The Ground-Based BIOMEX Experiment Verification Tests for Life Detection on Mars
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation and Cultivation Conditions
2.2. Test Facilities and Exposure Conditions
2.3. DNA Integrity Assay
2.3.1. DNA Extraction and Quantification
2.3.2. DNA Amplification
2.3.3. Statistical Analysis
2.4. Fungal Melanin Assay
2.4.1. Pigment Extraction and Quantification
2.4.2. Pigment Detection
2.5. Fungal Organic Compound Detection
3. Results
3.1. DNA Damage Assessment
3.2. Pigment Characterization by UV-Vis Spectrophotometry
3.3. Pigment Characterization by Raman Spectroscopy
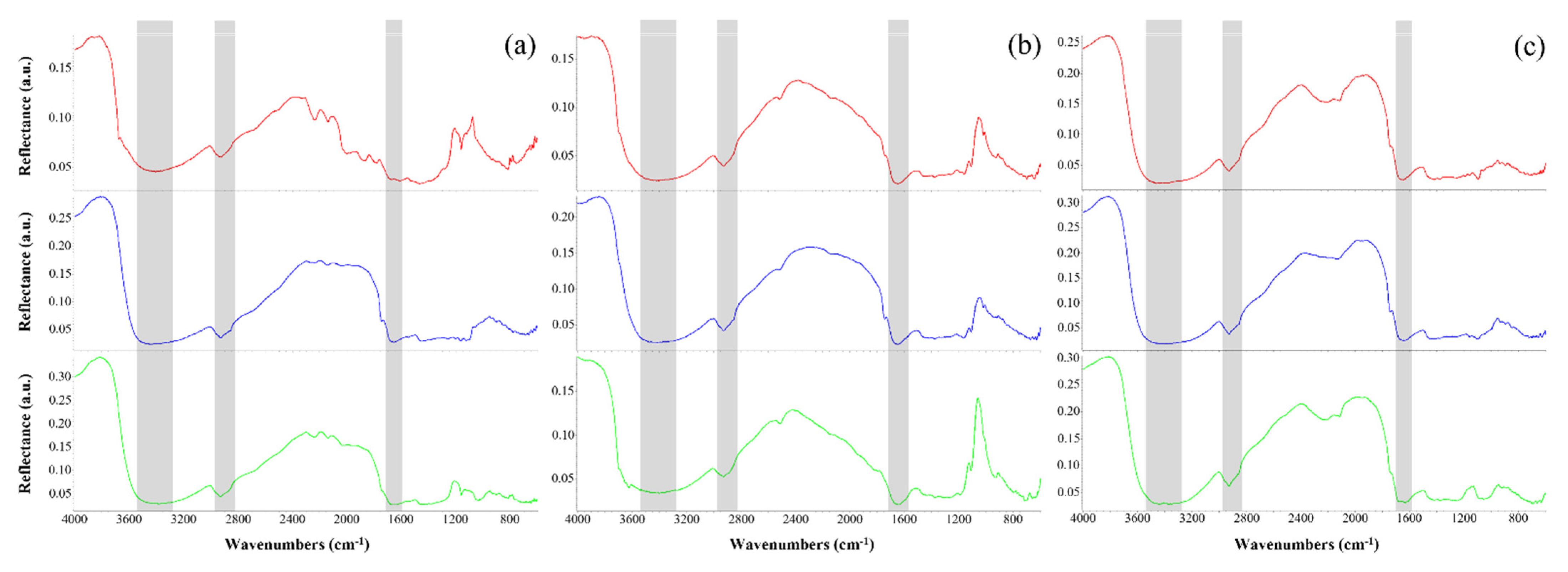
3.4. Organic Compounds Characterization by FT-IR Spectroscopy
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aerts, J.W.; Röling, W.F.; Elsaesser, A.; Ehrenfreund, P. Biota and biomolecules in extreme environments on Earth: Implications for life detection on Mars. Life 2014, 4, 535–565. [Google Scholar] [CrossRef] [PubMed]
- Engel, M.H.; Macko, S.A. (Eds.) Organic Geochemistry: Principles and Applications; Springer US: New York, NY, USA, 1993. [Google Scholar] [CrossRef]
- Simoneit, B.R.; Summons, R.E.; Jahnke, L.L. Biomarkers as tracers for life on early Earth and Mars. Orig. Life Evol. Biosph. 1998, 28, 475–483. [Google Scholar] [CrossRef]
- Cockell, C.S.; Schuerger, A.C.; Billi, D.; Friedmann, E.I.; Panitz, C. Effects of a simulated martian UV flux on the cyanobacterium, Chroococcidiopsis sp.029. Astrobiology 2005, 5, 127–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dartnell, L.R.; Patel, M.R.; Storrie-Lombardi, M.C.; Ward, J.M.; Muller, J.P. Experimental determination of photostability and fluorescence-based detection of PAHs on the Martian surface. Meteorit. Planet. Sci. 2012, 47, 806–819. [Google Scholar] [CrossRef] [Green Version]
- Dartnell, L.R.; Patel, M.R. Degradation of microbial fluorescence biosignatures by solar ultraviolet radiation on Mars. Int. J. Astrobiol. 2014, 13, 112–123. [Google Scholar] [CrossRef]
- Poch, O.; Noblet, A.; Stalport, F.; Correia, J.J.; Grand, N.; Szopa, C.; Coll, P. Chemical evolution of organic molecules under Mars-like UV radiation conditions simulated in the laboratory with the “Mars organic molecule irradiation and evolution” (MOMIE) setup. Planet. Space Sci. 2013, 85, 188–197. [Google Scholar] [CrossRef]
- Friedmann, E.I.; Ocampo-Friedmann, R. The Antarctic cryptoendolithic ecosystem: Relevance to exobiology. Orig. Life 1984, 14, 771–776. [Google Scholar] [CrossRef]
- Patel, M.R.; Zarnecki, J.C.; Catling, D.C. Ultraviolet radiation on the surface of Mars and the Beagle 2 UV sensor. Planet. Space Sci. 2002, 50, 915–927. [Google Scholar] [CrossRef]
- Rothschild, L.J.; Cockell, C.S. Radiation: Microbial evolution, ecology, and relevance to Mars missions. Mutat. Res. Fundam. Mol. Mech. Mutagen. 1999, 430, 281–291. [Google Scholar] [CrossRef]
- Gómez-Elvira, J.; Armiens, C.; Carrasco, I.; Genzer, M.; Gómez, F.; Haberle, R.; Hamilton, V.E.; Harri, A.-M.; Kahanpää, H.; Kemppinen, O.; et al. Curiosity’s rover environmental monitoring station: Overview of the first 100 sols. J. Geophys. Res. Planets 2014, 119, 1680–1688. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, W.R.; Atreya, S.K. Solar radiation incident on the Martian surface. J. Mol. Evol. 1979, 14, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Schuerger, A.C.; Mancinelli, R.L.; Kern, R.G.; Rothschild, L.J.; McKay, C.P. Survival of endospores of Bacillus subtilis on spacecraft surfaces under simulated Martian environments: Implications for the forward contamination of Mars. Icarus 2003, 165, 253–276. [Google Scholar] [CrossRef]
- Stalport, F.; Coll, P.; Szopa, C.; Cottin, H.; Raulin, F. Investigating the photostability of carboxylic acids exposed to Mars surface ultraviolet radiation conditions. Astrobiology 2009, 9, 543–549. [Google Scholar] [CrossRef] [Green Version]
- Carrier, B.L.; Kounaves, S.P. The origins of perchlorate in the Martian soil. Geophys. Res. Lett. 2015, 42, 3739–3745. [Google Scholar] [CrossRef]
- Encrenaz, T.; Greathouse, T.K.; Lefèvre, F.; Atreya, S.K. Hydrogen peroxide on Mars: Observations, interpretation and future plans. Planet. Space Sci. 2012, 68, 3–17. [Google Scholar] [CrossRef]
- Glavin, D.P.; Freissinet, C.; Miller, K.E.; Eigenbrode, J.L.; Brunner, A.E.; Buch, A.; Sutter, B.; Archer, P.D., Jr.; Atreya, S.K.; Brinckerhoff, W.B.; et al. Evidence for perchlorates and the origin of chlorinated hydrocarbons detected by SAM at the Rocknest aeolian deposit in Gale Crater. J. Geophys. Res. Planets 2013, 118, 1955–1973. [Google Scholar] [CrossRef]
- Ming, D.W.; Archer, P.D., Jr.; Glavin, D.P.; Eigenbrode, J.L.; Franz, H.B.; Sutter, B.; Brunner, A.E.; Stern, J.C.; Freissinet, C.; McAdam, A.C.; et al. Volatile and organic compositions of sedimentary rocks in Yellowknife Bay, Gale Crater, Mars. Science 2014, 343, 1245267. [Google Scholar] [CrossRef]
- Quinn, R.C.; Martucci, H.F.; Miller, S.R.; Bryson, C.E.; Grunthaner, F.J.; Grunthaner, P.J. Perchlorate radiolysis on Mars and the origin of martian soil reactivity. Astrobiology 2013, 13, 515–520. [Google Scholar] [CrossRef] [Green Version]
- Hassler, D.M.; Zeitlin, C.; Wimmer-Schweingruber, R.F.; Ehresmann, B.; Rafkin, S.; Eigenbrode, J.L.; Brinza, D.E.; Weigle, G.; Böttcher, S.; Böhm, E. Mars’ Surface Radiation Environment Measured with the Mars Science Laboratory’s Curiosity Rover. Science 2014, 343, 1244797. [Google Scholar] [CrossRef] [Green Version]
- Moores, J.E.; Smith, P.H.; Tanner, R.; Schuerger, A.C.; Venkateswaran, K.J. The shielding effect of small-scale Martian surface geometry on ultraviolet flux. Icarus 2007, 192, 417–433. [Google Scholar] [CrossRef]
- Patel, M.R.; Bérces, A.; Kolb, C.; Lammer, H.; Rettberg, P.; Zarnecki, J.C.; Selsis, F. Seasonal and diurnal variations in Martian surface ultraviolet irradiation: Biological and chemical implications for the Martian regolith. Int. J. Astrobiol. 2003, 2, 21–34. [Google Scholar] [CrossRef] [Green Version]
- Schuerger, A.C.; Moores, J.E.; Clausen, C.A.; Barlow, N.G.; Britt, D.T. Methane from UV-irradiated carbonaceous chondrites under simulated Martian conditions. J. Geophys. Res. Planets 2012, 117, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Reyes, G.; Pilorget, C.; Moral, A.G.; Manrique, J.A.; Sanz, A.; Berrocal, A.; Veneranda, M.; Rull, F.; Medina, J.; Hamm, V.; et al. Raman Laser Spectrometer (RLS) calibration target design to allow onboard combined science between the RLS and MicrOmega instruments on the ExoMars rover. J. Raman Spectrosc. 2020, 51, 1718–1730. [Google Scholar] [CrossRef]
- Kinch, K.M.; Madsen, M.B.; Bell, J.F.; Maki, J.N.; Bailey, Z.J.; Hayes, A.G.; Jensen, O.B.; Merusi, M.; Bernt, M.H.; Sørensen, A.N.; et al. Radiometric calibration targets for the Mastcam-Z Camera on the Mars 2020 Rover mission. Space Sci. Rev. 2020, 216, 1–51. [Google Scholar] [CrossRef]
- De Vera, J.-P.; Alawi, M.; Backhaus, T.; Baqué, M.; Billi, D.; Böttger, U.; Berger, T.; Bohmeier, M.; Cockell, C.S.; Demets, R.; et al. Limits of life and the habitability of Mars: The ESA space experiment BIOMEX on the ISS. Astrobiology 2019, 19, 145–157. [Google Scholar] [CrossRef] [Green Version]
- Rabbow, E.; Rettberg, P.; Parpart, A.; Panitz, C.; Schulte, W.; Molter, F.; Schulte, W.; Jaramillo, E.; Demets, R.; Weiß, P.; et al. EXPOSE R2: The astrobiological ESA mission on board of the International Space Station. Front. Microbiol. 2017, 8, 1533. [Google Scholar] [CrossRef] [PubMed]
- Selbmann, L.; de Hoog, G.S.; Mazzaglia, A.; Friedmann, E.I.; Onofri, S. Fungi at the edge of life: Cryptoendolithic black fungi from Antarctic desert. Stud. Mycol. 2005, 51, 1–32. [Google Scholar]
- Böttger, U.; de Vera, J.-P.; Fritz, J.; Weber, I.; Hübers, H.W.; Schulze-Makuch, D. Optimizing the detection of carotene in cyanobacteria in a Martian regolith analogue with a Raman spectrometer for the ExoMars mission. Planet. Space Sci. 2012, 60, 356–362. [Google Scholar] [CrossRef]
- Cockell, C.S.; Catling, D.C.; Davis, W.L.; Snook, K.; Kepner, R.L.; Lee, P.; McKay, C.P. The ultraviolet environment of Mars: Biological implications past, present, and future. Icarus 2000, 146, 343–359. [Google Scholar] [CrossRef] [Green Version]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [Green Version]
- Hopple, J.S., Jr.; Vilgalys, R. Phylogenetic relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: Divergent domains, outgroups, and monophyly. Mol. Phylogenet. Evol. 1999, 13, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Selbmann, L.; Isola, D.; Zucconi, L.; Onofri, S. Resistance to UV-B induced DNA damage in extreme-tolerant cryptoendolithic Antarctic fungi: Detection by PCR assays. Fungal Biol. 2011, 115, 937–944. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; Computer Software, Version 3.5.2; R Foundation for Statistical Computing: Vienna, Austria, 2018; Available online: http://www.R-project.org/ (accessed on 8 August 2019).
- RStudio Team. RStudio: Integrated Development for R; Computer Software, Version 1.1.423; RStudio, PBC: Boston, MA, USA, 2016; Available online: http://www.rstudio.org/ (accessed on 8 August 2019).
- Pacelli, C.; Cassaro, A.; Maturilli, A.; Timperio, A.M.; Gevi, F.; Cavalazzi, B.; Stefan, M.; Ghica, D.; Onofri, S. Multidisciplinary characterization of melanin pigments from the black fungus Cryomyces antarcticus. Appl. Microbiol. Biotechnol. 2020, 104, 6385–6395. [Google Scholar] [CrossRef] [PubMed]
- Raman, N.M.; Ramasamy, S. Genetic validation and spectroscopic detailing of DHN-melanin extracted from an environmental fungus. Biochem. Biophys. Rep. 2017, 12, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Pacelli, C.; Cassaro, A.; Baqué, M.; Selbmann, L.; Zucconi, L.; Maturilli, A.; Onofri, S. Fungal biomarkers are detectable in Martian rock-analogues after space exposure: Implications for the search of life on Mars. Int. J. Astrobiol. 2021, 20, 1–14. [Google Scholar] [CrossRef]
- Menges, F. Spectragryph—Optical Spectroscopy Software. Computer Software, Version 1.2.14. 2019. Available online: http://www.effemm2.de/spectragryph/ (accessed on 7 February 2020).
- Bell, A.A.; Wheeler, M.H. Biosynthesis and functions of fungal melanins. Annu. Rev. Phytopathol. 1986, 24, 411–451. [Google Scholar] [CrossRef]
- Cockell, C.S.; Knowland, J. Ultraviolet radiation screening compounds. Biol. Rev. 1999, 74, 311–345. [Google Scholar] [CrossRef]
- Centeno, S.A.; Shamir, J. Surface enhanced Raman scattering (SERS) and FTIR characterization of the sepia melanin pigment used in works of art. J. Mol. Struct. 2008, 873, 149–159. [Google Scholar] [CrossRef]
- Culka, A.; Jehlička, J.; Ascaso, C.; Artieda, O.; Casero, C.M.; Wierzchos, J. Raman microspectrometric study of pigments in melanized fungi from the hyperarid Atacama desert gypsum crust. J. Raman Spectrosc. 2017, 48, 1487–1493. [Google Scholar] [CrossRef]
- De Gussem, K.; Vandenabeele, P.; Verbeken, A.; Moens, L. Raman spectroscopic study of Lactarius spores (Russulales, Fungi). Spectrochim. Acta A Mol. Biomol. Spectrosc. 2005, 61, 2896–2908. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Lui, H.; Chen, M.X.; Alajlan, A.; McLean, D.I.; Zeng, H. Raman spectroscopy of in vivo cutaneous melanin. J. Biomed. Opt. 2004, 9, 1198–1206. [Google Scholar] [CrossRef] [PubMed]
- Maquelin, K.; Choo-Smith, L.P.; Kirschner, C.; Ngo-Thi, N.A.; Naumann, D.; Puppels, G.J. Vibrational spectroscopic studies of microorganisms. Handb. Vib. Spectrosc. 2002, 5, 3308–3334. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Rösch, P.; Harz, M.; Peschke, K.D.; Ronneberger, O.; Burkhardt, H.; Popp, J. Identification of single eukaryotic cells with micro-Raman spectroscopy. Biopolymers 2006, 82, 312–316. [Google Scholar] [CrossRef]
- Samokhvalov, A.; Liu, Y.; Simon, J.D. Characterization of the Fe (III)-binding Site in Sepia Eumelanin by Resonance Raman Confocal Microspectroscopy. Photochem. Photobiol. 2004, 80, 84–88. [Google Scholar] [CrossRef]
- Saif, F.A.; Yaseen, S.A.; Alameen, A.S.; Mane, S.B.; Undre, P.B. Identification and characterization of Aspergillus species of fruit rot fungi using microscopy, FT-IR, Raman and UV–Vis spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 246, 119010. [Google Scholar] [CrossRef] [PubMed]
- Hou, R.; Liu, X.; Xiang, K.; Chen, L.; Wu, X.; Lin, W.; Zheng, M.; Fu, J. Characterization of the physicochemical properties and extraction optimization of natural melanin from Inonotus hispidus mushroom. Food Chem. 2019, 277, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Jones, R.A. Physicochemical Properties of Pyrroles. Adv. Heterocycl. Chem. 1970, 20, 383–472. [Google Scholar] [CrossRef]
- Magarelli, M.; Passamonti, P.; Renieri, C. Purification, characterization and analysis of sepia melanin from commercial sepia ink (Sepia Officinalis). Rev. CES Med. Vet. Zootec. 2010, 5, 18–29. [Google Scholar]
- Movasaghi, Z.; Rehman, S.; Rehman, D.I. Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Safar, M.; Bertrand, D.; Robert, P.; Devaux, M.F.; Genot, C. Characterization of edible oils, butters and margarines by Fourier transform infrared spectroscopy with attenuated total reflectance. J. Am. Oil Chem. Soc. 1994, 71, 371–377. [Google Scholar] [CrossRef]
- Coates, J. Interpretation of infrared spectra, a practical approach. In Encyclopedia of Analytical Chemistry: Applications, Theory and Instrumentation; Meyers, R.A., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2000; pp. 10815–10837. [Google Scholar]
- Schulz, H.; Baranska, M. Identification and quantification of valuable plant substances by IR and Raman spectroscopy. Vib. Spectrosc. 2007, 43, 13–25. [Google Scholar] [CrossRef]
- Stuart, B.H. Infrared Spectroscopy: Fundamentals and Applications; John Wiley & Sons, Ltd.: Chichester, UK, 2004; pp. 71–93. [Google Scholar]
- Kosa, G.; Kohler, A.; Tafintseva, V.; Zimmermann, B.; Forfang, K.; Afseth, N.K.; Tzimorotas, D.; Vuoristo, K.S.; Horn, S.J.; Mounier, J.; et al. Microtiter plate cultivation of oleaginous fungi and monitoring of lipogenesis by high-throughput FTIR spectroscopy. Microb. Cell Fact. 2017, 16, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Salman, A.; Shufan, E.; Tsror, L.; Moreh, R.; Mordechai, S.; Huleihel, M. Classification of Colletotrichum coccodes isolates into vegetative compatibility groups using infrared attenuated total reflectance spectroscopy and multivariate analysis. Methods 2014, 68, 325–330. [Google Scholar] [CrossRef]
- Shurvell, H.F. Spectra–structure correlations in the mid-and far-infrared. In Handbook of Vibrational Spectroscopy; Chalmers, J.M., Griffiths, P.R., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2006; Volume 1, pp. 1–34. [Google Scholar] [CrossRef]
- Guillén, M.D.; Cabo, N. Usefulness of the frequency data of the Fourier transform infrared spectra to evaluate the degree of oxidation of edible oils. J. Agric. Food Chem. 1999, 47, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Irudayaraj, J.; Sivakesava, S.; Kamath, S.; Yang, H. Monitoring chemical changes in some foods using Fourier transform photoacoustic spectroscopy. J. Food Sci. 2001, 66, 1416–1421. [Google Scholar] [CrossRef]
- Gladman, B.J.; Burns, J.A.; Duncan, M.; Lee, P.; Levison, H.F. The exchange of impact ejecta between terrestrial planets. Science 1996, 271, 1387–1392. [Google Scholar] [CrossRef]
- Melosh, H.J. The rocky road to panspermia. Nature 1988, 332, 687–688. [Google Scholar] [CrossRef]
- Mileikowsky, C.; Cucinotta, F.A.; Wilson, J.W.; Gladman, B.; Horneck, G.; Lindegren, L.; Melosh, J.; Rickman, H.; Valtonen, M.; Zheng, J.Q. Natural transfer of viable microbes in space: 1. From Mars to Earth and Earth to Mars. Icarus 2000, 145, 391–427. [Google Scholar] [CrossRef]
- Pavlov, A.A.; Vasilyev, G.; Ostryakov, V.M.; Pavlov, A.K.; Mahaffy, P. Degradation of the organic molecules in the shallow subsurface of Mars due to irradiation by cosmic rays. Geophys. Res. Lett. 2012, 39, L13202. [Google Scholar] [CrossRef] [Green Version]
- Hays, L.E.; Graham, H.V.; Des Marais, D.J.; Hausrath, E.M.; Horgan, B.; McCollom, T.M.; Parenteau, M.N.; Potter-McIntyre, S.L.; Williams, A.J.; Lynch, K.L. Biosignature preservation and detection in Mars analog environments. Astrobiology 2017, 17, 363–400. [Google Scholar] [CrossRef]
- Williams, A.J.; Craft, K.L.; Millan, M.; Johnson, S.S.; Knudson, C.A.; Juarez Rivera, M.; McAdam, A.C.; Tobler, D.; Skok, J.R. Fatty Acid Preservation in Modern and Relict Hot-Spring Deposits in Iceland, with Implications for Organics Detection on Mars. Astrobiology 2021, 21, 60–82. [Google Scholar] [CrossRef]
- National Research Council. An Astrobiology Strategy for the Exploration of Mars; The National Academies Press: Washington, DC, USA, 2007. [Google Scholar] [CrossRef]
- Summons, R.E.; Amend, J.P.; Bish, D.; Buick, R.; Cody, G.D.; Des Marais, D.J.; Dromart, G.; Eigenbrode, J.L.; Knoll, A.H.; Sumner, D.Y. Preservation of Martian organic and environmental records: Final report of the Mars Biosignature Working Group. Astrobiology 2011, 11, 157–181. [Google Scholar] [CrossRef]
- Maggiori, C.; Stromberg, J.; Blanco, Y.; Goordial, J.; Cloutis, E.; García-Villadangos, M.; Parro, V.; Whyte, L. The Limits, Capabilities, and Potential for Life Detection with MinION Sequencing in a Paleochannel Mars Analog. Astrobiology 2020, 20, 375–393. [Google Scholar] [CrossRef] [PubMed]
- Pacelli, C.; Bryan, R.A.; Onofri, S.; Selbmann, L.; Shuryak, I.; Dadachova, E. Melanin is effective in protecting fast and slow growing fungi from various types of ionizing radiation. Environ. Microbiol. 2017, 19, 1612–1624. [Google Scholar] [CrossRef] [PubMed]
- Pacelli, C.; Cassaro, A.; Aureli, L.; Moeller, R.; Fujimori, A.; Onofri, S. The Responses of the Black Fungus Cryomyces antarcticus to High Doses of Accelerated Helium Ions Radiation within Martian Regolith Simulants and Their Relevance for Mars. Life 2020, 10, 130. [Google Scholar] [CrossRef]
- Meeßen, J.; Sánchez, F.J.; Sadowsky, A.; de la Torre, R.; Ott, S.; de Vera, J.P. Extremotolerance and Resistance of Lichens: Comparative Studies on Five Species Used in Astrobiological Research II. Secondary Lichen Compounds. Orig. Life Evol. Biosph. 2013, 43, 501–526. [Google Scholar] [CrossRef] [Green Version]
- Pal, A.K.; Gajjar, D.U.; Vasavada, A.R. DOPA and DHN pathway orchestrate melanin synthesis in Aspergillus species. Med. Mycol. 2013, 52, 10–18. [Google Scholar] [CrossRef] [Green Version]
- Suryanarayanan, T.S.; Ravishankar, J.P.; Venkatesan, G.; Murali, T.S. Characterization of the melanin pigment of a cosmopolitan fungal endophyte. Mycol. Res. 2004, 108, 974–978. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Burleigh, S.H.; Dawson, J.O. Melanin biosynthesis by Frankia strain CeI5. Physiol. Plant. 2007, 131, 180–190. [Google Scholar] [CrossRef]
- Mach, H.; Volkin, D.B.; Burke, C.J.; Middaugh, C.R. Ultraviolet Absorption Spectroscopy. In Protein Stability and Folding. Methods in Molecular Biology; Shirley, B.A., Ed.; Humana Press: Totowa, NJ, USA, 1995; Volume 40, pp. 91–114. [Google Scholar] [CrossRef]
- Baqué, M.; Hanke, F.; Böttger, U.; Leya, T.; Moeller, R.; de Vera, J.-P. Protection of cyanobacterial carotenoids’ Raman signatures by Martian mineral analogues after high-dose gamma irradiation. J. Raman Spectrosc. 2018, 49, 1617–1627. [Google Scholar] [CrossRef]
- Dartnell, L.R.; Storrie-Lombardi, M.C.; Mullineaux, C.W.; Ruban, A.V.; Wright, G.; Griffiths, A.D.; Muller, J.P.; Ward, J.M. Degradation of cyanobacterial biosignatures by ionizing radiation. Astrobiology 2011, 11, 997–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibring, J.P.; Hamm, V.; Pilorget, C.; Vago, J.L. The micrOmega investigation onboard ExoMars. Astrobiology 2017, 17, 621–626. [Google Scholar] [CrossRef]
- De Sanctis, M.C.; Altieri, F.; Ammannito, E.; Biondi, D.; De Angelis, S.; Meini, M.; Mondello, G.; Novi, S.; Paolinetti, R.; Soldani, M.; et al. Ma_MISS on ExoMars: Mineralogical characterization of the Martian subsurface. Astrobiology 2017, 17, 612–620. [Google Scholar] [CrossRef]
- Rull, F.; Maurice, S.; Hutchinson, I.; Moral, A.; Perez, C.; Diaz, C.; Colombo, M.; Belenguer, T.; Lopez-Reyes, G.; Parot, Y.; et al. The Raman laser spectrometer for the ExoMars rover mission to Mars. Astrobiology 2017, 17, 627–654. [Google Scholar] [CrossRef] [Green Version]
- Vago, J.L.; Westall, F.; Pasteur Instrument Teams; Landing Site Selection Working Group; Other Contributors. Habitability on early Mars and the search for biosignatures with the ExoMars Rover. Astrobiology 2017, 17, 471–510. [Google Scholar] [CrossRef]
- Kendall, C.; Isabelle, M.; Bazant-Hegemark, F.; Hutchings, J.; Orr, L.; Babrah, J.; Baker, R.; Stone, N. Vibrational spectroscopy: A clinical tool for cancer diagnostics. Analyst 2009, 134, 1029–1045. [Google Scholar] [CrossRef]
- McCreery, R.L. Raman spectroscopy for chemical analysis. Meas. Sci. Technol. 2001, 12, 5–653. [Google Scholar] [CrossRef] [Green Version]
- Bonner, T.G.; Duncan, A. Infra-red spectra of some melanins. Nature 1962, 194, 1078–1079. [Google Scholar] [CrossRef]
- Kannan, P.; Ganjewala, D. Preliminary characterization of melanin isolated from fruits and seeds of Nyctanthes arbor-tristis. J. Sci. Res. 2009, 1, 655–661. [Google Scholar] [CrossRef] [Green Version]
- Sajjan, S.; Kulkarni, G.; Yaligara, V.; Kyoung, L.; Karegoudar, T.B. Purification and physiochemical characterization of melanin pigment from Klebsiella sp. GSK. J. Microbiol. Biotechnol. 2010, 20, 1513–1520. [Google Scholar] [CrossRef] [Green Version]
- Paim, S.; Linhares, L.F.; Mangrich, A.S.; Martin, J.P. Characterization of fungal melanins and soil humic acids by chemical analysis and infrared spectroscopy. Biol. Fertil. Soils 1990, 10, 72–76. [Google Scholar] [CrossRef]
- Sun, S.; Zhang, X.; Sun, S.; Zhang, L.; Shan, S.; Zhu, H. Production of natural melanin by Auricularia auricula and study on its molecular structure. Food Chem. 2016, 190, 801–807. [Google Scholar] [CrossRef] [PubMed]
- Forfang, K.; Zimmermann, B.; Kosa, G.; Kohler, A.; Shapaval, V. FTIR spectroscopy for evaluation and monitoring of lipid extraction efficiency for oleaginous fungi. PLoS ONE 2017, 12, e0170611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivshin, V.P.; Artamonova, S.D.; Ivshina, T.N.; Sharnina, F.F. Methods for isolation of chitin-glucan complexes from higher fungi native biomass. Polym. Sci. Ser. B 2007, 49, 305–310. [Google Scholar] [CrossRef]
- Rinaudo, M. Chitin and chitosan: Properties and applications. Prog. Polym. Sci. 2006, 31, 603–632. [Google Scholar] [CrossRef]
- Böttger, U.; Meeßen, J.; Martínez-Frías, J.; Hübers, H.W.; Rull, F.; Sánchez, F.J.; de la Torre, R.; de Vera, J.P. Raman spectroscopic analysis of the calcium oxalate producing extremotolerant lichen Circinaria gyrosa. Int. J. Astrobiol. 2014, 13, 19–27. [Google Scholar] [CrossRef] [Green Version]
- Briggs, D.E.; Summons, R.E. Ancient biomolecules: Their origins, fossilization, and role in revealing the history of life. BioEssays 2014, 36, 482–490. [Google Scholar] [CrossRef]
- Ivarsson, M.; Broman, C.; Holmström, S.J.; Ahlbom, M.; Lindblom, S.; Holm, N.G. Putative fossilized fungi from the lithified volcaniclastic apron of Gran Canaria, Spain. Astrobiology 2011, 11, 633–650. [Google Scholar] [CrossRef]
- Cockell, C.S.; Raven, J.A. Zones of photosynthetic potential on Mars and the early Earth. Icarus 2004, 169, 300–310. [Google Scholar] [CrossRef]
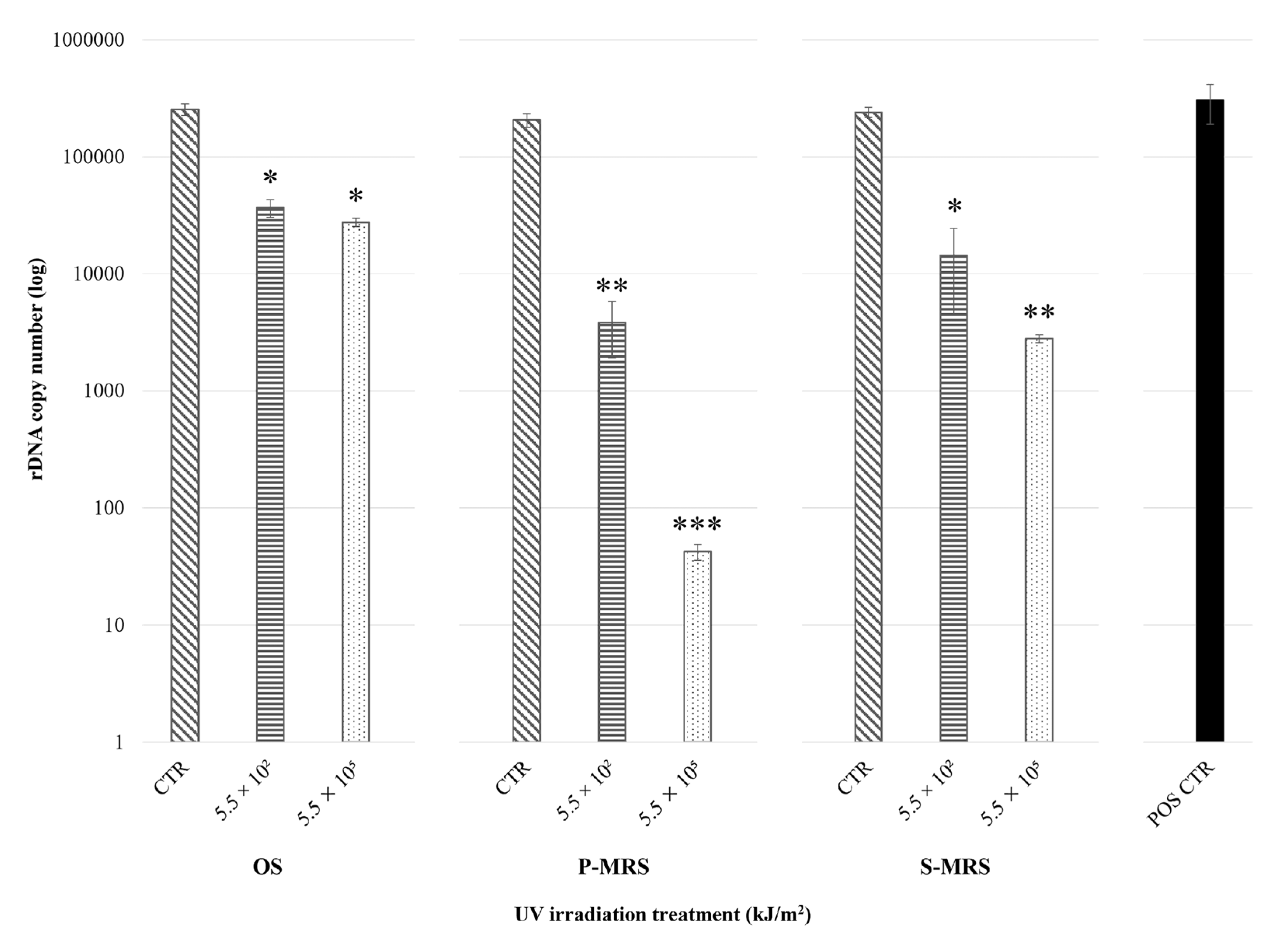
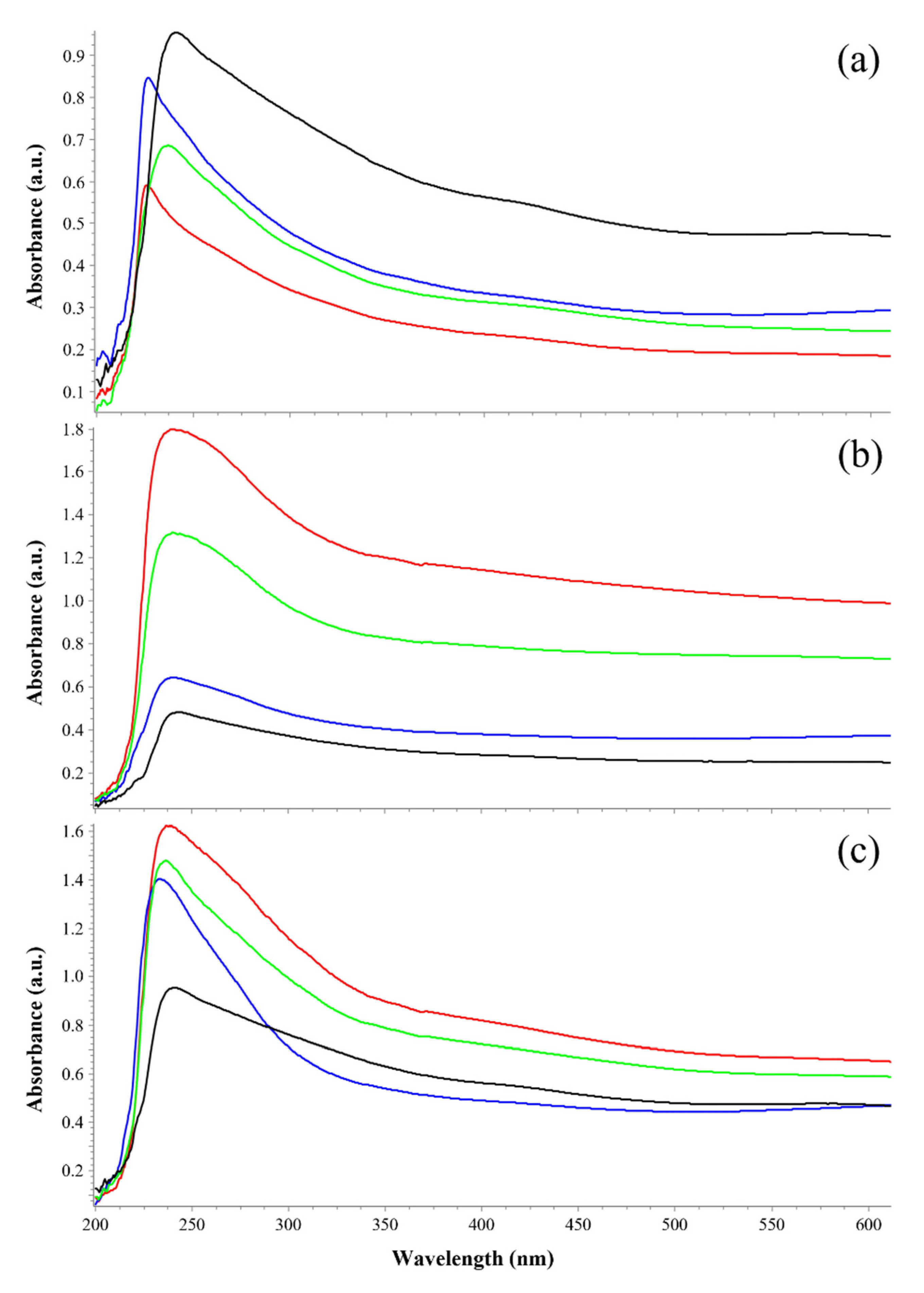
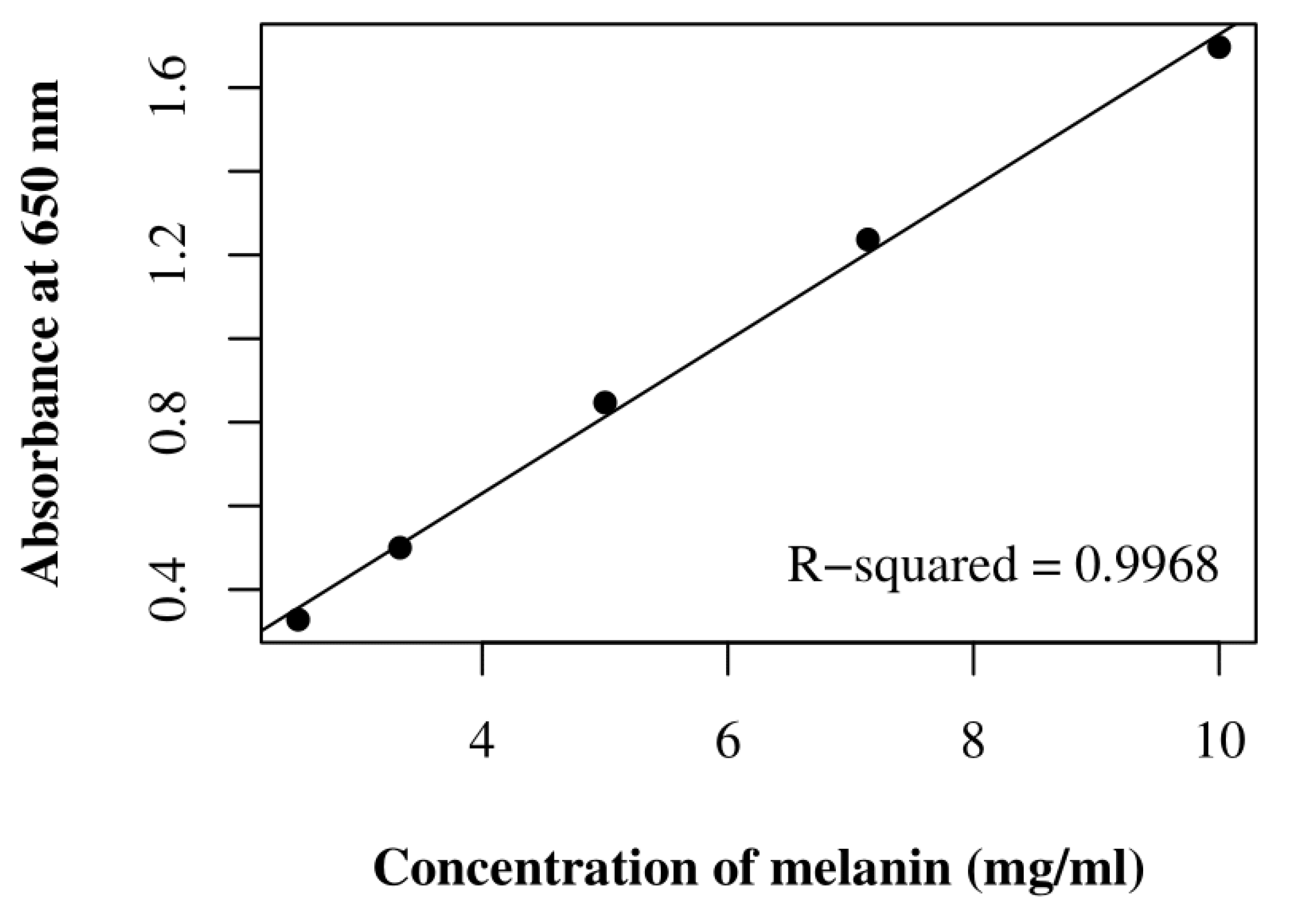
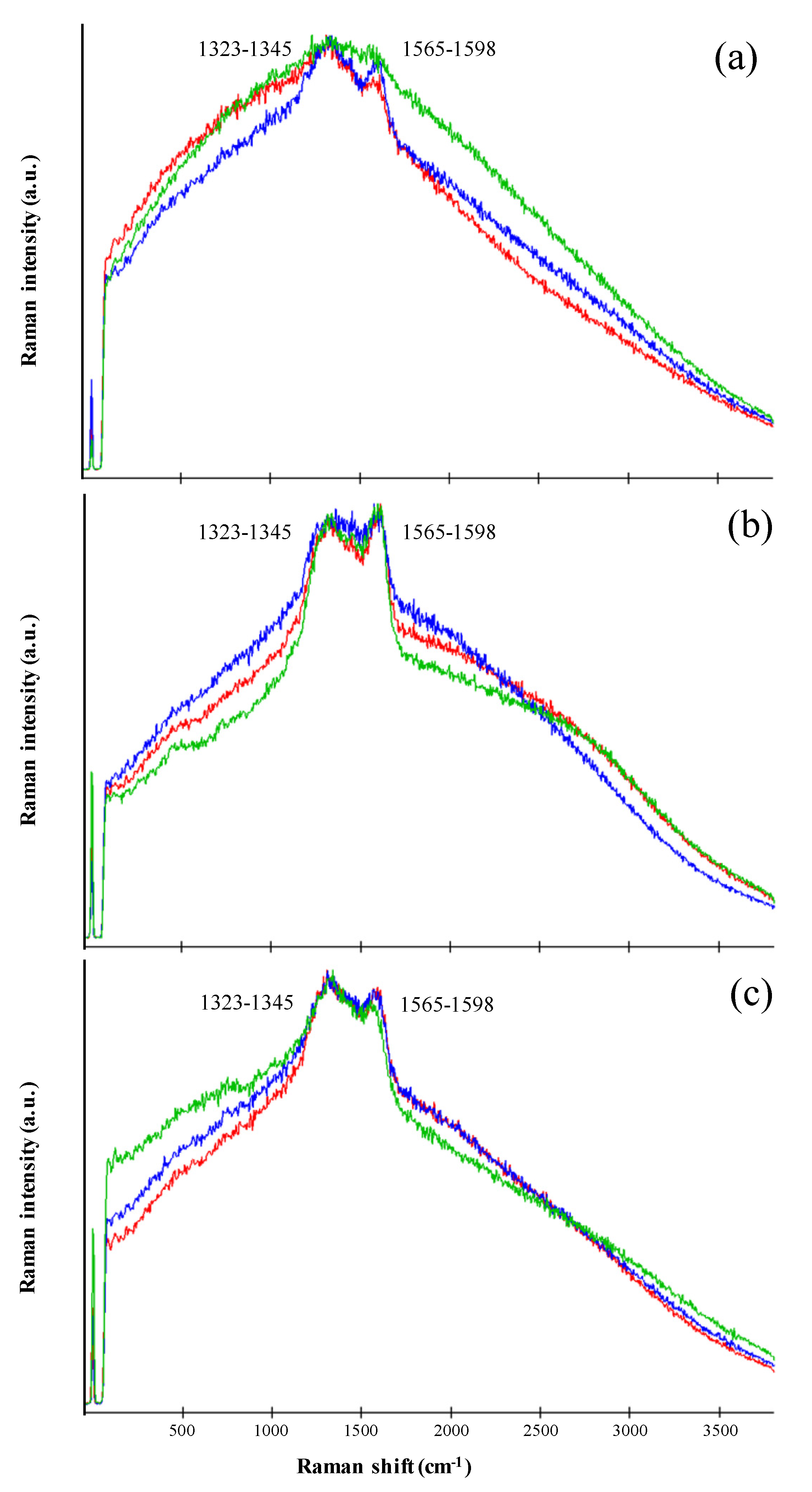
| Polychromatic UV Irradiance with SOL2000 (200–400 nm) | Irradiation Time/Sample | Resulting Fluence/Sample 1 | Simulated Martian Days 2 | Sample Set Substrate |
|---|---|---|---|---|
| 0 J/(s·m2) → CTR (laboratory control) | 0 s | 0 kJ/m2 | - | MEA + OS/ MEA + P-MRS/ MEA + S-MRS |
| 0 J/(s·m2) → Dark (transport control) | 0 s | 0 kJ/m2 | - | |
| 1271 J/(s·m2) | 7 min, 12 s | 5.5 × 102 kJ/m2 (0.1% ND 3) | 0.37 | |
| 1271 J/(s·m2) | 1 h, 12 min | 5.5 × 103 kJ/m2 (1.0% ND 3) | 3.70 | |
| 1271 J/(s·m2) | 30 h | 1.4 × 105 kJ/m2 | 94.15 | |
| 1271 J/(s·m2) | 60 h | 2.7 × 105 kJ/m2 | 181.57 | |
| 1271 J/(s·m2) | 120 h | 5.5 × 105 kJ/m2 | 369.87 | |
| Physiological conditions → POS CTR (positive control) | 0 s | 0 kJ/m2 | - | MEA |
| Sample | Concentration (mg/mL) | Absorption Peak (nm) |
|---|---|---|
| OS CTR | 5.271 (±2.705) | 228 (±4) |
| OS 5.5 × 102 kJ/m2 | 12.116 (±3.708) | 230 (±6) |
| OS 5.5 × 105 kJ/m2 | 6.758 (±7.760) | 233 (±7) |
| P-MRS CTR | 322.409 (±103.428) | 234 (±9) |
| P-MRS 5.5 × 102 kJ/m2 | 100.973 (±22.709) | 236 (±6) |
| P-MRS 5.5 × 105 kJ/m2 | 237.980 (±134.799) | 234 (±9) |
| S-MRS CTR | 24.005 (±15.427) | 234 (±6) |
| S-MRS 5.5 × 102 kJ/m2 | 19.811 (±3.545) | 232 (±7) |
| S-MRS 5.5 × 105 kJ/m2 | 22.096 (±14.595) | 233 (±6) |
| POS CTR | 20.136 (±5.752) | 233 (±6) |
| Wavenumber (cm−1) | Molecular Bond Interpretation | References |
|---|---|---|
| 3461–3372 | NH2/OH stretching of phenols and aromatic amines in indolic and pyrrolic systems | [53] |
| 2931–2926 | CH2 asymmetric stretching of saturated aliphatic groups in fatty acids | [55,56] |
| 2858–2851 | CH2 symmetric stretching of saturated aliphatic groups in fatty acids | [55,56] |
| 2515–2511 | OH stretching in carboxylic acids | [58,61] |
| 2249 | Acetylenic C≡C/aliphatic nitrile C≡N stretching | [56,58] |
| 2233–2211 | Acetylenic C≡C/aromatic nitrile C≡N stretching | [56,58] |
| 2136–2114 | Acetylenic C≡C/aliphatic isonitrile N≡C stretching | [56,58] |
| 2037–1788 | Several vibrations from overtones and combinations of substituted benzene rings | [61] |
| 1743–1738 | C=O stretching of aliphatic esters in triglycerides | [58,62] |
| 1669–1632 | Alkenyl C=C/C=O/C=N stretching of amide I in proteins | [56,58] |
| 1462–1445 | CH2 scissoring/CH3 asymmetric bending of aliphatic compounds in proteins or fatty acids | [58,63] |
| 1422–1415 | OH in-plane bending in carboxylic acids | [61] |
| 1380–1374 | CH3 symmetric bending of aliphatic compounds in proteins or fatty acids | [58,63] |
| 1254–1252 | P=O asymmetric/CO stretching of aliphatic phosphorus compounds, aromatic ethers and esters in carbohydrates or lipids | [58,59] |
| 1193–1190 | PO/CN/CO stretching of aromatic amines, phosphates, and phenols in carbohydrates or lipids | [58,61] |
| 1164–1158 | CO stretching of aliphatic ethers, esters, and alcohols in carbohydrates | [58,61] |
| 1122–1085 | CN/COC asymmetric/CO stretching of aliphatic amines, ethers, esters, and alcohols in carbohydrates | [57,61] |
| 1061–1018 | POC asymmetric/CO stretching of aliphatic phosphorus compounds, ethers, and alcohols in carbohydrates | [57,58] |
| 928–921 | COH out-of-plane bending in carboxylic acids | [58] |
| 901–814 | CH out-of-plane bending of benzenes | [57,61] |
| 721–712 | –(CH2)n– rocking of methylene chains in hydrocarbons | [58,63] |
| 700–687 | Olefinic cis-CH/phenolic OH out-of-plane bending | [61] |
| 675–642 | Acetylenic C=CH bending | [56] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pacelli, C.; Cassaro, A.; Catanzaro, I.; Baqué, M.; Maturilli, A.; Böttger, U.; Rabbow, E.; de Vera, J.-P.P.; Onofri, S. The Ground-Based BIOMEX Experiment Verification Tests for Life Detection on Mars. Life 2021, 11, 1212. https://doi.org/10.3390/life11111212
Pacelli C, Cassaro A, Catanzaro I, Baqué M, Maturilli A, Böttger U, Rabbow E, de Vera J-PP, Onofri S. The Ground-Based BIOMEX Experiment Verification Tests for Life Detection on Mars. Life. 2021; 11(11):1212. https://doi.org/10.3390/life11111212
Chicago/Turabian StylePacelli, Claudia, Alessia Cassaro, Ilaria Catanzaro, Mickael Baqué, Alessandro Maturilli, Ute Böttger, Elke Rabbow, Jean-Pierre Paul de Vera, and Silvano Onofri. 2021. "The Ground-Based BIOMEX Experiment Verification Tests for Life Detection on Mars" Life 11, no. 11: 1212. https://doi.org/10.3390/life11111212
APA StylePacelli, C., Cassaro, A., Catanzaro, I., Baqué, M., Maturilli, A., Böttger, U., Rabbow, E., de Vera, J.-P. P., & Onofri, S. (2021). The Ground-Based BIOMEX Experiment Verification Tests for Life Detection on Mars. Life, 11(11), 1212. https://doi.org/10.3390/life11111212









