Genetics and Cognitive Vulnerability to Sleep Deprivation in Healthy Subjects: Interaction of ADORA2A, TNF-α and COMT Polymorphisms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Study Design and Testing Conditions
2.3. Procedures
2.4. Neurobehavioral Testing
2.4.1. Karolinska Sleeping Scale (KSS) for Sleepiness
2.4.2. Psychomotor Vigilance Task (PVT) for Sustained Attention
2.4.3. The Go/NoGo Executive Task for Inhibition and Impulsivity
2.5. Saliva DNA Extract and Genotyping
2.6. Statistical Analysis
3. Results
3.1. Genotypes Frequencies
3.2. SNPs Distribution Associations
3.3. Correlations between Tests and Questionnaire Results
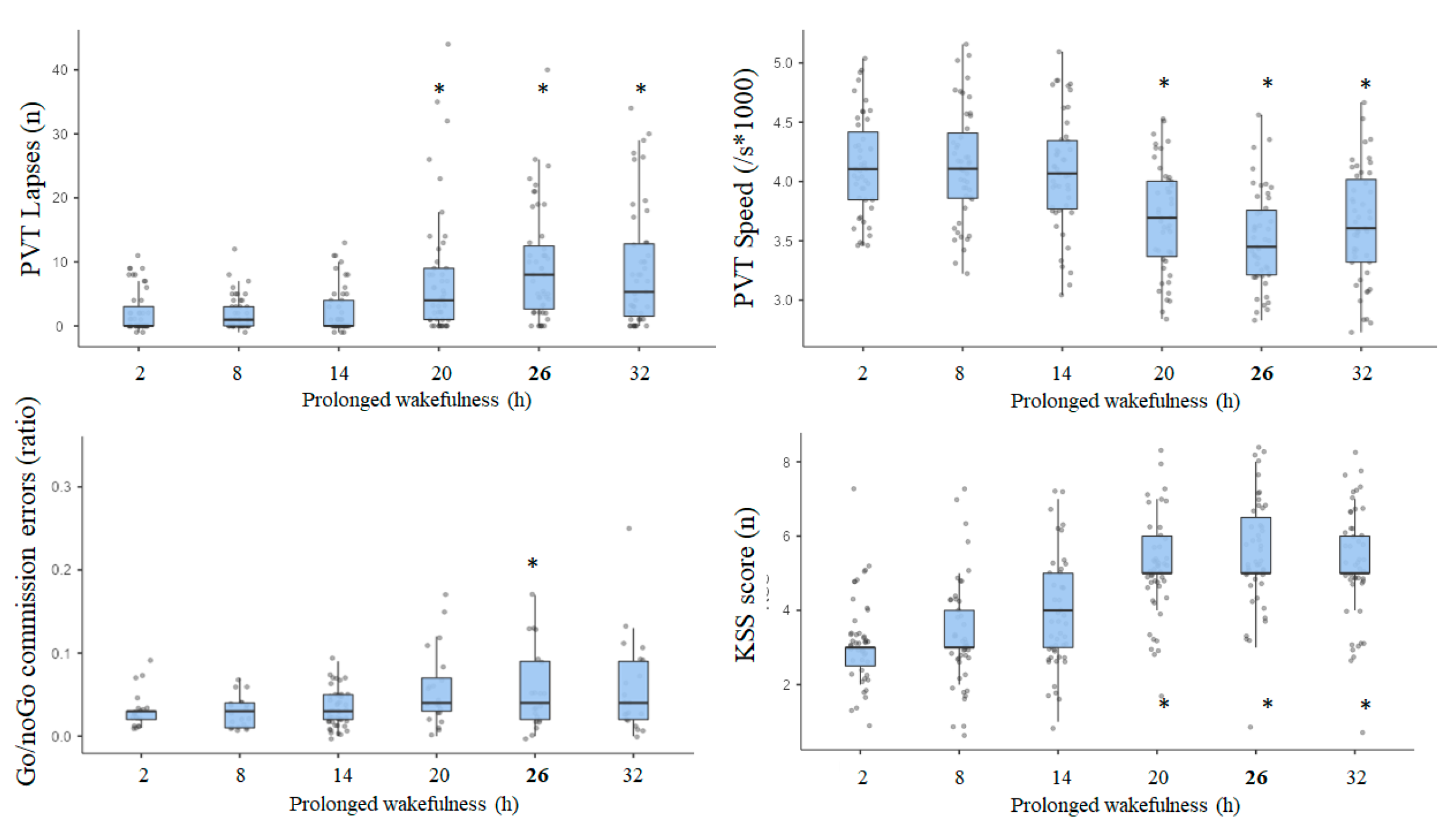
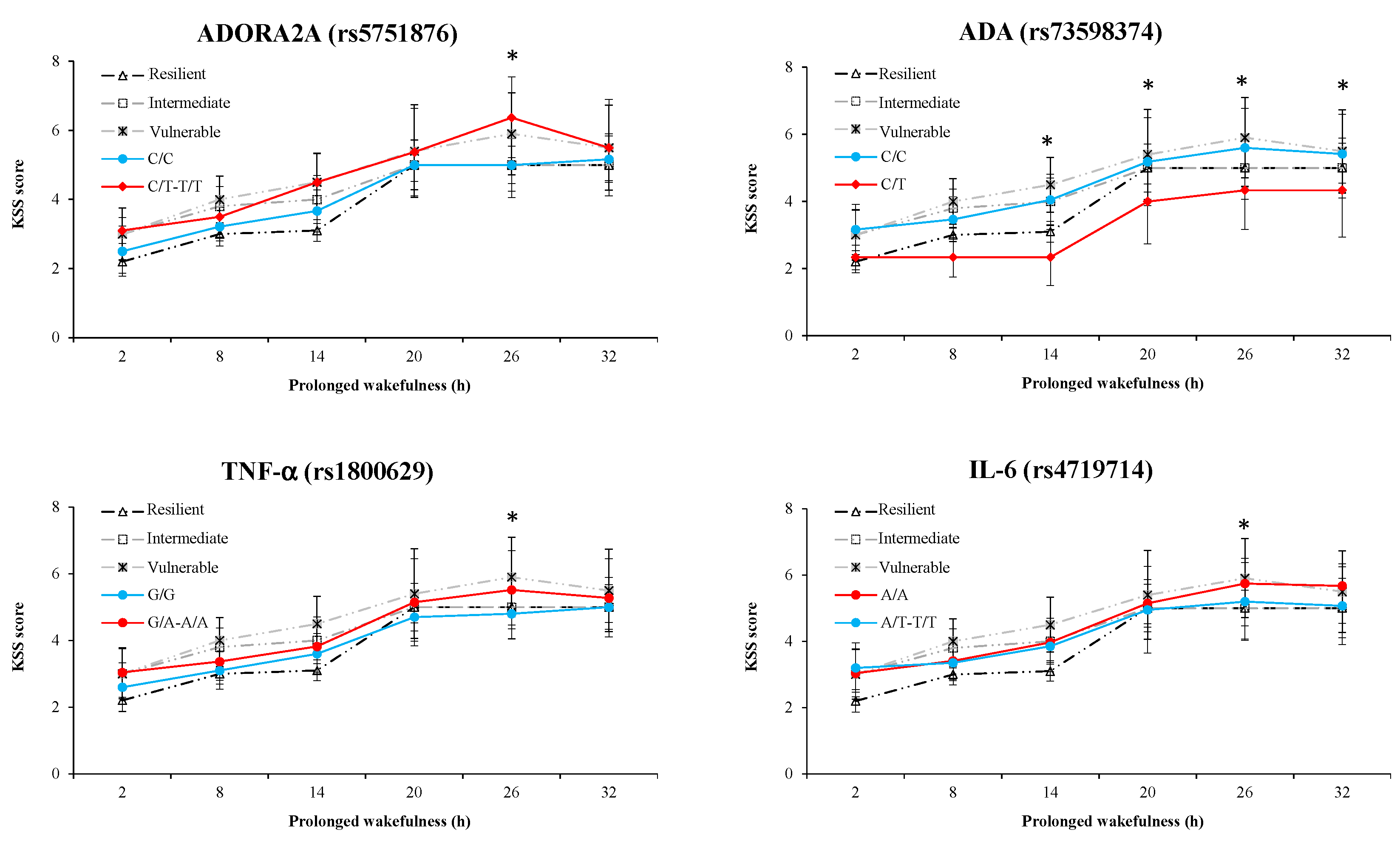
3.4. Results of Awakening Effect on PVT, Go/NoGo, and KSS Parameters
3.5. Results of Effects Size (ES) Analysis at Nadir Time Point (08:30) on Day 2 (i.e., 26 h of Continuous Awaking)
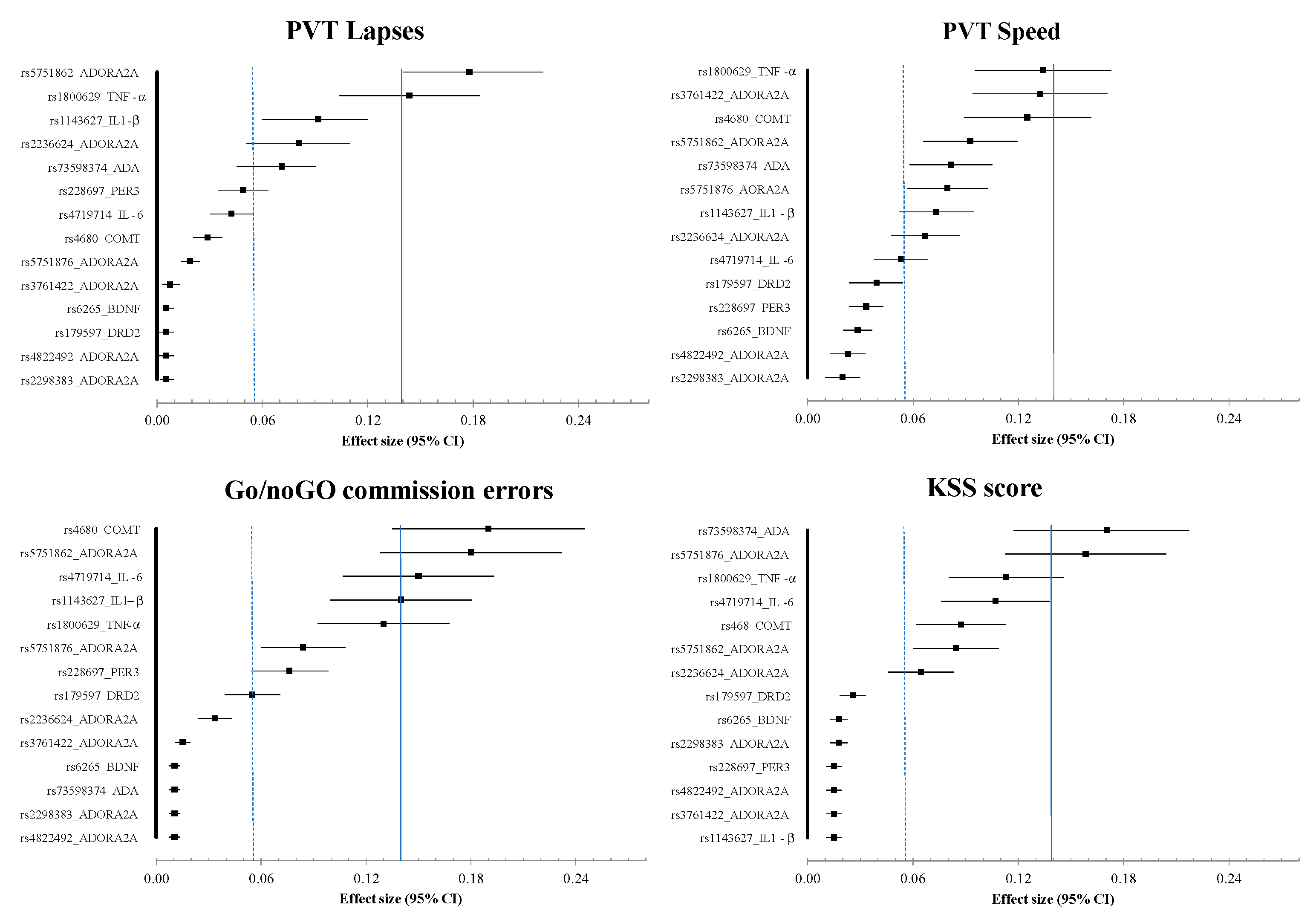
3.6. Results of Polymorphism Effect and Interaction with Awakening (SNP × TSD) on PVT, Go/noGo, and KSS Parameters
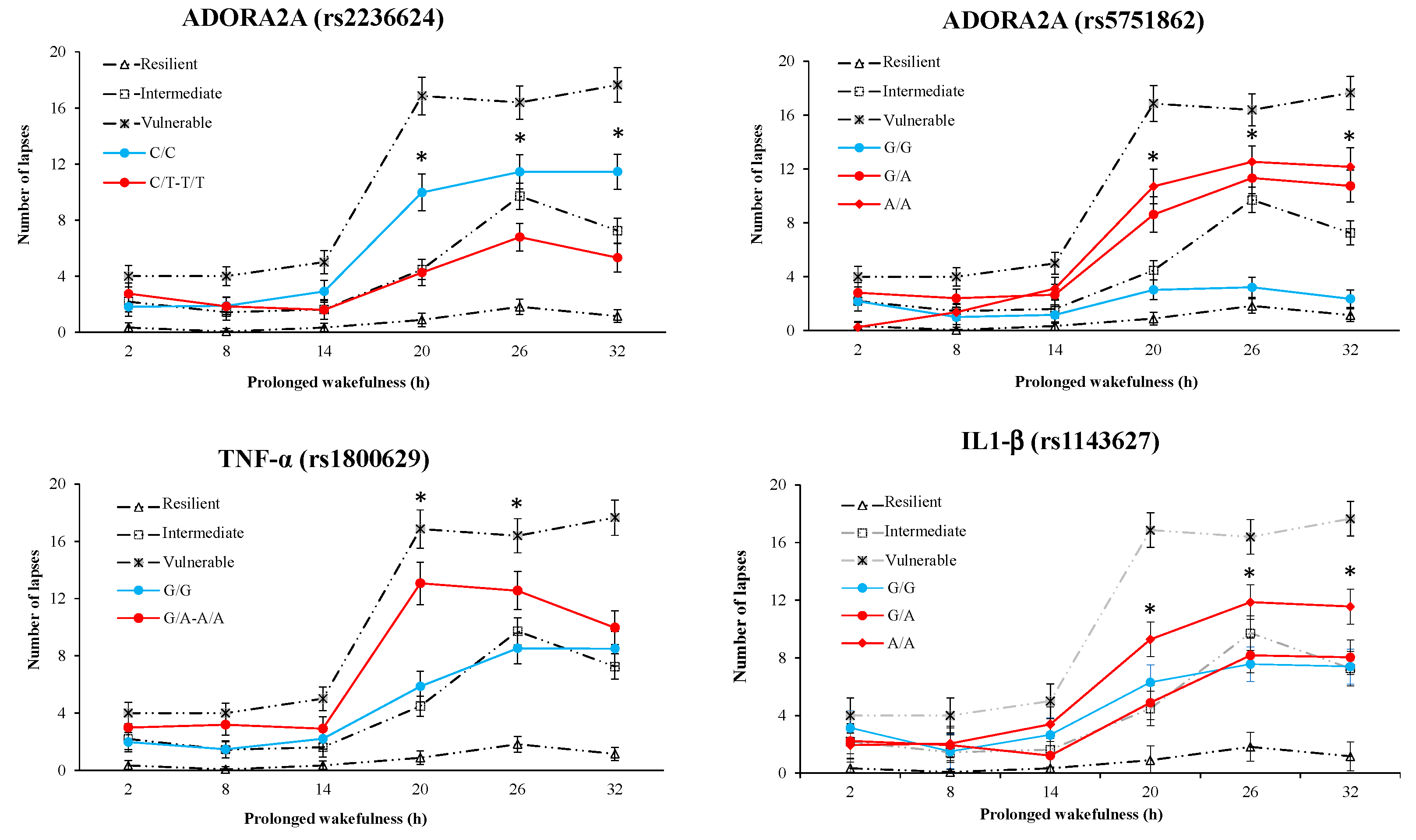
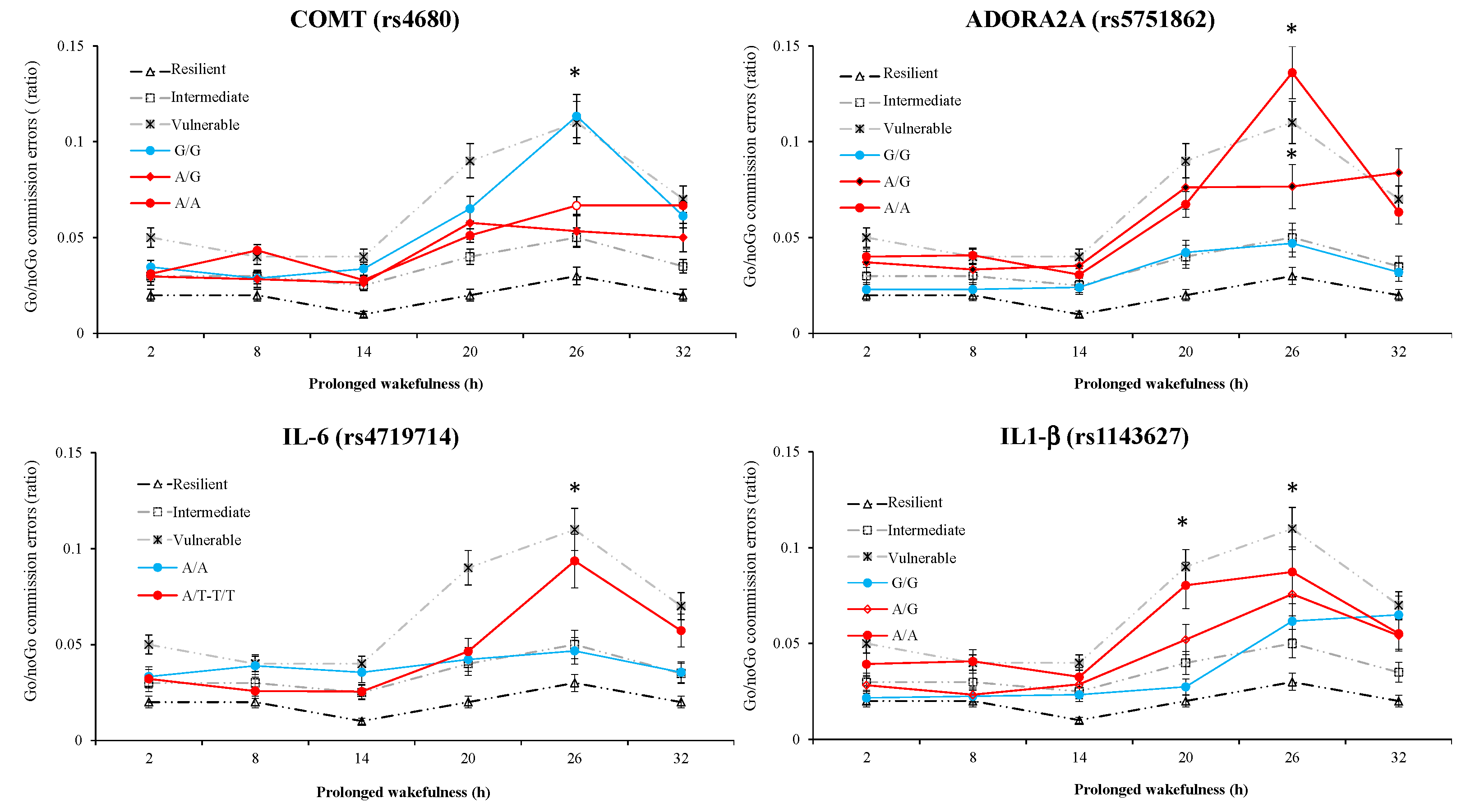
3.7. Results of Polymorphism Combination Analysis on PVT, Go/NoGo, and KSS Parameters
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Connor, J.; Whitlock, G.; Norton, R.; Jackson, R. The role of driver sleepiness in car crashes: A systematic review of epidemiological studies. Accid. Anal. Prev. 2001, 33, 31–41. [Google Scholar] [CrossRef]
- Goel, N.; Rao, H.; Durmer, J.S.; Dinges, D.F. Neurocognitive consequences of sleep deprivation. Semin. Neurol. 2009, 29, 320–339. [Google Scholar] [CrossRef] [Green Version]
- Killgore, W.D.S. Effects of sleep deprivation on cognition. In Progress in Brain Research; Elsevier: Amsterdam, The Netherlands, 2010; Volume 185, pp. 105–129. ISBN 9780444537027. [Google Scholar]
- Troxel, W.M.; Shih, R.A.; Pedersen, E.R.; Geyer, L.; Fisher, M.P.; Griffin, B.A.; Haas, A.C.; Kurz, J.; Steinberg, P.S. Sleep in the military: Promoting healthy sleep among 629 US servicemembers. Rand Health Q. 2015, 5, 19. [Google Scholar] [PubMed]
- Van Dongen, H.P.A.; Maislin, G.; Mullington, J.M.; Dinges, D.F. The cumulative cost of additional wakefulness: Dose-response effects on neurobehavioral functions and sleep physiology from chronic sleep restriction and total sleep deprivation. Sleep 2003, 26, 117–126. [Google Scholar] [CrossRef]
- Basner, M.; Dinges, D.F. Maximizing sensitivity of the psychomotor vigilance test (PVT) to sleep loss. Sleep 2011, 34, 581–591. [Google Scholar] [CrossRef]
- Arnal, P.J.; Sauvet, F.; Leger, D.; Van Beers, P.; Bayon, V.; Bougard, C.; Rabat, A.; Millet, G.Y.; Chennaoui, M. Benefits of sleep extension on sustained attention and sleep pressure before and during total sleep deprivation and recovery. Sleep 2015, 38, 1935–1943. [Google Scholar] [CrossRef]
- Van Dongen, H.P.A.; Maislin, G.; Dinges, D.F. Dealing with inter-individual differences in the temporal dynamics of fatigue and performance: Importance and techniques. Aviat. Space Environ. Med. 2004, 75, A147–A154. [Google Scholar]
- Tkachenko, O.; Dinges, D.F. Interindividual variability in neurobehavioral response to sleep loss: A comprehensive review. Neurosci. Biobehav. Rev. 2018, 89, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Rétey, J.V.; Adam, M.; Gottselig, J.M.; Khatami, R.; Dürr, R.; Achermann, P.; Landolt, H.-P. Adenosinergic mechanisms contribute to individual differences in sleep deprivation-induced changes in neurobehavioral function and brain rhythmic activity. J. Neurosci. 2006, 26, 10472–10479. [Google Scholar] [CrossRef] [PubMed]
- Goel, N. Neurobehavioral effects and biomarkers of sleep loss in healthy adults. Curr. Neurol. Neurosci. Rep. 2017, 17, 89. [Google Scholar] [CrossRef]
- Satterfield, B.C.; Stucky, B.; Landolt, H.-P.; Van Dongen, H.P.A. Unraveling the genetic underpinnings of sleep deprivation-induced impairments in human cognition. Prog. Brain Res. 2019, 246, 127–158. [Google Scholar] [CrossRef]
- Porkka-Heiskanen, T. Adenosine in sleep and wakefulness. Ann. Med. 1999, 31, 125–129. [Google Scholar] [CrossRef]
- Logue, S.F.; Gould, T.J. The neural and genetic basis of executive function: Attention, cognitive flexibility, and response inhibition. Pharmacol. Biochem. Behav. 2014, 123, 45–54. [Google Scholar] [CrossRef] [Green Version]
- Erblang, M.; Drogou, C.; Gomez-Merino, D.; Metlaine, A.; Boland, A.; Deleuze, J.F.; Thomas, C.; Sauvet, F.; Chennaoui, M. The impact of genetic variations in ADORA2A in the association between caffeine consumption and sleep. Genes 2019, 10, 1021. [Google Scholar] [CrossRef] [Green Version]
- Bachmann, V.; Klaus, F.; Bodenmann, S.; Schäfer, N.; Brugger, P.; Huber, S.; Berger, W.; Landolt, H.-P. Functional ADA polymorphism increases sleep depth and reduces vigilant attention in humans. Cereb. Cortex 2012, 22, 962–970. [Google Scholar] [CrossRef] [Green Version]
- Bodenmann, S.; Hohoff, C.; Freitag, C.; Deckert, J.; Rétey, J.V.; Bachmann, V.; Landolt, H.-P. Polymorphisms of ADORA2A modulate psychomotor vigilance and the effects of caffeine on neurobehavioural performance and sleep eeg after sleep deprivation. Br. J. Pharmacol. 2012, 165, 1904–1913. [Google Scholar] [CrossRef] [Green Version]
- Reichert, C.F.; Maire, M.; Gabel, V.; Hofstetter, M.; Viola, A.U.; Kolodyazhniy, V.; Strobel, W.; Goetz, T.; Bachmann, V.; Landolt, H.-P.; et al. The circadian regulation of sleep: Impact of a functional ada-polymorphism and its association to working memory improvements. PLoS ONE 2014, 9, e113734. [Google Scholar] [CrossRef] [Green Version]
- Erblang, M.; Sauvet, F.; Drogou, C.; Quiquempoix, M.; Van Beers, P.; Guillard, M.; Rabat, A.; Trignol, A.; Bourrilhon, C.; Erkel, M.-C.; et al. Genetic determinants of neurobehavioral responses to caffeine administration during sleep deprivation: A randomized, cross over study (NCT03859882). Genes 2021, 12, 555. [Google Scholar] [CrossRef]
- Satterfield, B.C.; Wisor, J.P.; Field, S.A.; Schmidt, M.A.; Van Dongen, H.P.A. TNFα G308A polymorphism is associated with resilience to sleep deprivation-induced psychomotor vigilance performance impairment in healthy young adults. Brain Behav. Immun. 2015, 47, 66–74. [Google Scholar] [CrossRef] [Green Version]
- Krueger, J. The role of cytokines in sleep regulation. Curr. Pharm. Des. 2008, 14, 3408–3416. [Google Scholar] [CrossRef] [Green Version]
- Knisely, M.R.; Maserati, M.; Heinsberg, L.W.; Shah, L.L.; Li, H.; Zhu, Y.; Ma, Y.; Graves, L.Y.; Merriman, J.D.; Conley, Y.P. Symptom science: Advocating for inclusion of functional genetic polymorphisms. Biol. Res. Nurs. 2019, 21, 349–354. [Google Scholar] [CrossRef]
- Louis, E.; Franchimont, D.; Piron, A.; Gevaert, Y.; Schaaf-Lafontaine, N.; Roland, S.; Mahieu, P.; Malaise, M.; De Groote, D.; Belaiche, J. Tumour necrosis factor (TNF) gene polymorphism influences TNF-alpha production in lipopolysaccharide (LPS)-stimulated whole blood cell culture in healthy humans. Clin. Exp. Immunol. 1998, 113, 401–406. [Google Scholar] [CrossRef]
- Wilson, A.G.; Symons, J.A.; McDowell, T.L.; McDevitt, H.O.; Duff, G.W. Effects of a polymorphism in the human tumor necrosis factor promoter on transcriptional activation. Proc. Natl. Acad. Sci. USA 1997, 94, 3195–3199. [Google Scholar] [CrossRef] [Green Version]
- Satterfield, B.C.; Hinson, J.M.; Whitney, P.; Schmidt, M.A.; Wisor, J.P.; Van Dongen, H.P.A. Catechol-O-methyltransferase (COMT) genotype affects cognitive control during total sleep deprivation. Cortex 2018, 99, 179–186. [Google Scholar] [CrossRef] [Green Version]
- Holst, S.C.; Müller, T.; Valomon, A.; Seebauer, B.; Berger, W.; Landolt, H.-P. Functional polymorphisms in dopaminergic genes modulate neurobehavioral and neurophysiological consequences of sleep deprivation. Sci. Rep. 2017, 7, 45982. [Google Scholar] [CrossRef] [Green Version]
- Whitney, P.; Hinson, J.M.; Satterfield, B.C.; Grant, D.A.; Honn, K.A.; Van Dongen, H.P.A. Sleep deprivation diminishes attentional control effectiveness and impairs flexible adaptation to changing conditions. Sci. Rep. 2017, 7, 16020. [Google Scholar] [CrossRef] [Green Version]
- Maire, M.; Reichert, C.F.; Gabel, V.; Viola, A.U.; Strobel, W.; Krebs, J.; Landolt, H.P.; Bachmann, V.; Cajochen, C.; Schmidt, C. Sleep ability mediates individual differences in the vulnerability to sleep loss: Evidence from a PER3 polymorphism. Cortex 2014, 52, 47–59. [Google Scholar] [CrossRef]
- Lo, J.C.; Groeger, J.A.; Santhi, N.; Arbon, E.L.; Lazar, A.S.; Hasan, S.; Von Schantz, M.; Archer, S.N.; Dijk, D.-J. Effects of partial and acute total sleep deprivation on performance across cognitive domains, individuals and circadian phase. PLoS ONE 2012, 7, e45987. [Google Scholar] [CrossRef] [Green Version]
- Bachmann, V.; Klein, C.; Bodenmann, S.; Schäfer, N.; Berger, W.; Brugger, P.; Landolt, H.P. The BDNF Val66Met polymorphism modulates sleep intensity: EEG frequency- and state-specificity. Sleep 2012, 35, 335–344. [Google Scholar] [CrossRef]
- Grant, L.K.; Cain, S.W.; Chang, A.-M.; Saxena, R.; Czeisler, C.A.; Anderson, C. Impaired cognitive flexibility during sleep deprivation among carriers of the Brain Derived Neurotrophic Factor (BDNF) Val66Met allele. Behav. Brain Res. 2018, 338, 51–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cunha, C.; Brambilla, R.; Thomas, K.L. A simple role for BDNF in learning and memory? Front. Mol. Neurosci. 2010, 3, 1. [Google Scholar] [CrossRef] [Green Version]
- Komar, A.A. Single Nucleotide Polymorphisms—Methods and Protocols, 2nd ed.; Komar, A.A., Ed.; Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar] [CrossRef] [Green Version]
- Drogou, C.; Sauvet, F.; Erblang, M.; Detemmerman, L.; Derbois, C.; Erkel, M.C.; Boland, A.; Deleuze, J.F.; Gomez-Merino, D.; Chennaoui, M. Genotyping on blood and buccal cells using loop-mediated isothermal amplification in healthy humans. Biotechnol. Rep. 2020, 26, e00468. [Google Scholar] [CrossRef] [PubMed]
- Zigmond, A.S.; Snaith, R.P. The hospital anxiety and depression scale. Acta Psychiatr. Scand. 1983, 67, 361–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johns, M.W. A new method for measuring daytime sleepiness: The epworth sleepiness scale. Sleep 1991, 14, 540–545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buysse, D.J.; Reynolds, C.F.; Monk, T.H.; Berman, S.R.; Kupfer, D.J. The Pittsburgh sleep quality index: A new instrument for psychiatric practice and research. Psychiatry Res. 1989, 28, 193–213. [Google Scholar] [CrossRef]
- Horne, J.A.; Östberg, O. A self-assessment questionnaire to determine morningness-eveningness in human circadian rhythms. Int. J. Chronobiol. 1976, 4, 97–110. [Google Scholar]
- Åkerstedt, T.; Gillberg, M. Subjective and objective sleepiness in the active individual. Int. J. Neurosci. 1990, 52, 29–37. [Google Scholar] [CrossRef]
- Rabat, A.; Gomez-Merino, D.; Roca-Paixao, L.; Bougard, C.; Van Beers, P.; Dispersyn, G.; Guillard, M.; Bourrilhon, C.; Drogou, C.; Arnal, P.J.; et al. Differential kinetics in alteration and recovery of cognitive processes from a chronic sleep restriction in young healthy men. Front. Behav. Neurosci. 2016, 10, 95. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J. Statistical Power Analysis for the Behavioral Sciences; Academic Press: Cambridge, MA, USA, 2013. [Google Scholar]
- Richardson, J.T.E. Eta squared and partial eta squared as measures of effect size in educational research. Educ. Res. Rev. 2011, 6, 135–147. [Google Scholar] [CrossRef]
- Solé, X.; Guinó, E.; Valls, J.; Iniesta, R.; Moreno, V. SNPStats: A web tool for the analysis of association studies. Bioinformatics 2006, 15, 1928–1929. [Google Scholar] [CrossRef] [Green Version]
- Drummond, S.; Paulus, M.P.; Tapert, S.F. Effects of two nights sleep deprivation and two nights recovery sleep on response inhibition. J. Sleep Res. 2006, 15, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Horn, N.R.; Dolan, M.; Elliot, R.; Deakin, J.F.W.; Woodruuf, P.R.W. Response inhibition and impulsivity: An FMRI study. Neuropsychologia 2003, 41, 1959–1966. [Google Scholar] [CrossRef]
- Childs, E.; Hohoff, C.; Deckert, J.; Xu, K.; Badner, J.; De Wit, H. Association between ADORA2A and DRD2 polymorphisms and caffeine-induced anxiety. Neuropsychopharmacology 2008, 33, 2791–2800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hohoff, C.; Garibotto, V.; Elmenhorst, D.; Baffa, A.; Kroll, T.; Hoffmann, A.; Schwarte, K.; Zhang, W.; Arolt, V.; Deckert, J.; et al. Association of adenosine receptor gene polymorphisms and in vivo adenosine A1 receptor binding in the human brain. Neuropsychopharmacology 2014, 39, 2989–2999. [Google Scholar] [CrossRef] [Green Version]
- Hohoff, C.; Kroll, T.; Zhao, B.; Kerkenberg, N.; Lang, I.; Schwarte, K.; Elmenhorst, D.; Elmenhorst, E.M.; Aeschbach, D.; Zhang, W.; et al. ADORA2A variation and adenosine A1 receptor availability in the human brain with a focus on anxiety-related brain regions: Modulation by ADORA1 variation. Transl. Psychiatry 2020, 10, 406. [Google Scholar] [CrossRef]
- Huin, V.; Dhaenens, C.M.; Homa, M.; Carvalho, K.; Buée, L.; Sablonnière, B. Neurogenetics of the human adenosine receptor genes: Genetic structures and involvement in brain diseases. J. Caffeine Adenosine Res. 2019, 9, 73–88. [Google Scholar] [CrossRef] [Green Version]
- Barrett, L.W.; Fletcher, S.; Wilton, S.D. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell. Mol. Life Sci. 2012, 69, 3613–3634. [Google Scholar] [CrossRef] [Green Version]
- Chennaoui, M.; Arnal, P.J.; Drogou, C.; Leger, D.; Sauvet, F.; Gomez-Merino, D. Leukocyte expression of type 1 and type 2 purinergic receptors and pro-inflammatory cytokines during total sleep deprivation and/or sleep extension in healthy subjects. Front. Neurosci. 2017, 11, 240. [Google Scholar] [CrossRef]
- Chennaoui, M.; Sauvet, F.; Drogou, C.; Van Beers, P.; Langrume, C.; Guillard, M.; Gourby, B.; Bourrilhon, C.; Florence, G.; Gomez-Merino, D. Effect of one night of sleep loss on changes in tumor necrosis factor alpha (TNF-α) levels in healthy men. Cytokine 2011, 56, 318–324. [Google Scholar] [CrossRef]
- Kroeger, K.M.; Steer, J.H.; Joyce, D.A.; Abraham, L.J. Effects of stimulus and cell type on the expression of the- 308 tumour necrosis factor promoter polymorphism. Cytokine 2000, 12, 110–119. [Google Scholar] [CrossRef]
- Wesensten, N.J.; Belenky, G.; Kautz, M.A.; Thorne, D.R.; Reichardt, R.M.; Balkin, T.J. Maintaining alertness and performance during sleep deprivation: Modafinil versus caffeine. Psychopharmacology 2002, 159, 238–247. [Google Scholar] [CrossRef] [PubMed]
- Tunbridge, E.M.; Harrison, P.J. Importance of the COMT gene for sex differences in brain function and predisposition to psychiatric disorders. Curr. Top. Behav. Neurosci. 2011, 8, 119–140. [Google Scholar] [PubMed]
- Tunbridge, E.M.; Narajos, M.; Harrison, C.H.; Beresford, C.; Cipriani, A.; Harrison, P.J. Which dopamine polymorphisms are functional? Systematic review and meta-analysis of COMT, DAT, DBH, DDC, DRD1–5, MAOA, MAOB, TH, VMAT1, and VMAT. Biol. Psychiatry 2019, 86, 608–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vgontzas, A.N.; Papanicolaou, D.A.; Bixler, E.O.; Lotsikas, A.; Zachman, K.; Kales, A.; Prolo, P.; Wong, M.L.; Licinio, J.; Gold, P.W.; et al. Circadian interleukin-6 secretion and quantity and depth of sleep. J. Clin. Endocrinol. Metab. 1999, 84, 2603–2607. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.J.; D’Aiuto, F.; Palmen, J.; Cooper, J.A.; Samuel, J.; Thompson, S.; Sanders, J.; Donos, N.; Nibali, L.; Brull, D.; et al. Association of serum interleukin-6 concentration with a functional IL6- 6331T> C polymorphism. Clin. Chem. 2008, 54, 841–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rétey, J.V.; Adam, M.; Honegger, E.; Khatami, R.; Luhmann, U.F.O.; Jung, H.H.; Berger, W.; Landolt, H.P. A functional genetic variation of adenosine deaminase affects the duration and intensity of deep sleep in humans. Proc. Natl. Acad. Sci. USA 2005, 102, 15676–15681. [Google Scholar] [CrossRef] [Green Version]
- Sauvet, F.; Arnal, P.J.; Tardo-Dino, P.-E.; Drogou, C.; Van Beers, P.; Erblang, M.; Guillard, M.; Rabat, A.; Malgoyre, A.; Bourrilhon, C.; et al. Beneficial effects of exercise training on cognitive performances during total sleep deprivation in healthy subjects. Sleep Med. 2020, 65, 26–35. [Google Scholar] [CrossRef] [PubMed]

| Parameters | Values |
|---|---|
| Age (years) | 37.0 ± 1.1 |
| Weight (kg) | 71.1 ± 1.4 |
| Height (m) | 1.7 ± 0.1 |
| BMI (kg.m−2) | 24.3 ± 0.7 |
| Daily caffeine consumption (mg) | 288 ± 17 |
| Total sleep time (TST) (h) 1 | 7.1 ± 0.4 |
| Physical exercise (h/week) | 3.7 ± 0.3 |
| Epworth sleepiness scale | 7.8 ± 0.7 |
| Genetic Polymorphisms | Genotypes | Observed Count N (%) | Genotypes (Considered for Analysis) a | N (%) | Expected Count from 1000 Genomes b |
|---|---|---|---|---|---|
| (%) | |||||
| rs1800629_TNF-α | G/G (ancestral) | 36 (76.6%) | G/G (ancestral) | 36 (76.6%) | 74.4% |
| (6:31.575.254) | G/A | 10 (21.3%) | G/A–A/A | 11 (23.4%) | 24.5% |
| A/A | 1 (2.1%) | 1.2% | |||
| rs1143627_IL1-β | G/G (ancestral) | 8 (17.4%) | 12.3% | ||
| (2:112.836.810) | G/A | 18 (39.1%) | 45.7% | ||
| A/A | 20 (43.5%) | 41.9% | |||
| rs4719714_IL-6 | A/A (ancestral) | 27 (57.4%) | A/A (ancestral) | 27 (57.4%) | 59.8% |
| (7:22.721.094) | A/T | 15 (31.9%) | A/T–T/T | 20 (42.6%) | 34.4% |
| T/T | 5 (10.6%) | 5.8% | |||
| rs5751862_ADORA2A | G/G (ancestral) | 12 (25.5%) | 25.0% | ||
| (22:24.406.596) | G/A | 27 (57.4%) | 50.5% | ||
| A/A | 8 (17.1%) | 24.5% | |||
| rs2298383_ADORA2A | C/C (ancestral) | 7 (15.2%) | 17.1% | ||
| (22:24.429.543) | C/T | 25 (54.3%) | 48.5% | ||
| T/T | 14 (30.5%) | 34.4% | |||
| rs3761422_ADORA2A | C/C (ancestral) | 16 (34.0%) | C/C (ancestral) | 16 (24.0%) | 38.4% |
| (22:24.430.704) | C/T | 27 (57.5%) | C/T–T/T | 31 (36.0%) | 46.3% |
| T/T | 4 (8.5%) | 15.3% | |||
| rs2236624_ADORA2A | C/C (ancestral) | 27 (57.4%) | C/C (ancestral) | 27 (57.4%) | 54.7% |
| (22:24.440.056) | C/T | 19 (40.4%) | C/T–T/T | 20 (42.6%) | 36.8% |
| T/T | 1 (2.2%) | 8.5% | |||
| rs5751876_ADORA2A | C/C (ancestral) | 15 (32.6%) | C/C (ancestral) | 15 (32.6%) | 37.4% |
| (22:24.441.33) | C/T | 27 (58.7%) | C/T–T/T | 31 (67.4%) | 47.1% |
| T/T | 4 (8.7%) | 15.5% | |||
| rs4822492_ADORA2A | C/C (ancestral) | 7 (14.9%) | 17.1% | ||
| (22:24.447.626) | C/G | 26 (55.3%) | 48.5% | ||
| G/G | 14 (29.8%) | 34.4% | |||
| rs73598374_ADA | C/C (ancestral) | 43 (93.5%) | 88.1% | ||
| (20:44.651.586) | C/T | 3 (6.5%) | 11.5% | ||
| rs228697_PER3 | C/C (ancestral) | 38 (80.9%) | C/C (ancestral) | 38 (80.9%) | 81.7% |
| (1:7.827.519) | C/G | 8 (17.0%) | C/G–G/G | 9 (19.1%) | 17.3% |
| G/G | 1 (2.1%) | 1.0% | |||
| rs4680_COMT | G/G (ancestral) | 17 (36.2%) | 26.4% | ||
| (22:19.963.748) | G/A | 18 (38.3%) | 47.1% | ||
| A/A | 12 (25.5%) | 26.4% | |||
| rs1079597_DRD2 | C/C (ancestral) | 32 (69.6%) | C/C (ancestral) | 32 (69.6%) | 73.4% |
| (11:113.425.564) | C/T | 13 (28.2%) | C/T–T/T | 14 (30.4%) | 23.5% |
| T/T | 1 (2.2%) | 3.2% | |||
| rs6265_BDNF | C/C (ancestral) | 32 (69.6%) | C/C (ancestral) | 32 (69.6%) | 64.2% |
| (11:27.658.369) | C/T | 13 (28.2%) | C/T–T/T | 14 (30.4%) | 32.2% |
| T/T | 1 (2.2%) | 3.6% |
| PVT_Lapses | PVT_Speed | Go/NoGo | KSS | |||||
|---|---|---|---|---|---|---|---|---|
| SNPs | SNP Effect | Interaction (SNP × TSD) | SNP Effect | Interaction (SNP × TSD) | SNP Effect | Interaction (SNP × TSD) | SNP Effect | Interaction (SNP × TSD) |
| rs1800629_TNF-α | p = 0.020 | p = 0.030 | p = 0.040 | p = 0.250 | p = 0.001 | p = 0250 | p = 0.010 | p = 0.001 |
| rs1143627_IL1-β | p = 0.030 | p = 0.260 | p = 0.400 | p = 0.570 | p = 0.600 | p = 0.400 | p = 0.080 | p = 0.320 |
| rs4719714_IL-6 | p = 0.040 | p = 0.930 | p = 0.040 | p = 0.570 | p = 0.010 | p = 0.280 | p = 0.040 | p = 0.470 |
| rs5751876_ADORA2A | p = 0.200 | p = 0.020 | p = 0.390 | p = 0.440 | p = 0.001 | p = 0.670 | p = 0.010 | p = 0.940 |
| rs5751862_ADORA2A | p = 0.001 | p = 0.018 | p = 0.170 | p = 0.210 | p = 0.390 | p = 0.090 | p = 0.240 | p = 0.430 |
| rs4822492_ADORA2A | p = 0.040 | p = 0.680 | p = 0.500 | p = 0.170 | p = 0.510 | p = 0.060 | p = 0.690 | p = 0.450 |
| rs2236624_ADORA2A | p = 0.006 | p = 0.007 | p = 0.390 | p = 0.240 | p = 0.010 | p = 0.060 | p = 0.990 | p = 0.450 |
| rs3761422_ADORA2A | p = 0.430 | p = 0.620 | p = 0.390 | p = 0.250 | p = 0.790 | p = 0.170 | p = 0.640 | p = 0.510 |
| rs2298383_ADORA2A | p = 0.050 | p = 0.680 | p = 0.500 | p = 0.810 | p = 0.560 | p = 0.060 | p = 0.720 | p = 0.400 |
| rs73598374_ADA | p = 0.460 | p = 0.790 | p = 0.620 | p = 0.770 | p < 0.001 | p = 0.520 | p = 0.002 | p = 0.610 |
| rs4680_COMT | p = 0.110 | p = 0.030 | p = 0.350 | p = 0.230 | p = 0.350 | p = 0.050 | p = 0.020 | p = 0.520 |
| rs1079597_DRD2 | p = 0.430 | p = 0.760 | p = 0.590 | p = 0.040 | p = 0.710 | p = 0.520 | p = 0.630 | p = 0.220 |
| rs6265_BDNF | p = 0.430 | p = 0.520 | p = 0.380 | p = 0.040 | p = 0.270 | p = 0.710 | p = 0.540 | p = 0.210 |
| rs228697_PER3 | p = 0440 | p = 0.001 | p = 0.400 | p = 0.810 | p = 0.007 | p = 0.030 | p = 0.550 | p = 0.880 |
| PVT Lapses | ||||||||
| rs1143627 IL-1β | rs1800629 TNF-α | rs5751862 ADORA2A | rs2236624 ADORA2A | Frequency (%) | Value (mean ± SD) | Difference(95% CI) | p-Value | |
| 1 | A | G | A | C | 25.3 | 12.2 ± 1.1 | --- | |
| 2 | A | G | G | C | 16.2 | 10.3 ± 0.5 | −2.8 (−8.5–2.8) | 0.32 |
| 3 | G | G | G | C | 15.2 | 7.8 ± 0.7 | −5.4 (−1.7–0.8) | 0.09 |
| 4 | G | G | A | C | 11.4 | 13.1 ± 0.8 | −0.1 (−6.9–6.6) | 0.97 |
| 5 | A | G | G | T | 9.6 | 5.4 ± 0.4 | −7.7 (−14.0–−0.5) | 0.04 |
| 6 | G | G | G | T | 9.1 | 4.1 ± 1.5 | −9.1 (−15.2–−0.9) | 0.02 |
| 7 | A | A | A | C | 9.1 | 14.9 ± 1.1 | 2.7 (−4.5–9.9) | 0.48 |
| 8 | A | A | G | T | 2.3 | 10.4 ± 0.8 | −2.8 (−5.2–9.6) | 0.66 |
| 9 | G | A | G | T | 1.6 | 11.4 ± 0.8 | 1.8 (−5.82–19.4) | 0.84 |
| Go/noGo Commission Errors | ||||||||
| rs1143627 IL-1β | rs4719714 IL-6 | rs4680 COMT | rs5751862 ADORA2A | Frequency(%) | Value (mean ± SD) | Difference (95% CI) | p-Value | |
| 1 | A | A | G | G | 14.3 | 0.18 ± 0.03 | ||
| 2 | G | A | G | G | 13.0 | 0.11 ± 0.03 | −0.08 (−0.13–−0.02) | <0.01 |
| 3 | A | A | G | A | 11.9 | 0.16 ± 0.02 | −0.03 (−0.12–0.04) | 0.49 |
| 4 | A | A | A | A | 10.2 | 0.08 ± 0.01 | −0.10 (−0.17–−0.03) | <0.01 |
| 5 | G | A | G | A | 8.8 | 0.14 ± 0.02 | −0.04 (−0.11–0.03) | 0.29 |
| 6 | A | T | A | A | 8.3 | 0.13 ± 0.01 | −0.05 (−0.11–0.01) | 0.09 |
| 7 | A | T | G | G | 7.4 | 0.07 ± 0.01 | −0.10 (−0.17–-0.03) | <0.01 |
| 8 | G | A | A | A | 6.6 | 0.13 ± 0.02 | −0.05 (−0.13–0.02) | 0.16 |
| 9 | G | T | A | G | 6.4 | 0.12 ± 0.03 | −0.06 (−0.12–0.01) | 0.11 |
| 10 | A | A | A | G | 6.3 | 0.11 ± 0.02 | −0.08 (−0.15–−0.01) | 0.03 |
| 11 | A | T | A | G | 4.5 | 0.11 ± 0.01 | −0.07 (−0.16–0.01) | 0.08 |
| KSS Score | ||||||||
| rs1800629 TNF-α | rs4719714 IL-6 | rs73598374 ADA | rs5751876 ADORA2A | Frequency(%) | Value (mean ± SD) | Difference (95% CI) | p-Value | |
| 1 | G | A | C | C | 50.6 | 5.7 ± 0.5 | 0 | --- |
| 2 | G | T | C | C | 17.8 | 5.3 ± 0.4 | −0.4 (−1.2–0.4) | 0.36 |
| 3 | G | A | C | T | 8.8 | 5.1 ± 0.4 | −0.6 (−1.9–0.6) | 0.31 |
| 4 | A | A | C | C | 8.7 | 4.8 ± 0.3 | −0.9 (−2.1–0.3) | 0.16 |
| 5 | G | T | C | T | 7.5 | 4.2 ± 0.4 | −1.5 (−2.8–-0.2) | 0.03 |
| 6 | A | A | C | T | 4.1 | 5.8 ± 0.5 | 0.1 (−1.8–1.9) | 0.92 |
| 7 | G | A | T | T | 1.3 | 2.8 ± 0.3 | −2.9 (−6.2–0.4) | 0.09 |
| 8 | G | T | T | C | 1.3 | 4.6 ± 0.3 | −1.15 (−3.9–1.6) | 0.42 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erblang, M.; Drogou, C.; Gomez-Merino, D.; Rabat, A.; Guillard, M.; Beers, P.V.; Quiquempoix, M.; Boland, A.; Deleuze, J.F.; Olaso, R.; et al. Genetics and Cognitive Vulnerability to Sleep Deprivation in Healthy Subjects: Interaction of ADORA2A, TNF-α and COMT Polymorphisms. Life 2021, 11, 1110. https://doi.org/10.3390/life11101110
Erblang M, Drogou C, Gomez-Merino D, Rabat A, Guillard M, Beers PV, Quiquempoix M, Boland A, Deleuze JF, Olaso R, et al. Genetics and Cognitive Vulnerability to Sleep Deprivation in Healthy Subjects: Interaction of ADORA2A, TNF-α and COMT Polymorphisms. Life. 2021; 11(10):1110. https://doi.org/10.3390/life11101110
Chicago/Turabian StyleErblang, Mégane, Catherine Drogou, Danielle Gomez-Merino, Arnaud Rabat, Mathias Guillard, Pascal Van Beers, Michael Quiquempoix, Anne Boland, Jean François Deleuze, Robert Olaso, and et al. 2021. "Genetics and Cognitive Vulnerability to Sleep Deprivation in Healthy Subjects: Interaction of ADORA2A, TNF-α and COMT Polymorphisms" Life 11, no. 10: 1110. https://doi.org/10.3390/life11101110
APA StyleErblang, M., Drogou, C., Gomez-Merino, D., Rabat, A., Guillard, M., Beers, P. V., Quiquempoix, M., Boland, A., Deleuze, J. F., Olaso, R., Derbois, C., Prost, M., Dorey, R., Léger, D., Thomas, C., Chennaoui, M., & Sauvet, F. (2021). Genetics and Cognitive Vulnerability to Sleep Deprivation in Healthy Subjects: Interaction of ADORA2A, TNF-α and COMT Polymorphisms. Life, 11(10), 1110. https://doi.org/10.3390/life11101110








