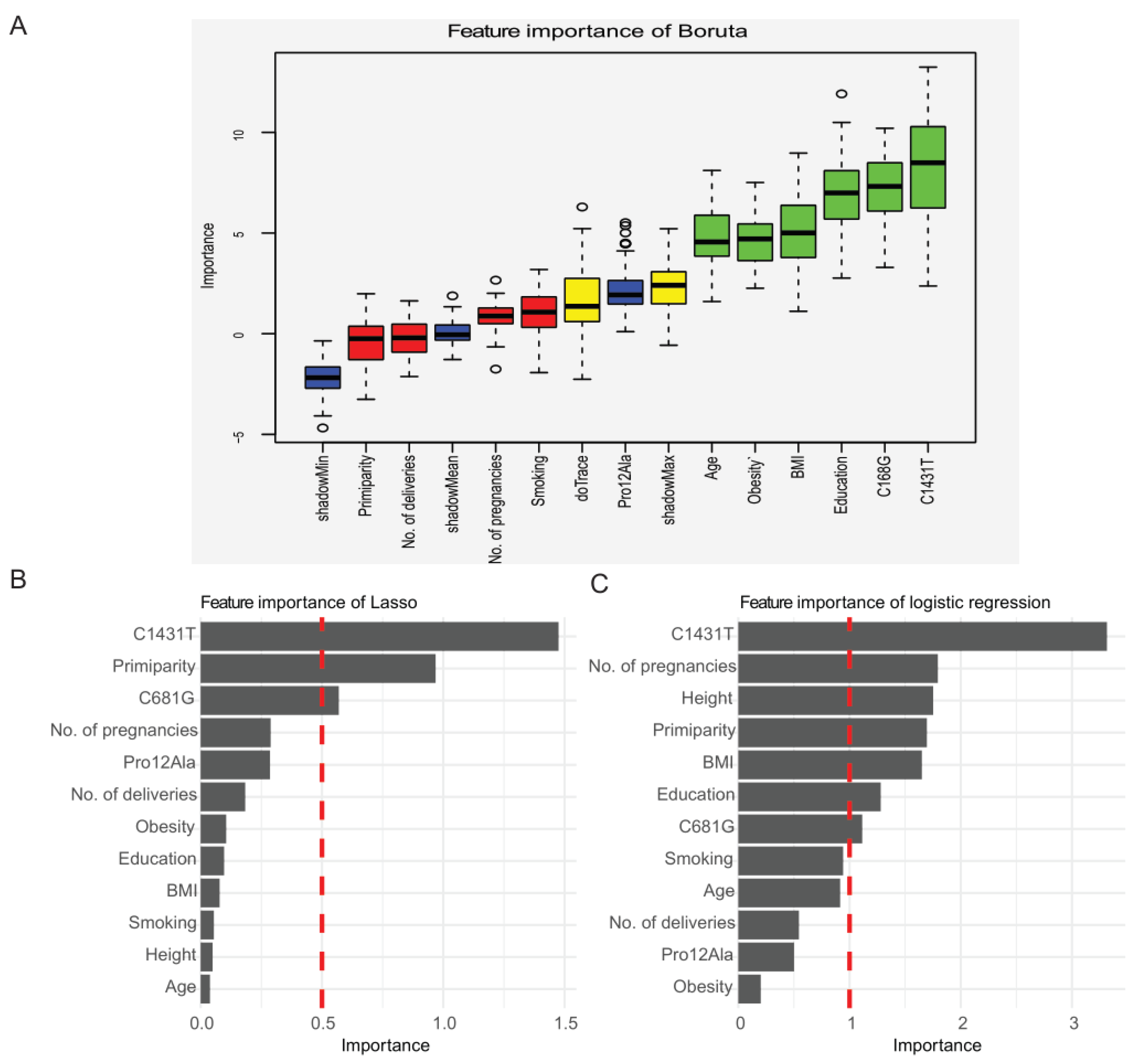
2.2. Selection of Candidate Features for the Prediction of Preeclampsia Using Three Methods: Boruta Algorithm, Lasso Regression, and Logistic Regression
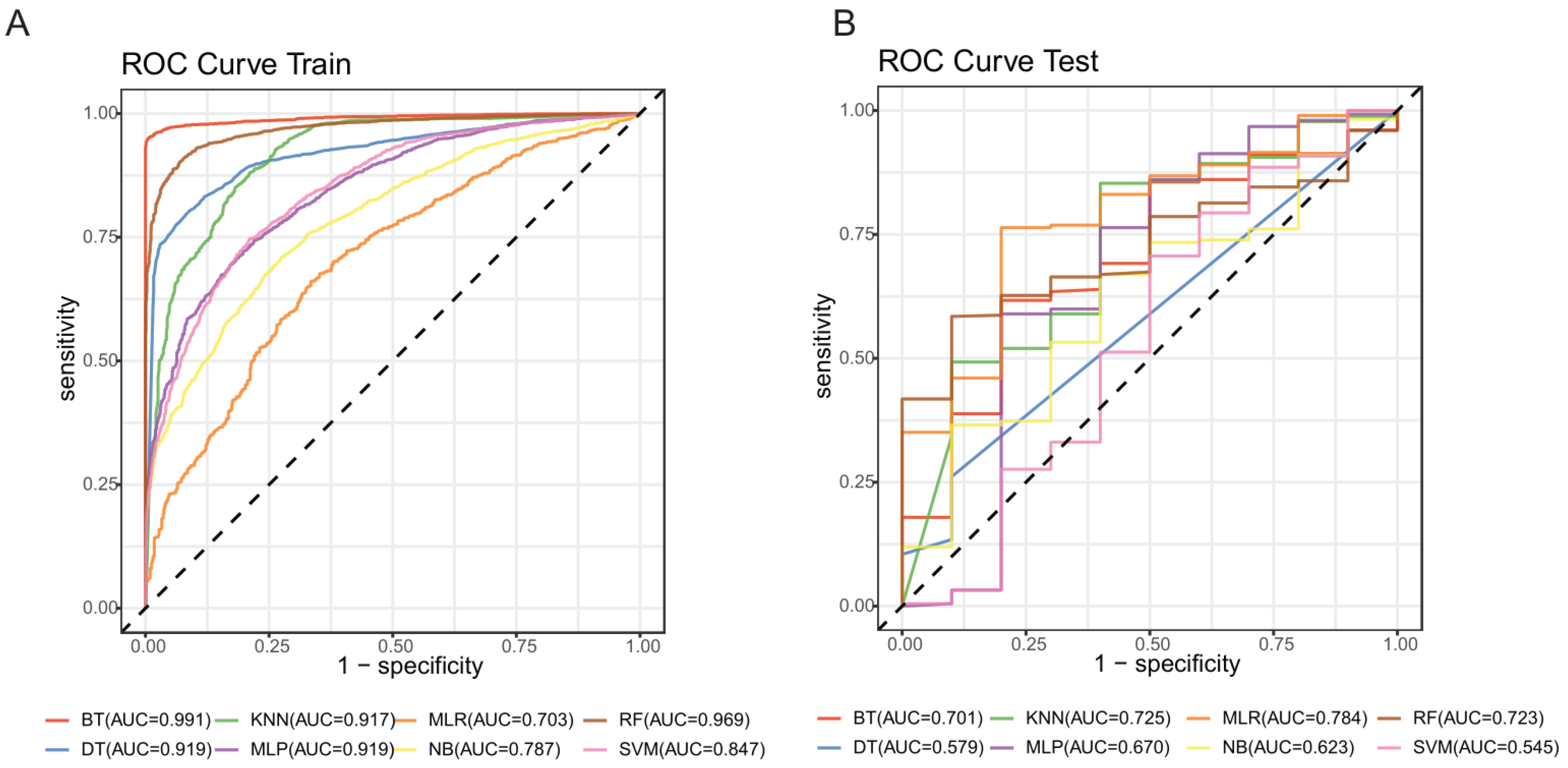
2.3. Modeling Based on Machine Learning
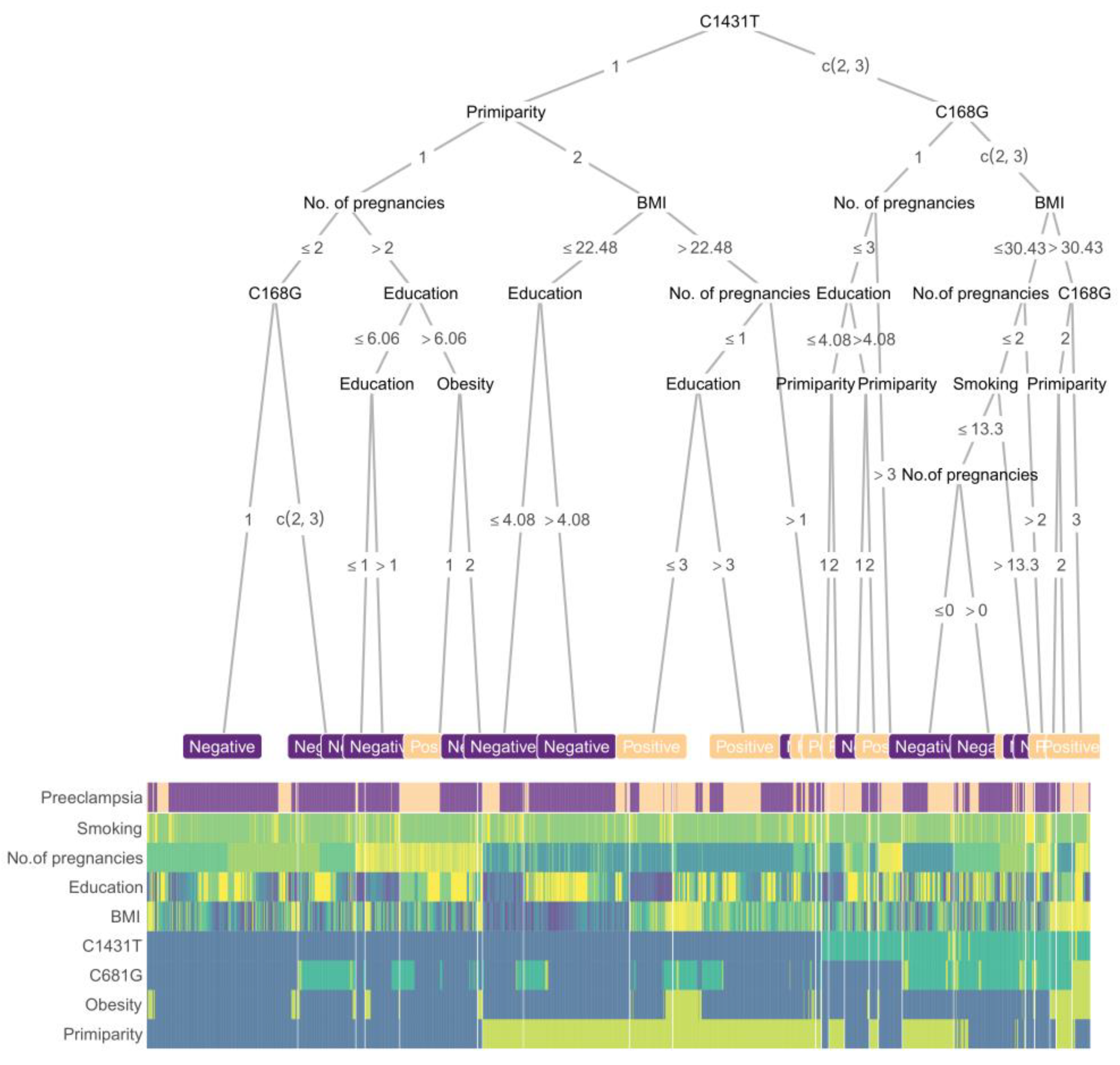
2.4. Prediction Procedures of Boost Tree
3. Discussion
4. Materials and Methods
4.1. Study Population
4.2. Clinical Features
4.3. Genotyping
4.4. Basic Statistical Analyses
4.5. Feature Selection
4.6. Modeling and Evaluation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- James, M.R.; Phyllis, A.A.; George, B.; John, R.B.; Ira, M.B.; Maurice, D.; Robert, R.G.; Joey, R.G.; Arun, J.; Donna, D.J.; et al. Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists’ task force on hypertension in pregnancy. Obstet. Gynecol. 2013, 122, 1122–1131. [Google Scholar]
- Kuklina, E.V.; Ayala, C.; Callaghan, W.M. Hypertensive disorders and severe obstetric morbidity in the United States. Obstet. Gynecol. 2009, 113, 1299–1306. [Google Scholar] [CrossRef] [PubMed]
- Hogan, M.C.; Foreman, K.J.; Naghavi, M.; Ahn, S.Y.; Wang, M.; Makela, S.M.; Lopez, A.D.; Lozano, R.; Murray, C.J. Maternal mortality for 181 countries, 1980–2008: A systematic analysis of progress towards Millennium Development Goal 5. Lancet 2010, 375, 1609–1623. [Google Scholar] [CrossRef]
- Olié, V.; Moutengou, E.; Deneux-Tharaux, C.; Kretz, S.; Vallée, A.; Blacher, J.; Tsatsaris, V.; Plu-Bureau, G. Prevalence of hypertensive disorders during pregnancy and post-partum in France. Arch. Cardiovasc. Dis. Suppl. 2020, 12, 155–156. [Google Scholar] [CrossRef]
- Bahado-Singh, R.; Poon, L.C.; Yilmaz, A.; Syngelaki, A.; Turkoglu, O.; Kumar, P.; Kirma, J.; Allos, M.; Accurti, V.; Li, J.; et al. Integrated proteomic and metabolomic prediction of term preeclampsia. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Kelly, R.S.; Croteau-Chonka, D.C.; Dahlin, A.; Mirzakhani, H.; Wu, A.C.; Wan, E.S.; McGeachie, M.J.; Qiu, W.; Sordillo, J.E.; Al-Garawi, A.; et al. Integration of metabolomic and transcriptomic networks in pregnant women reveals biological pathways and predictive signatures associated with preeclampsia. Metabolomics 2016, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Cerdeira, A.S.; Redman, C.; Vatish, M. Meta-analysis and systematic review to assess the role of soluble FMS-like tyrosine kinase-1 and placenta growth factor ratio in prediction of preeclampsia: The SaPPPhirE study. Hypertension 2018, 71, 306–316. [Google Scholar] [CrossRef]
- Verlohren, S.; Dröge, L.-A. The diagnostic value of angiogenic and antiangiogenic factors in differential diagnosis of preeclampsia. Am. J. Obstet. Gynecol. 2020, 20, S0002-9378. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Powe, C.E.; Salahuddin, S.; Verlohren, S.; Perschel, F.H.; Levine, R.J.; Lim, K.-H.; Wenger, J.B.; Thadhani, R.; Karumanchi, S.A. Angiogenic factors and the risk of adverse outcomes in women with suspected preeclampsia. Circulation 2012, 125, 911–919. [Google Scholar] [CrossRef]
- Romero, R.; Nien, J.K.; Espinoza, J.; Todem, D.; Fu, W.; Chung, H.; Kusanovic, J.P.; Gotsch, F.; Erez, O.; Mazaki-Tovi, S.; et al. A longitudinal study of angiogenic (placental growth factor) and anti-angiogenic (soluble endoglin and soluble vascular endothelial growth factor receptor-1) factors in normal pregnancy and patients destined to develop preeclampsia and deliver a small for gestational age neonate. J. Matern. Neonatal Med. 2008, 21, 9–23. [Google Scholar] [CrossRef]
- O’Gorman, N.; Wright, D.; Syngelaki, A.; Akolekar, R.; Wright, A.; Poon, L.C.; Nicolaides, K.H. Competing risks model in screening for preeclampsia by maternal factors and biomarkers at 11–13 weeks gestation. Am. J. Obstet. Gynecol. 2016, 214, 103.e1–103.e12. [Google Scholar] [CrossRef] [PubMed]
- O’Gorman, N.N.; Wright, D.; Poon, L.L.; Rolnik, D.L.; Syngelaki, A.A.; Wright, A.A.; Akolekar, R.R.; Cicero, S.S.; Janga, D.D.; Jani, J.; et al. Accuracy of competing-risks model in screening for pre-eclampsia by maternal factors and biomarkers at 11–13 weeks’ gestation. Ultrasound Obstet. Gynecol. 2017, 49, 751–755. [Google Scholar] [CrossRef] [PubMed]
- McGinnis, R.; Steinthorsdottir, V.; Williams, N.O.; Thorleifsson, G.; Shooter, S.; Hjartardottir, S.; Bumpstead, S.; Stefansdottir, L.; Hildyard, L.; Sigurdsson, J.K.; et al. Variants in the fetal genome near FLT1 are associated with risk of preeclampsia. Nat. Genet. 2017, 49, 1255–1260. [Google Scholar] [CrossRef] [PubMed]
- Gray, K.J.; Kovacheva, V.P.; Mirzakhani, H.; Bjonnes, A.C.; Almoguera, B.; DeWan, A.T.; Triche, E.W.; Saftlas, A.F.; Hoh, J.; Bodian, D.L.; et al. Gene-centric analysis of preeclampsia identifies maternal association at PLEKHG1. Hypertension 2018, 72, 408–416. [Google Scholar] [CrossRef]
- Meirhaeghe, A.; Fajas, L.; Helbecque, N.; Cottel, D.; Lebel, P.; Dallongeville, J.; Deeb, S.; Auwerx, J.; Amouyel, P. A genetic polymorphism of the peroxisome proliferator-activated receptor γ gene influences plasma leptin levels in obese humans. Hum. Mol. Genet. 1998, 7, 435–440. [Google Scholar] [CrossRef]
- Doney, A.; Fischer, B.; Frew, D.; Cumming, A.; Flavell, D.M.; World, M.; Montgomery, H.E.; Boyle, D.; Morris, A.; Palmer, C.N. Haplotype analysis of the PPARγ Pro12Ala and C1431T variants reveals opposing associations with body weight. BMC Genet. 2002, 3, 21. [Google Scholar] [CrossRef]
- Valve, R.; Sivenius, K.; Miettinen, R.; Pihlajamaki, J.; Rissanen, A.; Deeb, S.S.; Auwerx, J.; Uusitupa, M.; Laakso, M. Two polymorphisms in the peroxisome proliferator-activated receptor-γ gene are associated with severe overweight among obese women. J. Clin. Endocrinol. Metab. 1999, 84, 3708–3712. [Google Scholar] [CrossRef]
- Cai, G.; Zhang, X.; Weng, W.; Shi, G.; Xue, S.; Zhang, B. Associations between PPARG polymorphisms and the risk of essential hypertension. PLoS ONE 2017, 12, e0181644. [Google Scholar] [CrossRef]
- Chao, T.H.; Li, Y.H.; Chen, J.H.; Wu, H.L.; Shi, G.Y.; Liu, P.Y.; Tsai, W.C.; Guo, H.R. The 161TT genotype in the exon 6 of the peroxisome-proliferator-activated receptor γ gene is associated with premature acute myocardial infarction and increased lipid peroxidation in habitual heavy smokers. Clin. Sci. Lond. 2004, 107, 461–466. [Google Scholar] [CrossRef]
- Wang, X.L.; Oosterhof, J.; Duarte, N. Peroxisome proliferator-activated receptor γ C161 → T polymorphism and coronary artery disease. Cardiovasc. Res. 1999, 44, 588–594. [Google Scholar] [CrossRef]
- Lin, J.; Chen, Y.; Tang, W.-F.; Liu, C.; Zhang, S.; Guo, Z.-Q.; Chen, G.; Zheng, X.-W. PPARG rs3856806 C> T polymorphism increased the risk of colorectal cancer: A case-control study in Eastern Chinese Han population. Front. Oncol. 2019, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- Heude, B.; Pelloux, V.; Forhan, A.; Bedel, J.F.; Lacorte, J.M.; Clement, K.; Charles, M.A.; EDEN Mother-Child Cohort Study Group. Association of the Pro12Ala and C1431T variants of PPARγ and their haplotypes with susceptibility to gestational diabetes. J. Clin. Endocrinol. Metab. 2011, 96, E1656–E1660. [Google Scholar] [CrossRef] [PubMed]
- Gannoun, M.; Raguema, N.; Zitouni, H.; Mehdi, M.; Seda, O.; Mahjoub, T.; Lavoie, J. MMP-2 and MMP-9 polymorphisms and preeclampsia risk in Tunisian Arabs: A case-control study. J. Clin. Med. 2021, 10, 2647. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Souza, C.C.; Coeli-Lacchini, F.; Luizon, M.R.; Cavalli, R.C.; Lacchini, R.; Sandrim, V.C. Effects of arginase genetic polymorphisms on nitric oxide formation in healthy pregnancy and in preeclampsia. Nitric Oxide 2021, 109-110, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Gray, K.J.; Kovacheva, V.P.; Mirzakhani, H.; Bjonnes, A.C.; Almoguera, B.; Wilson, M.L.; Ingles, S.A.; Lockwood, C.J.; Hakonarson, H.; McElrath, T.F.; et al. Risk of pre-eclampsia in patients with a maternal genetic predisposition to common medical conditions: A case–control study. BJOG Int. J. Obstet. Gynaecol. 2021, 128, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Azhar, S. Peroxisome proliferator-activated receptors, metabolic syndrome and cardiovascular disease. Future Cardiol. 2010, 6, 657–691. [Google Scholar] [CrossRef]
- Peng, L.; Yang, H.; Ye, Y.; Ma, Z.; Kuhn, C.; Rahmeh, M.; Mahner, S.; Makrigiannakis, A.; Jeschke, U.; von Schönfeldt, V. Role of peroxisome proliferator-activated receptors (PPARs) in trophoblast functions. Int. J. Mol. Sci. 2021, 22, 433. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.Z.; Ivashchenko, C.Y.; Whitesall, S.E.; D’Alecy, L.G.; Duquaine, D.C.; Brosius, F.C., 3rd; Gonzalez, F.J.; Vinson, C.; Pierre, M.A.; Milstone, D.S.; et al. Hypotension, lipodystrophy, and insulin resistance in generalized PPARγ-deficient mice rescued from embryonic lethality. J. Clin. Investig. 2007, 117, 812–822. [Google Scholar] [CrossRef]
- Barak, Y.; Nelson, M.C.; Ong, E.S.; Jones, Y.Z.; Ruiz-Lozano, P.; Chien, K.R.; Koder, A.; Evans, R.M. PPAR γ is required for placental, cardiac, and adipose tissue development. Mol. Cell 1999, 4, 585–595. [Google Scholar] [CrossRef]
- McCarthy, F.P.; Drewlo, S.; English, F.A.; Kingdom, J.; Johns, E.J.; Kenny, L.C.; Walsh, S.K. Evidence implicating peroxisome proliferator-activated receptor-γ in the pathogenesis of preeclampsia. Hypertension 2011, 58, 882–887. [Google Scholar] [CrossRef] [PubMed]
- Almeida, S.M.; Furtado, J.M.; Mascarenhas, P.; Ferraz, M.E.; Ferreira, J.C.; Monteiro, M.; Vilanova, M.; Ferraz, F.P. Association between LEPR, FTO, MC4R, and PPARG-2 polymorphisms with obesity traits and metabolic phenotypes in school-aged children. Endocrine 2018, 60, 466–478. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Xie, Z.; Guo, J.; Wang, Y.; Liu, C.; Zhang, S.; Tang, W.; Chen, Y. Association of PPARG rs 1801282 C> G polymorphism with risk of colorectal cancer: From a case-control study to a meta-analysis. Oncotarget 2017, 8, 100558–100569. [Google Scholar] [CrossRef] [PubMed]
- Rocha, R.M.; Barra, G.; Rosa, É.C.; Garcia, É.C.; Amato, A.A.; Azevedo, M.F. Prevalence of the rs1801282 single nucleotide polymorphism of the PPARG gene in patients with metabolic syndrome. Arch. Endocrinol. Metab. 2015, 59, 297–302. [Google Scholar] [CrossRef]
- Ho, J.S.; Germer, S.; Tam, C.H.; So, W.-Y.; Martin, M.; Ma, R.C.; Chan, J.; Ng, M.C. Association of the PPARG Pro12Ala polymorphism with type 2 diabetes and incident coronary heart disease in a Hong Kong Chinese population. Diabetes Res. Clin. Pr. 2012, 97, 483–491. [Google Scholar] [CrossRef] [PubMed]
- Deeb, S.S.; Fajas, L.; Nemoto, M.; Pihlajamaki, J.; Mykkanen, L.; Kuusisto, J.; Laakso, M.; Fujimoto, W.; Auwerx, J. A Pro12Ala substitution in PPARγ2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat. Genet. 1998, 20, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Altshuler, D.; Hirschhorn, J.N.; Klannemark, M.; Lindgren, C.M.; Vohl, M.C.; Nemesh, J.; Lane, C.R.; Schaffner, S.F.; Bolk, S.; Brewer, C.; et al. The common PPARγ Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 2000, 26, 76–80. [Google Scholar] [CrossRef] [PubMed]
- Beamer, B.A.; Yen, C.-J.; Andersen, R.E.; Muller, D.; Elahi, D.; Cheskin, L.J.; Andres, R.; Roth, J.; Shuldiner, A. Association of the Pro12Ala variant in the peroxisome proliferator-activated receptor-γ2 gene with obesity in two Caucasian populations. Diabetes 1998, 47, 1806–1808. [Google Scholar] [CrossRef]
- Cecil, J.E.; Fischer, B.; Doney, A.S.F.; Hetherington, M.; Watt, P.; Wrieden, W.; Bolton-Smith, C.; Palmer, C.N.A. The Pro12Ala and C–681G variants of the PPARG locus are associated with opposing growth phenotypes in young schoolchildren. Diabetologia 2005, 48, 1496–1502. [Google Scholar] [CrossRef][Green Version]
- Heude, B.; Forhan, A.; Slama, R.; Douhaud, L.; Bedel, S.; Saurel-Cubizolles, M.-J.; Hankard, R.; Thiebaugeorges, O.; de Agostini, M.; Annesi-Maesano, I.; et al. Cohort profile: The EDEN mother-child cohort on the prenatal and early postnatal determinants of child health and development. Int. J. Epidemiol. 2016, 45, 353–363. [Google Scholar] [CrossRef]
- Permadi, W.; Mantilidewi, K.I.; Khairani, A.F.; Lantika, U.A.; Ronosulistyo, A.R.; Bayuaji, H. Differences in expression of peroxisome proliferator-activated receptor-γ in early-onset preeclampsia and late-onset preeclampsia. BMC Res. Notes 2020, 13, 181. [Google Scholar] [CrossRef]
- Kadam, L.; Kohan-Ghadr, H.R.; Drewlo, S. The balancing act—PPAR-γ’s roles at the maternal-fetal interface. Syst. Biol. Reprod. Med. 2015, 61, 65–71. [Google Scholar] [CrossRef]
- McCarthy, F.P.; Drewlo, S.; Kingdom, J.; Johns, E.J.; Walsh, S.K.; Kenny, L.C. Peroxisome proliferator-activated receptor-γ as a potential therapeutic target in the treatment of preeclampsia. Hypertension 2011, 58, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Ganss, R. Maternal metabolism and vascular adaptation in pregnancy: The PPAR link. Trends Endocrinol. Metab. 2017, 28, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Laasanen, J.; Heinonen, S.; Hiltunen, M.; Mannermaa, A.; Laakso, M. Polymorphism in the peroxisome proliferator-activated receptor-γ gene in women with preeclampsia. Early Hum. Dev. 2002, 69, 77–82. [Google Scholar] [CrossRef]
- Hoffman, M.K.; Ma, N.; Roberts, A. A machine learning algorithm for predicting maternal readmission for hypertensive disorders of pregnancy. Am. J. Obstet. Gynecol. MFM 2021, 3, 100250. [Google Scholar] [CrossRef]
- Sufriyana, H.; Wu, Y.-W.; Su, E.C.-Y. Artificial intelligence-assisted prediction of preeclampsia: Development and external validation of a nationwide health insurance dataset of the BPJS Kesehatan in Indonesia. EBioMedicine 2020, 54, 102710. [Google Scholar] [CrossRef]
- Bodnar, L.M.; Cartus, A.R.; Kirkpatrick, S.I.; Himes, K.P.; Kennedy, E.H.; Simhan, H.N.; Grobman, W.A.; Duffy, J.Y.; Silver, R.M.; Parry, S.; et al. Machine learning as a strategy to account for dietary synergy: An illustration based on dietary intake and adverse pregnancy outcomes. Am. J. Clin. Nutr. 2020, 111, 1235–1243. [Google Scholar] [CrossRef]
- Jhee, J.H.; Lee, S.; Park, Y.; Lee, S.E.; Kim, Y.A.; Kang, S.-W.; Kwon, J.-Y.; Park, J.T. Prediction model development of late-onset preeclampsia using machine learning-based methods. PLoS ONE 2019, 14, e0221202. [Google Scholar] [CrossRef]
- ACOG practice bulletin no. 202 summary: Gestational hypertension and preeclampsia. Obstet. Gynecol. 2019, 133, 1.
- Committee opinion no. 638: First-trimester risk assessment for early-onset preeclampsia. Obstet. Gynecol. 2015, 126, e25–e27.
- Brasier, A.; Victor, S.; Ju, H.; Busse, W.W.; Curran-Everett, U.; Bleecker, E.; Castro, M.; Chung, K.F.; Gaston, B.; Israel, E.; et al. Predicting intermediate phenotypes in asthma using bronchoalveolar lavage-derived cytokines. Clin. Transl. Sci. 2010, 3, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Dreiseitl, S.; Ohno-Machado, L. Logistic regression and artificial neural network classification models: A methodology review. J. Biomed. Inform. 2002, 35, 352–359. [Google Scholar] [CrossRef]
- Kuhle, S.; Maguire, B.; Zhang, H.; Hamilton, D.; Allen, A.C.; Joseph, K.S.; Allen, V.M. Comparison of logistic regression with machine learning methods for the prediction of fetal growth abnormalities: A retrospective cohort study. BMC Pregnancy Childbirth 2018, 18, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Luque, A.; Carrasco, A.; Martín, A.; de las Heras, A. The impact of class imbalance in classification performance metrics based on the binary confusion matrix. Pattern Recognit. 2019, 91, 216–231. [Google Scholar] [CrossRef]
- Banerjee, P.; Dehnbostel, F.O.; Preissner, R. Prediction is a balancing act: Importance of sampling methods to balance sensitivity and specificity of predictive models based on imbalanced chemical data sets. Front. Chem. 2018, 6, 362. [Google Scholar] [CrossRef] [PubMed]
- Gorman, N.O.; Wright, D.; Poon, L.C.; Rolnik, D.L.; Syngelaki, A.; de Alvarado, M.; Carbone, I.F.; Dutemeyer, V.; Fiolna, M.; Frick, A.; et al. Multicenter screening for pre-eclampsia by maternal factors and biomarkers at 11–13 weeks’ gestation: Comparison with NICE guidelines and ACOG recommendations. Ultrasound Obstet. Gynecol. 2017, 49, 756–760. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Salahuddin, S.; Mueller, A.; Berg, A.H.; Thadhani, R.I.; Karumanchi, S.A. Angiogenic biomarkers in triage and risk for preeclampsia with severe features. Pregnancy Hypertens. 2018, 13, 100–106. [Google Scholar] [CrossRef]
- Odibo, A.O.; Goetzinger, K.R.; Odibo, L.; Cahill, A.G.; Macones, G.A.; Nelson, D.M.; Dietzen, D.J. First-trimester prediction of preeclampsia using metabolomic biomarkers: A discovery phase study. Prenat. Diagn. 2011, 31, 990–994. [Google Scholar] [CrossRef] [PubMed]
- Bahado-Singh, R.O.; Syngelaki, A.; Akolekar, R.; Mandal, R.; Bjondahl, T.C.; Han, B.; Dong, E.; Bauer, S.; Alpay-Savasan, Z.; Graham, S.; et al. Validation of metabolomic models for prediction of early-onset preeclampsia. Am. J. Obstet. Gynecol. 2015, 213, 530.e1–530.e10. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.; Liu, L.; Zhuge, Q.-C.; Yu, Z.; Zhang, X.; Xie, J.; Hong, W.; Wang, S.; Yang, Y.; Chen, B. The meta-analysis of the association of PPARG P12A, C161T polymorphism and coronary heart disease. Wien. Klin. Wochenschr. 2012, 124, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Van Buuren, S.; Groothuis-Oudshoorn, K. Mice: Multivariate imputation by chained equations in R. J. Stat. Softw. 2011, 45, 1–67. [Google Scholar] [CrossRef]
- Stekhoven, D.J.; Buhlmann, P. MissForest—Non-parametric missing value imputation for mixed-type data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Cordón, I.; García, S.; Fernández, A.; Herrera, F. Imbalance: Oversampling algorithms for imbalanced classification in R. Knowl.-Based Syst. 2018, 161, 329–341. [Google Scholar] [CrossRef]
- Kursa, M.; Rudnicki, W. Feature selection with the Boruta package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef]

| Clinical Features | Controls N = 1613 | Preeclampsia N = 35 | P-Value |
|---|---|---|---|
| Maternal age at delivery (year) | 29.6 ± 4.8 | 28.3 ± 6.1 | 0.232 |
| Maternal height (cm) | 163.0 ± 6.23 | 165.0 ± 6.35 | 0.108 |
| Maternal education (level) | 6.55 ± 2.47 | 6.05 ± 2.86 | 0.358 |
| BMI (before pregnancy, kg/m2) | 23.2 ± 4.6 | 25.2 ± 5.8 | 0.067 |
| Primiparity | 44 (703) | 60 (21) | 0.079 |
| Number of pregnancies | 1.35 ± 1.49 | 1.37 ± 1.77 | 0.956 |
| Number of deliveries | 0.834 ± 0.971 | 0.657 ± 0.906 | 0.261 |
| Obesity | 9(137) | 18(6) | 0.109 |
| Cigarette use (no.) | 1.48 ± 3.44 | 1.09 ± 3.37 | 0.524 |
| Maternal SNPs (% (N)) | |||
| Pro12Ala | 19.7 (333) | 26.3 (10) | 0.493 |
| C1431T | 21.3 (349) | 42.9 (15) | 0.004 |
| C681G | 39.3 (663) | 39.5 (15) | 1 |
| Maternal SNPs (% (N)) | Controls N = 1613 | Preeclampsia N = 35 | p1 a | p2 b | p3 c | ||||
|---|---|---|---|---|---|---|---|---|---|
| c/c | c/t | t/t | c/c | c/t | t/t | ||||
| C1431T | 78.9 (1272) | 20.2 (326) | 0.9 (15) | 57.1 (20) | 40 (14) | 2.9 (1) | 0.004 | 0.29 | 0.008 |
| C681G | 60.3 (967) | 35.2 (565) | 4.5 (72) | 60 (21) | 34.3 (12) | 5.7 (2) | 1 | 0.67 | 1 |
| Pro12Ala | 80.4 (1297) | 18.7 (301) | 0.9 (15) | 74.2 (26) | 22.9 (8) | 2.9 (1) | 0.49 | 0.29 | 0.68 |
| Training Set | Testing Set | |||
|---|---|---|---|---|
| Accuracy | AUC | Accuracy | AUC | |
| Elastic Net Regression | 0.661 ± 0.005 | 0.695 ± 0.006 | 0.857 | 0.784 |
| Random Forest | 0.913 ± 0.006 | 0.969 ± 0.003 | 0.896 | 0.723 |
| Support Vector Machine | 0.772 ± 0.003 | 0.847 ± 0.004 | 0.862 | 0.545 |
| Decision Tree | 0.849 ± 0.007 | 0.919 ± 0.006 | 0.874 | 0.579 |
| K-Nearest Neighbor | 0.826 ± 0.006 | 0.917 ± 0.006 | 0.801 | 0.725 |
| Naïve Bayes | 0.693 ± 0.005 | 0.787 ± 0.007 | 0.930 | 0.619 |
| Boost Tree | 0.971 ± 0.002 | 0.991 ± 0.001 | 0.951 | 0.701 |
| Multilayer Perceptron | 0.899 ± 0.007 | 0.919 ± 0.006 | 0.811 | 0.670 |
| True Condition | |||
|---|---|---|---|
| Condition Positive | Condition Negative | ||
| Predicted condition | Predicted condition positive | True positive (TP) | False positive (FP) |
| Predicted condition negative | False negative (FN) | True negative (TN) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, F.; Rouault, C.; Clément, K.; Zhu, W.; Degrelle, S.A.; Charles, M.-A.; Heude, B.; Fournier, T. C1431T Variant of PPARγ Is Associated with Preeclampsia in Pregnant Women. Life 2021, 11, 1052. https://doi.org/10.3390/life11101052
Liu F, Rouault C, Clément K, Zhu W, Degrelle SA, Charles M-A, Heude B, Fournier T. C1431T Variant of PPARγ Is Associated with Preeclampsia in Pregnant Women. Life. 2021; 11(10):1052. https://doi.org/10.3390/life11101052
Chicago/Turabian StyleLiu, Fulin, Christine Rouault, Karine Clément, Wencan Zhu, Séverine A. Degrelle, Marie-Aline Charles, Barbara Heude, and Thierry Fournier. 2021. "C1431T Variant of PPARγ Is Associated with Preeclampsia in Pregnant Women" Life 11, no. 10: 1052. https://doi.org/10.3390/life11101052
APA StyleLiu, F., Rouault, C., Clément, K., Zhu, W., Degrelle, S. A., Charles, M.-A., Heude, B., & Fournier, T. (2021). C1431T Variant of PPARγ Is Associated with Preeclampsia in Pregnant Women. Life, 11(10), 1052. https://doi.org/10.3390/life11101052







