Co-Evolution of Opioid and Adrenergic Ligands and Receptors: Shared, Complementary Modules Explain Evolution of Functional Interactions and Suggest Novel Engineering Possibilities
Abstract
1. Introduction
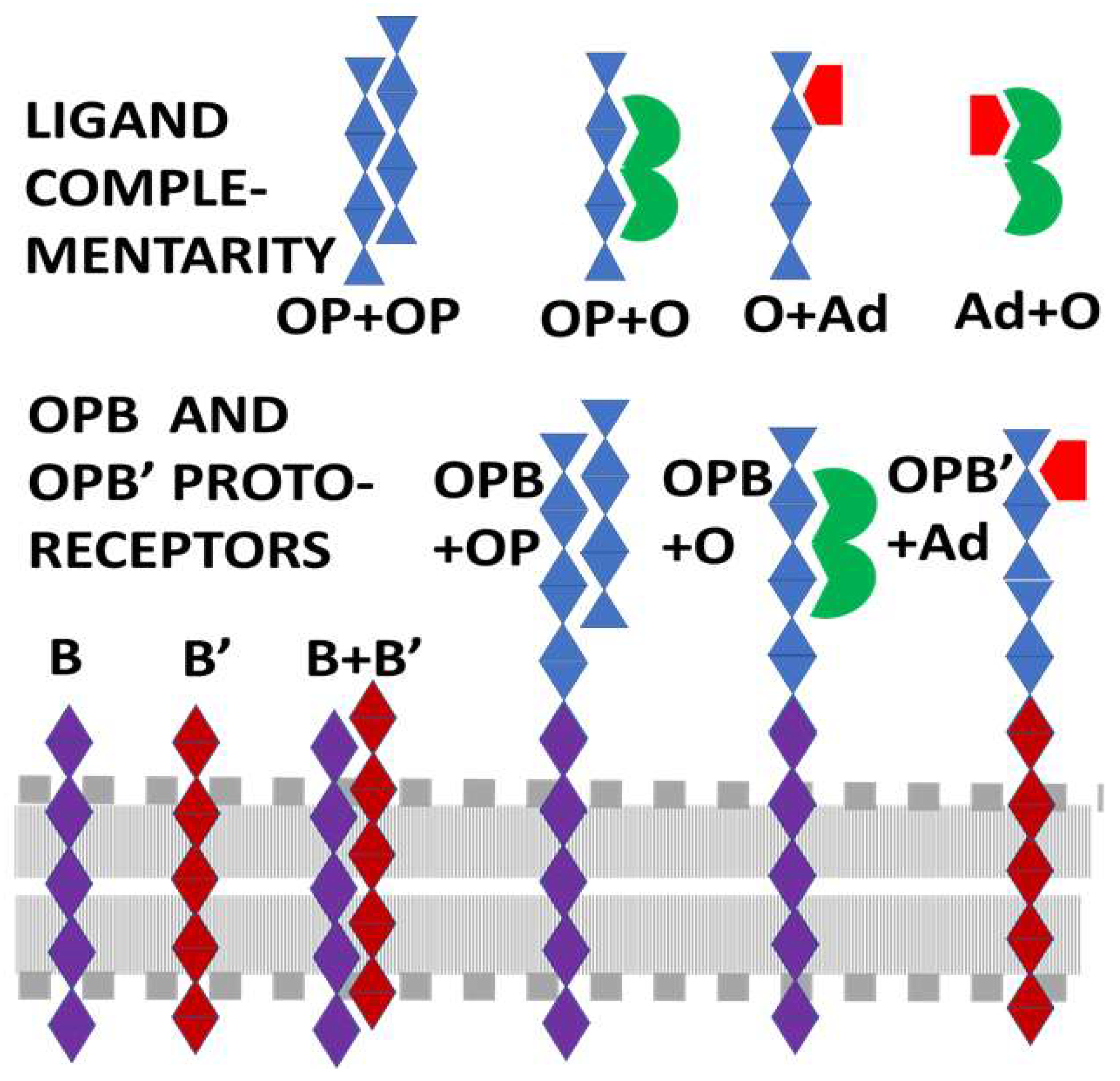
2. General Theory of Receptor–Ligand Evolution
- (1)
- Since adrenergic compounds modify the effects of opioid compounds and vice versa (see above), adrenergic compounds should be molecularly complementary to opioid compounds, which is to say that adrenergic compounds should bind specifically to opioids.
- (2)
- Opioid peptides that bind adrenergic compounds (hetero-complementarity) should provide the basis for the molecularly complementary modules upon which evolution has built receptors and transporters for adrenergic compounds so that adrenergic receptors should have opioid-like modules within their ligand binding regions.
- (3)
- Self-aggregation (homo-complementarity) of opioid peptides should provide the basis for the molecularly complementary modules which evolution has built receptors and transporters for opioid compounds.
- (4)
- Some of these opioid-like regions of opioid receptors should bind adrenergic compounds (heterocomplementarity) acting as allosteric modifiers of opioid receptor function.
- (5)
- Similarly, some of the opioid-like regions of adrenergic receptors should bind opioids (homocomplementarity again) and act as allosteric modulators of the receptor.
- (6)
- Opioid and adrenergic receptors will therefore be likely to share common, evolutionarily conserved modules.
- (7)
- Because opioid and adrenergic receptors can homodimerize as well as heterodimerize with each other, these conserved modules are likely to include some transmembrane regions of the receptors involved in receptor oligomerization.
- (8)
- The sum of these shared modules may help to explain how these receptors have integrated functions that result in mutual enhancement of function and how they manage their cross-talk.
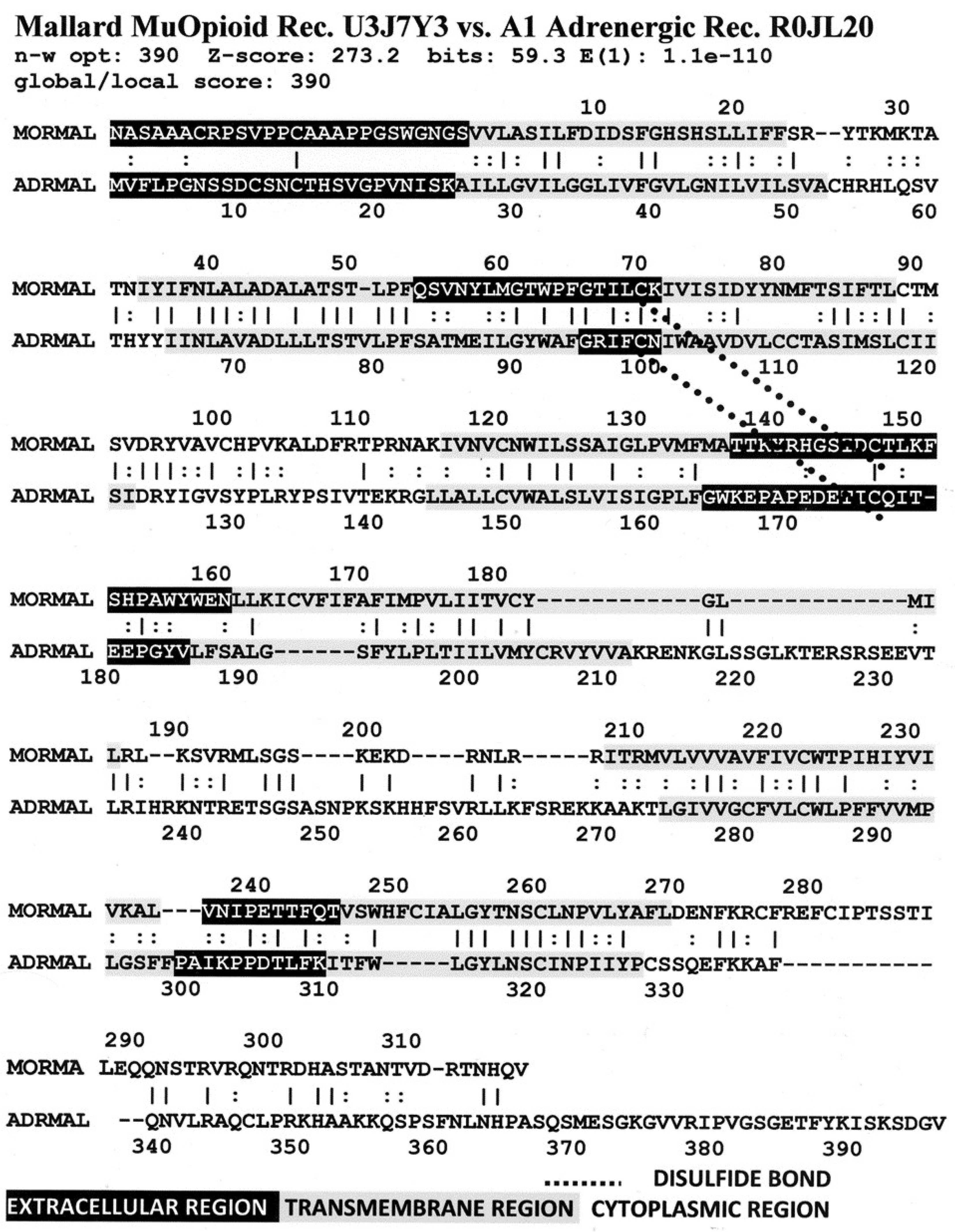
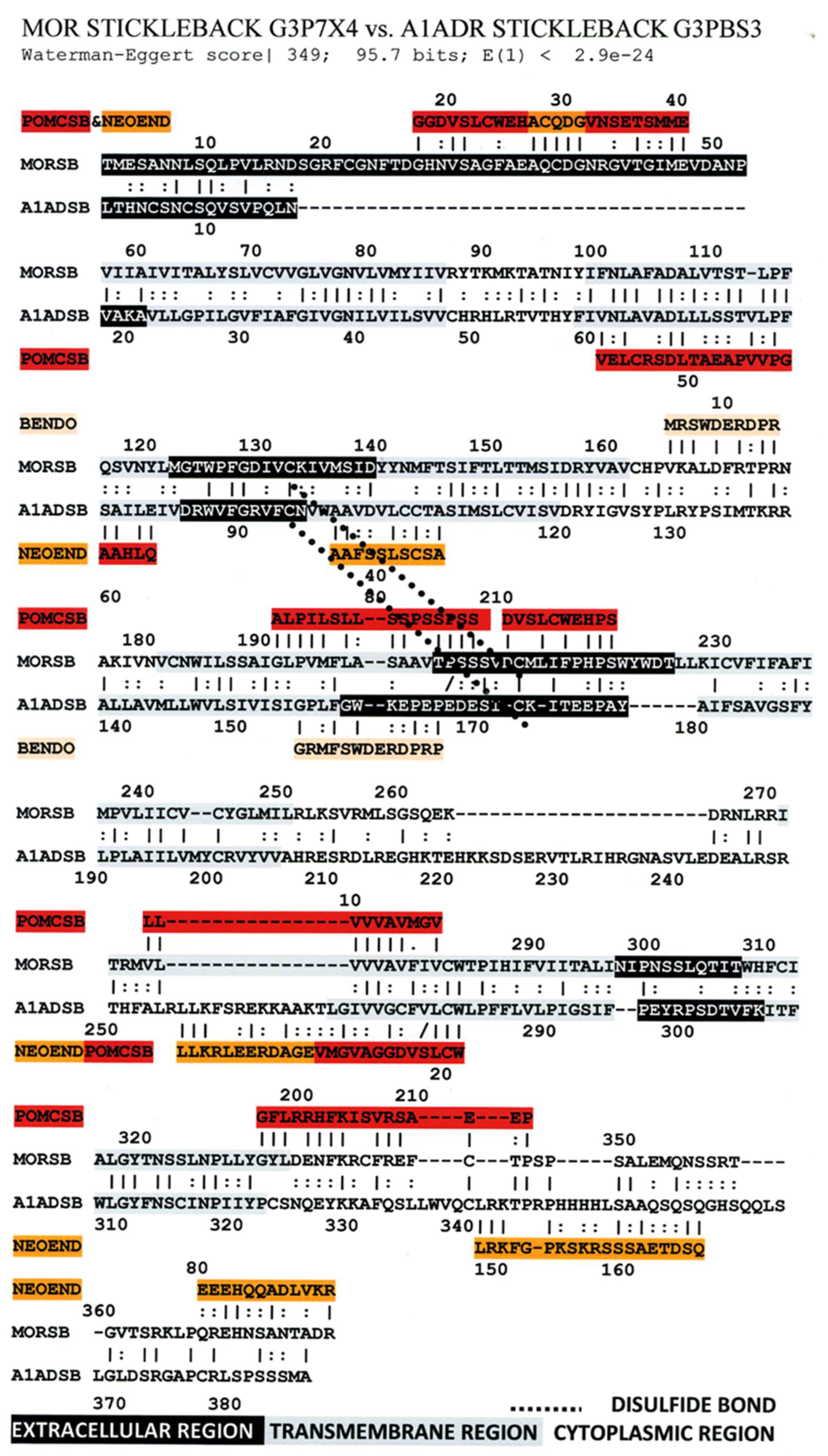
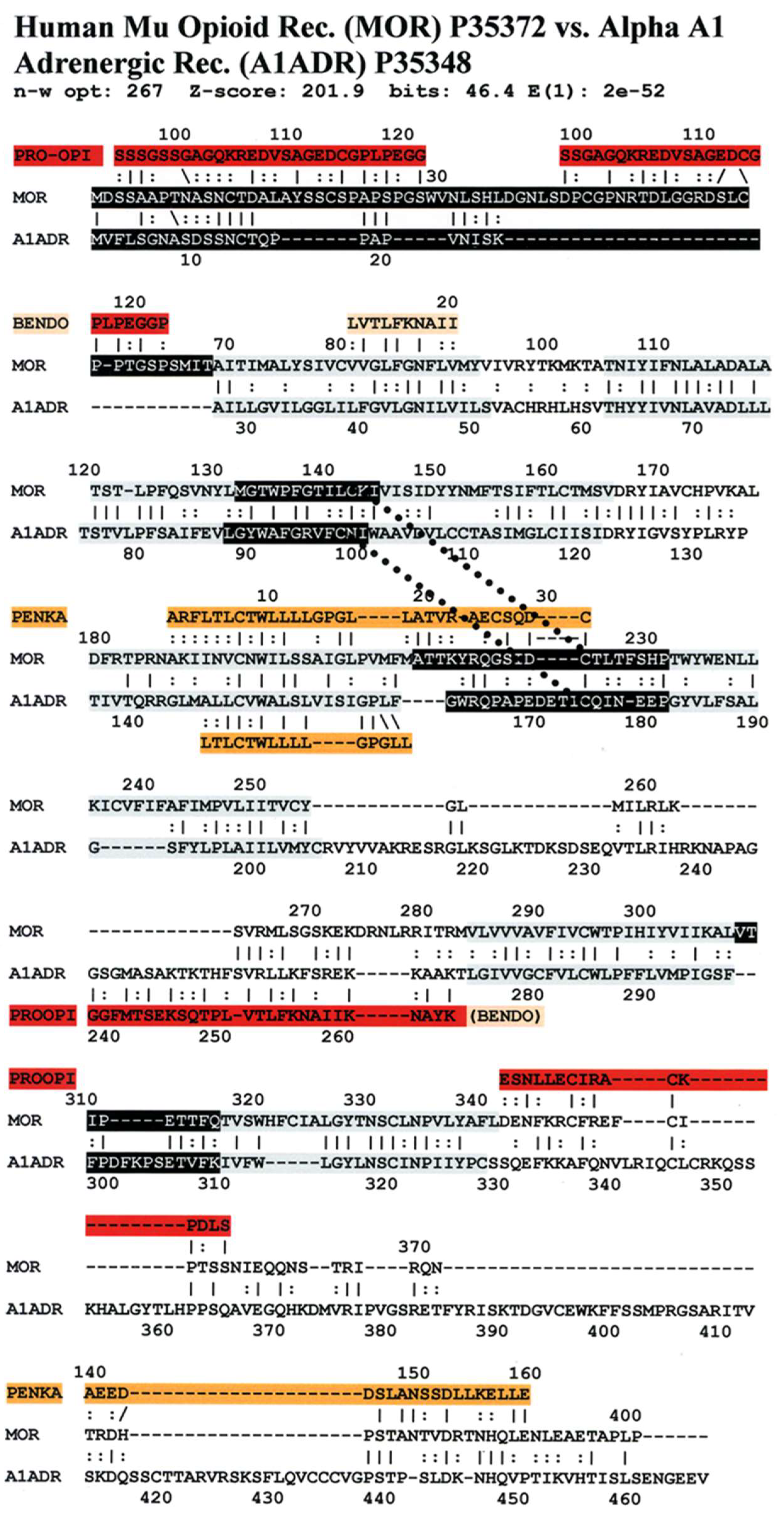
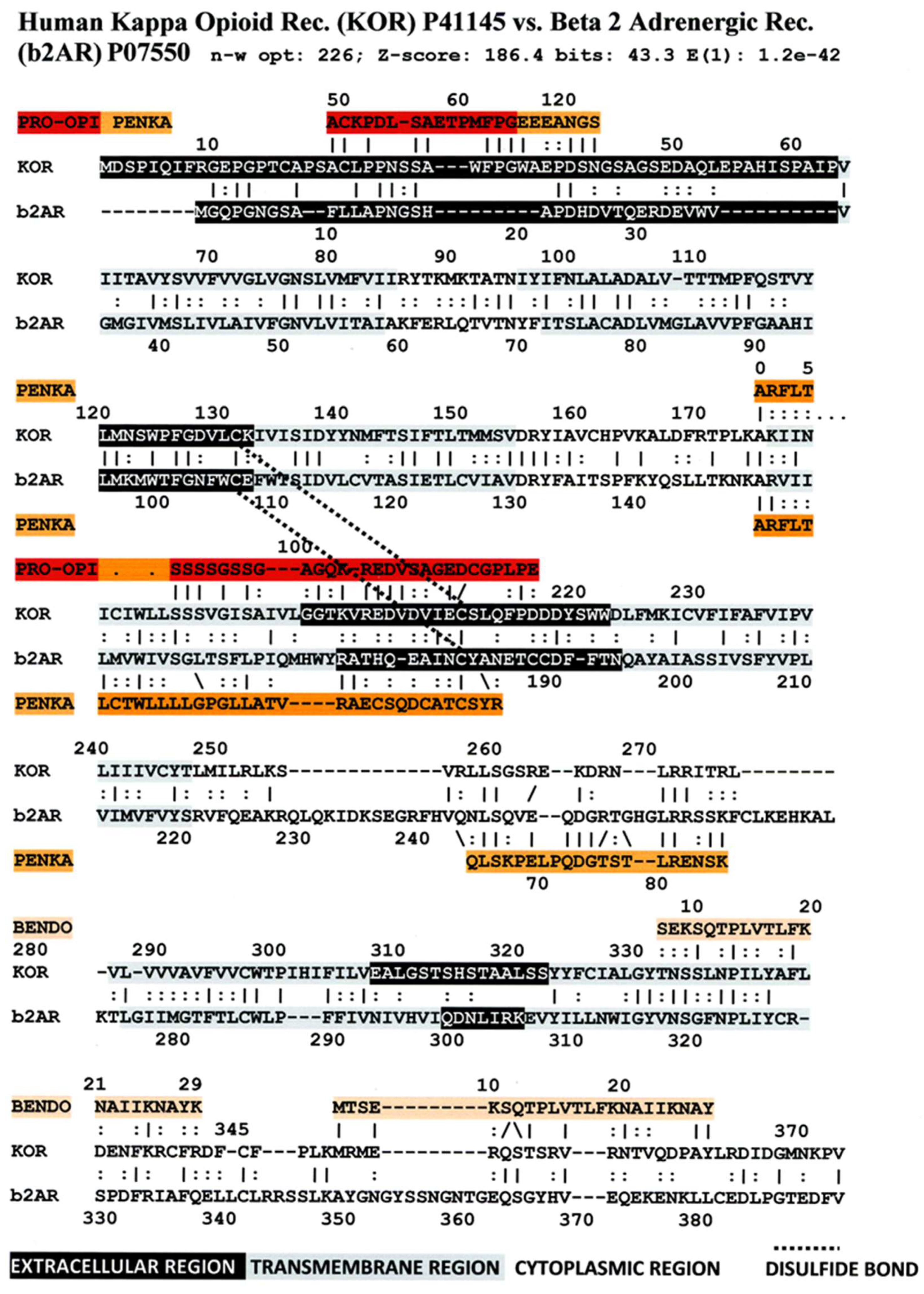
3. Literature Review of Results Relevant to Theoretical Predictions
Statistical Significance of the Results Reported Above
4. Discussion
4.1. Evolutionary Models of Results
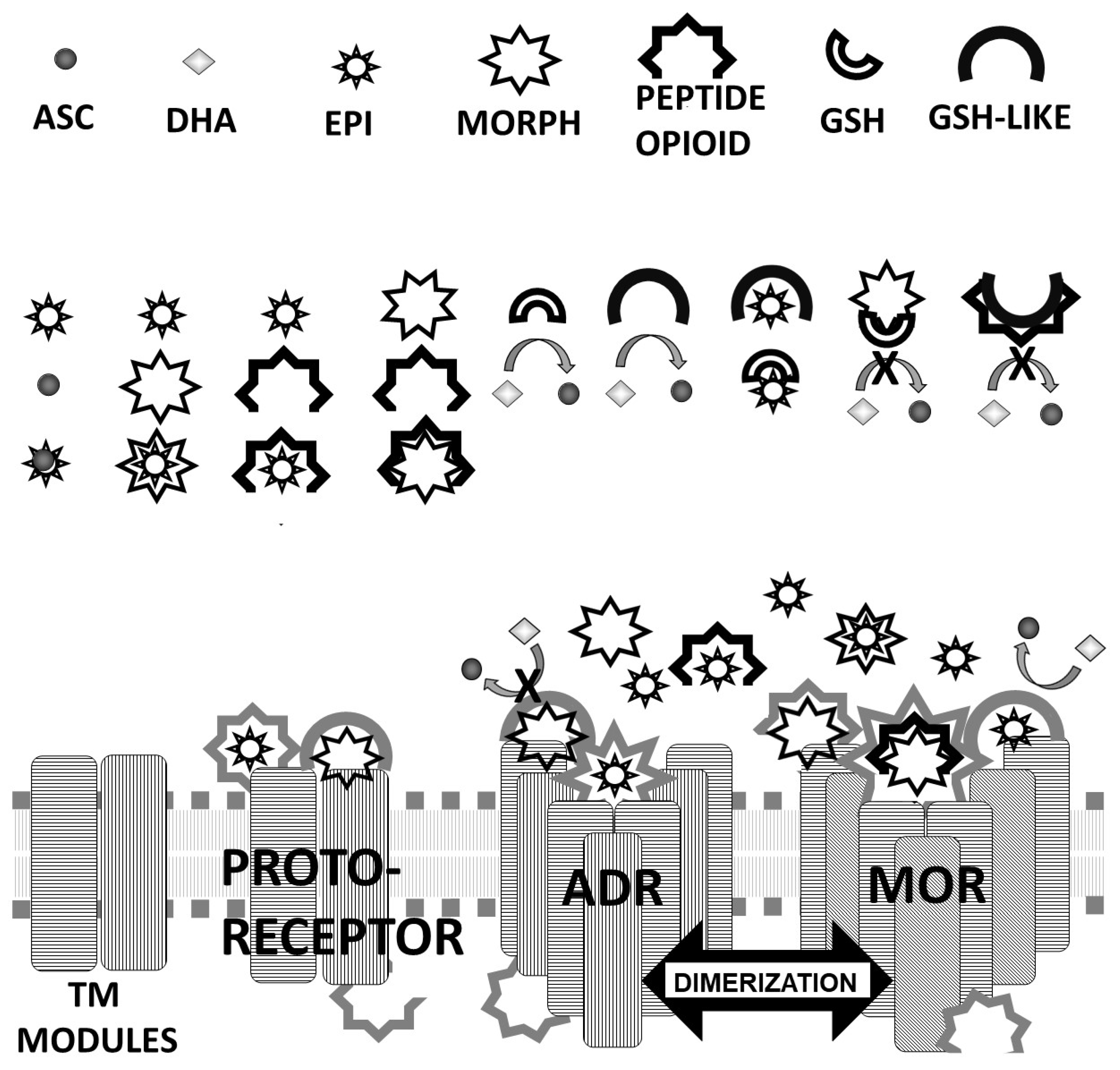
4.2. Model of ADR-OPR Co-Evolution Based on Molecularly Complementary Modules
4.2.1. Receptor Evolution as a Case Study in Interactome Emergence
4.2.2. Protein Engineering Opportunities Arising from Complementary Modularity
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Combination: | W-E Score: | 10 of 10 | 9 of 10 | 8 of 10 | 7 of 10 | 6 of 10 | 5 of 10 |
|---|---|---|---|---|---|---|---|
| Mu Opioid Receptor vs. Kappa Opioid Receptor | 1806 | ||||||
| Alpha-1A Adrenergic Receptor vs. Beta-2 Adrenergic Receptor | 786 | 0 | 0 | 3 | 0 | 6 | 9 |
| Mu Opioid Receptor vs. Alpha1a Adrenergic Receptor | 426 | 0 | 0 | 1 | 2 | 4 | 9 |
| Mu Opioid Receptor vs. Beta2 Adrenergic Receptor | 404 | 0 | 0 | 0 | 0 | 5 | 4 |
| Mu Opioid Receptor vs. Gastrin/Cholecystokinin Type B Receptor | 337 | 0 | 0 | 0 | 0 | 4 | 2 |
| Kappa Opiate Receptor vs. Beta2 Adrenergic Receptor | 327 | 0 | 0 | 0 | 0 | 3 | 1 |
| Kappa Opiate Receptor vs. Gastrin/Cholecytokinin Type B Receptor | 309 | 0 | 0 | 0 | 0 | 2 | 8 |
| Adrenocorticotropic Hormone Receptor vs. Alpha-1A Adrenergic Receptor | 292 | 0 | 0 | 0 | 1 | 5 | 8 |
| Gastrin Receptor vs. Beta-2 Adrenergic Receptor | 281 | 0 | 0 | 0 | 1 | 3 | 7 |
| Gastrin Receptor vs. Alpha 1-A Adrenergic Receptor | 264 | 0 | 0 | 0 | 1 | 2 | 4 |
| Adrenocorticotropic Hormone Receptor vs. Beta-2 Adrenergic Receptor | 207 | 0 | 0 | 0 | 2 | 4 | 4 |
| Mu Opioid Receptor vs. Adrenocoricotropic Hormone Receptor | 194 | 0 | 0 | 0 | 0 | 2 | 8 |
| Kappa Opiate Receptor vs. Adrenocorticotropic Hormone Receptor | 192 | 0 | 0 | 0 | 0 | 3 | 6 |
| Adrenocorticotropic Hormone Receptor vs. Gastrin Receptor | 98 | 0 | 0 | 0 | 0 | 2 | 5 |
| Insulin Receptor vs. Gastrin Receptor | 76 | 0 | 0 | 0 | 0 | 3 | 4 |
| Glucagon Receptor vs. Gastrin Receptor | 67 | 0 | 0 | 0 | 1 | 2 | 8 |
| Kappa Opiate Receptor vs. Glucagon Receptor | 58 | 0 | 0 | 0 | 0 | 0 | 5 |
| Mu Opioid Receptor vs. Glucagon Receptor | 58 | 0 | 0 | 0 | 0 | 1 | 5 |
| Glucagon Receptor vs. Insulin Receptor | 58 | 0 | 0 | 0 | 0 | 3 | 9 |
| Pro-Opiomelanocortin vs. Insulin Receptor | 57 | 0 | 0 | 0 | 0 | 3 | 4 |
| Insulin Receptor vs. Alpha-1A Adrenergic Receptor | 56 | 0 | 0 | 0 | 0 | 1 | 3 |
| Mu Opioid Receptor vs. Insulin Receptor | 55 | 0 | 0 | 0 | 0 | 0 | 2 |
| Glucagon Receptor vs. Beta-2 Adrenergic Receptor | 55 | 0 | 0 | 0 | 0 | 1 | 4 |
| Insulin Receptor vs. Adrenocorticotropic Hormone Receptor | 55 | 0 | 0 | 0 | 0 | 2 | 6 |
| Kappa Opiate Receptor vs. Insulin Receptor | 54 | 0 | 0 | 0 | 0 | 3 | 2 |
| Glucagon Receptor vs. Alpha-1A Adrenergic Receptor | 51 | 0 | 0 | 0 | 1 | 2 | 3 |
| Insulin Receptor vs. Beta-2 Adrenergic Receptor | 50 | 0 | 0 | 0 | 0 | 1 | 1 |
| Glucagon Receptor vs. Adrenocorticotropic Hormone Receptor | 49 | 0 | 0 | 0 | 0 | 2 | 4 |
Appendix B
| Combination | W-E Score | 10 of 10 | 9 of 10 | 8 of 10 | 7 of 10 | 6 of 10 | 5 of 10 |
|---|---|---|---|---|---|---|---|
| Pro-opiomelanocortin vs. Endorphin | 237 | 2 | 1 | 0 | 0 | 0 | 0 |
| Pro-opiomelanocortin vs. Proenkephalin-A | 92 | 0 | 0 | 0 | 6 | 2 | 0 |
| Proenkephalin-A vs. Alpha neo-endorphin | 59 | 0 | 0 | 0 | 0 | 0 | 7 |
| kappa opiate Receptor vs. Proenkephalin-A | 58 | 0 | 0 | 0 | 0 | 0 | 1 |
| Mu Opioid Receptor vs. Proenkephalin | 57 | 0 | 0 | 0 | 0 | 0 | 3 |
| Pro-opiomelanocortin vs. Insulin Receptor | 57 | 0 | 0 | 0 | 0 | 3 | 4 |
| Proenkephalin-A vs. Insulin Receptor | 56 | 0 | 0 | 0 | 0 | 2 | 2 |
| Proenkephalin-A vs. Gastrin/Cholecystokinin type B Receptor | 54 | 0 | 0 | 0 | 1 | 1 | 2 |
| Pro-opiomelanocortin vs. Glucagon Receptor | 52 | 0 | 0 | 0 | 2 | 4 | 7 |
| Proenkephalin-A vs. Adrenocorticotropic Hormone Receptor | 52 | 0 | 0 | 0 | 0 | 3 | 3 |
| Proenkephalin-A vs. Glucagon Receptor | 52 | 0 | 0 | 0 | 0 | 2 | 4 |
| Pro-opiomelanocortin vs. Adrenocorticotropic Hormone Receptor | 51 | 0 | 0 | 0 | 0 | 0 | 1 |
| Proenkephalin-A vs. Endorphin | 49 | 0 | 0 | 0 | 0 | 0 | 6 |
| Kappa Opiate Receptor vs. Pro-opiomelanocortin | 48 | 0 | 0 | 0 | 0 | 1 | 4 |
| Pro-opiomelanocortin vs. Alpha1A Adrenergic Receptor | 48 | 0 | 0 | 0 | 0 | 1 | 0 |
| Proenkephalin-A vs. Beta2 Adrenergic Receptor | 48 | 0 | 0 | 0 | 0 | 3 | 1 |
| Proenkephalin-A vs. alpha1A Adrenergic Receptor | 46 | 0 | 0 | 0 | 0 | 0 | 5 |
| Pro-opiomelanocortin vs. Gastrin/Cholecystokinin type B Receptor | 45 | 0 | 0 | 0 | 0 | 1 | 9 |
| Pro-opiomelanocortin vs. Alpha Neo-Endorphin | 42 | 0 | 0 | 0 | 0 | 0 | 0 |
| Endorphin vs. Alpha Neo-Endorphin | 42 | 0 | 0 | 0 | 0 | 0 | 0 |
| Endorphin vs. adrenocorticotropic Hormone Receptor | 41 | 0 | 0 | 0 | 0 | 0 | 0 |
| mu opioid Receptor vs. Pro-opiomelanocortin | 40 | 0 | 0 | 0 | 0 | 0 | 1 |
| Pro-opiomelanocortin vs. Beta2 Adrenergic Receptor | 40 | 0 | 0 | 0 | 0 | 0 | 2 |
| Endorphin vs. Insulin Receptor | 38 | 0 | 0 | 0 | 0 | 0 | 0 |
| Endorphin vs. alpha1A Adrenergic Receptor | 36 | 0 | 0 | 0 | 0 | 0 | 1 |
| Endorphin vs. glucagon Receptor | 36 | 0 | 0 | 0 | 0 | 0 | 0 |
| Alpha neo-Endorphin vs. Insulin Receptor | 34 | 0 | 0 | 0 | 0 | 0 | 0 |
| Endorphin vs. Gastrin/cholecystokinin type B Receptor | 33 | 0 | 0 | 0 | 0 | 0 | 0 |
| mu opioid Receptor vs. Alpha Neo-Endorphin | 32 | 0 | 0 | 0 | 0 | 0 | 1 |
| Endorphin vs. Beta2 Adrenergic Receptor | 30 | 0 | 0 | 0 | 0 | 0 | 1 |
| Alpha neo-Endorphin vs. Alpha1A Adrenergic Receptor | 26 | 0 | 0 | 0 | 0 | 0 | 0 |
| Alpha neo-Endorphin vs. Glucagon Receptor | 24 | 0 | 0 | 0 | 0 | 0 | 0 |
| Alpha neo-Endorphin vs. Gastrin/Cholecystokinin type B Receptor | 23 | 0 | 0 | 0 | 0 | 0 | 0 |
| Alpha neo-Endorphin vs. Adrenocorticotropic Hormone Receptor | 22 | 0 | 0 | 0 | 0 | 0 | 0 |
| Alpha neo-Endorphin vs. Beta2 Adrenergic Receptor | 22 | 0 | 0 | 0 | 0 | 0 | 0 |
| kappa opiate Receptor vs. Alpha Neo-Endorphin | 19 | 0 | 0 | 0 | 0 | 0 | 0 |
References
- Kanigel, R. Apprentice to Genius; Macmillan: New York, NY, USA, 1986; pp. 171–201. [Google Scholar]
- Munro, T.A.; Huang, X.-P.; Inglese, C.; Perrone, M.G.; Van’t Veer, A.; Carroll, F.I.; Béguin, C.; Carlezon, W.A., Jr.; Colabufo, N.A.; Cohen, B.M.; et al. Selective κ Opioid Antagonists nor-BNI, GNTI and JDTic Have Low Affinities for Non-Opioid Receptors and Transporters. PLoS ONE 2013, 8, e70701. [Google Scholar] [CrossRef] [PubMed]
- Tagaya, E.; Tamaoki, J.; Chiyotani, A.; Konno, K. Stimulation of opioid mu-receptors potentiates β adrenoceptor-mediated relaxation of canine airway smooth muscle. J. Pharmacol. Exp. Ther. 1995, 275, 1288–1292. [Google Scholar]
- He, J.-R.; Molnar, J.; Barraclough, C.A. Morphine amplifies norepinephrine (NE)-induced LH release but blocks NE-stimulated increases in LHRH mRNA levels: Comparison of responses obtained in ovariectomized, estrogen-treated normal and androgen-sterilized rats. Mol. Brain Res. 1993, 20, 71–78. [Google Scholar] [CrossRef]
- Kindman, L.A.; Kates, R.E.; Ginsburg, R. Opioids potentiate contractile response of rabbit myocardium to the β adrenergic agonist isoproterenol. J. Cardiovasc. Pharmacol. 1991, 17, 61–67. [Google Scholar] [CrossRef] [PubMed]
- Lechner, R.B.; Gurll, N.J.; Reynolds, D.G. Naloxone potentiates the cardiovascular effects of catecholamines in canine hemorrhagic shock. Circ. Shock 1985, 16, 347–361. [Google Scholar]
- He, J.-R.; Barraclough, C.A. Morphine but not naloxone enhances luteinizing hormone-releasing hormone neuronal responsiveness to norepinephrine. J. Neuroendocrinol. 1992, 4, 92–99. [Google Scholar] [CrossRef]
- Allgood, S.C.; Gurll, N.J.; Reynolds, D.G. Naloxone requires circulating catecholamines to attenuate the cardiovascular suppression of endotoxic shock. J. Surg. Res. 1988, 44, 73–81. [Google Scholar] [CrossRef]
- Caffrey, J.L.; Hathorne, L.F.; Carter, G.C.; Sinclair, R.J. Naloxone potentiates contractile responses to epinephrine in isolated canine arteries. Circ. Shock 1990, 31, 317–332. [Google Scholar]
- Caffrey, J.L.; Stoll, S.T.; Sinclair, R.J.; Barron, B.A. (+) naloxone enhances vascular contractile responses to added epinephrine. Prog. Clin. Biol. Res. 1990, 328, 375–378. [Google Scholar]
- Lechner, R.B. Naloxone potentiates inotropic but not chronotropic effects of isoproterenol in vitro. Circ. Shock 1993, 39, 226–230. [Google Scholar] [CrossRef]
- Gu, H.; Gaugl, J.F.; Barron, B.A.; Caffrey, J.L. Naloxone enhances cardiac contractile responses to epinephrine without altering epinephrine uptake from plasma. Circ. Shock 1990, 32, 257–271. [Google Scholar] [PubMed]
- McCubbin, J.A.; Surwit, R.S.; Kuhn, C.M.; Cochrane, C.; Feinglos, M.N. Naltrexone potentiates glycemic responses during stress and epinephrine challenge in genetically obese mice. Psychosom. Med. 1989, 51, 441–448. [Google Scholar] [CrossRef]
- Parra, L.; Pérez-Vizcaíno, F.; Alsasua, A.; Martín, M.I.; Tamargo, J. Mu- and delta-opioid receptor-mediated contractile effects on rat aortic vascular smooth muscle. Eur J Pharmacol. 1995, 277, 99–105. [Google Scholar] [CrossRef]
- Lee, C.H.; Berkowitz, B.A. Stereoselective and calcium-dependent contractile effects of narcotic antagonist analgesics in the vascular smooth muscle of the rat. J. Pharmacol. Exp. Ther. 1976, 198, 347–356. [Google Scholar] [PubMed]
- Root-Bernstein, R.S.; Dillon, P.F. A common molecular motif characterizes extracellular allosteric enhancers of GPCR aminergic receptors and suggests their mechanism of action. Curr. Med. Chem. 2014, 21, 3673–3686. [Google Scholar] [CrossRef]
- Root-Bernstein, R.; Fewins, J.; Dillon, P.F. Tartaric Acid Enhances Adrenergic Receptor Activity: Test of a General Theory of Extracellular Aminergic GPCR Enhancer Discovery. Curr. Drug Discov. Technol. 2014, 11, 293–307. [Google Scholar] [CrossRef]
- Abel, S.; Harris, S.C. Morphine-benzedrine analgesia in obstetrics. Fed. Proc. 1947, 6, 67. [Google Scholar]
- Milosevic, M.P. Effect of adrenaline on the analgesic response of mice to morphine and related drugs. Arch. Int. Pharmacodyn. Ther. 1955, 104, 50–56. [Google Scholar]
- Goyagi, T.; Nishikawa, T. The addition of epinephrine enhances postoperative analgesia by intrathecal morphine. Anesth. Analg. 1995, 81, 508–513. [Google Scholar]
- Goyagi, T.; Nishikawa, T. Oral clonidine premedication enhances the quality of postoperative analgesia by intrathecal morphine. Anesth. Analg. 1996, 82, 1192–1196. [Google Scholar]
- Sasson, S.; Unterwald, E.M.; Kornetsky, C. Potentiation of morphine analgesia by d-amphetamine. Psychopharmacology 1986, 90, 163–165. [Google Scholar] [CrossRef]
- Izenwasser, S.; Kornetsky, C. Potentiation of morphine analgesia by d-amphetamine is mediated by norepinephrine and not dopamine. Pain 1988, 33, 363–368. [Google Scholar] [CrossRef]
- Huang, K.S.; Tseng, C.H.; Cheung, K.S.; Hui, Y.L.; Tan, P.P. Influence of epinephrine as an adjuvant to epidural morphine for postoperative analgesia. Ma Zui Xue Za Zhi 1993, 31, 245–248. [Google Scholar]
- Sierralta, F.; Naquira, D.; Pinardi, G.; Miranda, H.F. α-Adrenoceptor and opioid receptor modulation of clonidine-induced antinociception. Br. J. Pharmacol. 1996, 119, 551–554. [Google Scholar] [CrossRef]
- Wu, Y.W.; Seah, Y.S.; Chung, K.T.; Liu, M.D. Postoperative pain relief in primigravida caesarean section patients--combination of intrathecal morphine and epinephrine. Acta Anaesthesiol. Sin. 1999, 37, 111–114. [Google Scholar]
- Gulati, A.; Bhalla, S.; Matwyshyn, G.; Zhang, Z.; Andurkar, S.V. Determination of adrenergic and imidazoline receptor involvement in augmentation of morphine and oxycodone analgesia by clonidine and BMS182874. Pharmacology 2009, 83, 45–58. [Google Scholar] [CrossRef]
- Fairbanks, C.A.; Posthumus, I.J.; Kitto, K.F.; Stone, L.S.; Wilcox, G.L. Moxonidine, a selective imidazoline/α2 adrenergic receptor agonist, synergizes with morphine and deltorphin II to inhibit substance P-induced behavior in mice. Pain 2000, 84, 13–20. [Google Scholar] [CrossRef]
- Gupta, S.; Raval, D.; Patel, M.; Patel, N.; Shah, N. Addition of epidural Clonidine enhances postoperative analgesia: A double-blind study in total knee- replacement surgeries. Anesth. Essays Res. 2010, 4, 70–74. [Google Scholar] [CrossRef]
- Engelman, E.; Marsala, C. Efficacy of adding clonidine to intrathecal morphine in acute postoperative pain: Meta-analysis. Br. J. Anaesth. 2013, 110, 21–27. [Google Scholar] [CrossRef]
- Katz, D.; Hamburger, J.; Gutman, D.; Wang, R.; Lin, H.M.; Marotta, M.; Zahn, J.; Beilin, Y. The effect of adding subarachnoid epinephrine to hyperbaric bupivacaine and morphine for repeat cesarean delivery: A double-blind prospective randomized control trial. Anesth. Analg. 2018, 127, 171–178. [Google Scholar] [CrossRef]
- Milne, B.; Sutak, M.; Cahill, C.M.; Jhamandas, K. Low doses of α 2-adrenoceptor antagonists augment spinal morphine analgesia and inhibit development of acute and chronic tolerance. Br. J. Pharmacol. 2008, 155, 1264–1278. [Google Scholar] [CrossRef]
- Satarian, L.; Javan, M.; Fathollahi, Y. Epinephrine inhibits analgesic tolerance to intrathecal administrated morphine and increases the expression of calcium-calmodulin-dependent protein kinase IIα. Neurosci. Lett. 2008, 430, 213–217. [Google Scholar] [CrossRef]
- Heimans, R.L. Catecholamines and the actions of morphine on the guinea-pig ileum. Arch. Int. Pharmacodyn. Ther. 1975, 216, 11–18. [Google Scholar]
- Ferri, S.; Reina, R.; Santagostino, A. Dopamine and the depressant action of morphine on stimulated guinea-pig ileum. Br. J. Pharmacol. 1977, 59, 25–28. [Google Scholar] [CrossRef]
- Goldstein, A.; Schulz, R. Morphine-tolerant longitudinal muscle strip from guinea-pig ileum. Br. J. Pharmacol. 1973, 48, 655–666. [Google Scholar] [CrossRef]
- Sarto, G. Modulation of the effect of morphine on the isolated guinea pig intestine by noradrenaline and serotonin. Boll. Soc. Ital. Biol. Sper. 1981, 57, 394–400. [Google Scholar]
- Park, W.K.; Chang, C.H.; Chae, J.E.; Kim, M.H.; Cho, Y.L.; Ahn, D.S. Phosphodiesterase inhibition by naloxone augments the inotropic actions of beta-adrenergic stimulation. Acta Anaesthesiol. Scand 2009, 53, 1043–1051. [Google Scholar] [CrossRef]
- Root-Bernstein, R.S.; Dillon, P.F. Fostering adventure research. A case study of the discovery that ascorbic acid enhances adrenergic drug activity. Drug Dev. Res. 2002, 57, 58–74. [Google Scholar] [CrossRef]
- Lengyel, I.; Toth, F.; Biyashev, D.; Szatmari, I.; Monory, K.; Tomboly, C.; Toth, G.; Benyhe, S.; Borsodi, A. A novel non-opioid binding site for endomorphin-1. J. Physiol. Pharmacol. 2016, 67, 605–616. [Google Scholar]
- Root-Bernstein, R.; Turke, M.; Subhramanyam, U.K.T.; Churchill, B.; Labahn, J. Adrenergic Agonists Bind to Adrenergic-Receptor-Like Regions of the Mu Opioid Receptor, Enhancing Morphine and Methionine-Enkephalin Binding: A New Approach to “Biased Opioids”? Int. J. Mol. Sci. 2018, 19, 272. [Google Scholar] [CrossRef]
- Monroe, P.J.; Perschke, S.E.; Crisp, T.; Smith, D.J. Evaluation of the interactions of serotonergic and adrenergic drugs with mu, delta, and kappa opioid binding sites. Neurosci. Lett. 1991, 133, 229–232. [Google Scholar] [CrossRef]
- Dillon, P.F.; Root-Bernstein, R.; Robinson, N.E.; Abraham, W.M.; Berney, C. Receptor-mediated enhancement of β adrenergic drug activity by ascorbate in vitro and in vivo. PLoS ONE 2010, 5, e15130. [Google Scholar] [CrossRef] [PubMed]
- Root-Bernstein, R.S.; Rhinesmith, T.; Koch, A.; Dillon, P.F. Enzymatic recycling of ascorbate by glutathione-like peptides derived from aminergic G-protein coupled receptors. J. Mol. Recog. 2016, 29, 296–302. [Google Scholar] [CrossRef]
- Montel, H.; Starke, K. Effects of narcotic analgesics and their antagonists on the rabbit isolated heart and its adrenergic nerves. Br. J. Pharmacol. 1973, 49, 628–641. [Google Scholar] [CrossRef]
- Llobel, F.; Laorden, M.L. Effects of morphine on atrial preparations obtained from nonfailing and failing human hearts. Br. J. Anaesth. 1996, 76, 106–110. [Google Scholar] [CrossRef]
- Carr, D.J.; Gebhardt, B.M.; Paul, D. αAdrenergic and μ-2 opioid receptors are involved in morphine-induced suppression of splenocyte natural killer activity. J. Pharmacol. Exp. Ther. 1993, 264, 1179–1186. [Google Scholar]
- Jordan, B.A.; Trapaidze, N.; Gomes, I.; Nivarthi, R.; Devi, L.A. Oligomerization of opioid receptors with β 2-adrenergic receptors: A role in trafficking and mitogen-activated protein kinase activation. Proc. Natl. Acad. Sci. USA 2001, 98, 343–348. [Google Scholar]
- Jordan, B.A.; Gomes, I.; Rios, C.; Filipovska, J.; Devi, L.A. Functional interactions between mu opioid and α 2A-adrenergic receptors. Mol. Pharmacol. 2003, 64, 1317–1324. [Google Scholar] [CrossRef]
- Rozenfeld, R.; Devi, L.A. Exploring a role for heteromerization in GPCR signaling specificity. Biochem. J. 2011, 433, 11–18. [Google Scholar] [CrossRef]
- Vilardaga, J.-P.; Nikolaev, O.V.; Lorentz, K.; Ferrandon, S.; Zhuang, Z.; Lohse, M.J. Direct inhibition of G protein signaling by cross-conformational switches between a2A-adrenergic and µ-opioid receptors. Nat. Chem. Biol. 2008, 4, 126–131. [Google Scholar] [CrossRef]
- Vilardaga, J.-P.; Agnati, L.F.; Fuxe, K.; Ciruela, F. G-protein-coupled receptor heteromer dynamics. J. Cell Sci. 2010, 123, 4215–4220. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Q.; Limbird, L.E. Hetero-oligomers of α2A-adrenergic and μ-opioid receptors do not lead to transactivation of G-proteins or altered endocytosis profiles. Biochem. Soc. Trans. 2004, 32, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Fujita, W.; Gomes, I.; Devi, L.A. Revolution in GPCR signalling: Opioid receptor heteromers as novel therapeutic targets: IUPHAR review 10. Br. J. Pharmacol. 2014, 171, 4155–4176. [Google Scholar] [CrossRef] [PubMed]
- Chabot-Doré, A.J.; Millecamps, M.; Naso, L.; Devost, D.; Trieu, P.; Piltonen, M.; Diatchenko, L.; Fairbanks, C.A.; Wilcox, G.L.; Hébert, T.E.; et al. Dual allosteric modulation of opioid antinociceptive potency by α2A-adrenoceptors. Neuropharmacology 2015, 99, 285–300. [Google Scholar] [CrossRef]
- Pepe, S.; van den Brink, O.W.; Lakatta, E.G.; Xiao, R.P. Cross-talk of opioid peptide receptor and beta-adrenergic receptor signalling in the heart. Cardiovasc. Res. 2004, 63, 414–422. [Google Scholar] [CrossRef]
- Dzimiri, N. Receptor crosstalk. Implications for cardiovascular function, disease and therapy. Eur. J. Biochem. 2002, 269, 4713–4730. [Google Scholar] [CrossRef]
- Vilardaga, J.P.; Nikolaev, V.O.; Lorenz, K.; Ferrandon, S.; Zhuang, Z.; Lohse, M.J. Conformational cross-talk between alpha2A-adrenergic and mu-opioid receptors controls cell signaling. Nat. Chem. Biol. 2008, 4, 126–131. [Google Scholar] [CrossRef]
- Dwyer, D.S. Amino acid sequence homology between ligands and their receptors: Potential identification of binding sites. Life Sci. 1989, 45, 421–429. [Google Scholar] [CrossRef]
- Root-Bernstein, R. Peptide self-aggregation and peptide complementarity as bases for the evolution of peptide receptors: A review. J. Mol. Recog. 2005, 18, 40–49. [Google Scholar] [CrossRef]
- Marinou, M.; Tzartos, S.J. Identification of regions involved in the binding of alpha-bungarotoxin to the human alpha7 neuronal nicotinic acetylcholine receptor using synthetic peptides. Biochem, J. 2003, 372 Pt 2, 543–554. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.T.; Hawrot, E.; Lentz, T.L. Distribution of alpha-bungarotoxin binding sites over residues 173-204 of the alpha subunit of the acetylcholine receptor. Mol. Pharmacol. 1988, 34, 643–650. [Google Scholar]
- Root-Bernstein, R. Molecular complementarity III. peptide complementarity as a basis for peptide receptor evolution: A bioinformatic case study of insulin, glucagon and gastrin. J. Theor. Biol. 2002, 218, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Root-Bernstein, R.S.; Vonck, J. The insulin receptor binds glucose altering the mutual affinity of insulin for its receptor. Cell. Molec. Life Sci. 2009, 6, 2721–2732. [Google Scholar] [CrossRef] [PubMed]
- Root-Bernstein, R.S.; Vonck, J. Modularity in receptor evolution. Insulin- and glucagon-like peptide modules as binding sites for insulin and glucose in the insulin receptor. J. Recept. Ligand Channel Res. 2010, 3, 87–96. [Google Scholar] [CrossRef]
- Root-Bernstein, R. A modular insulin-like basis for the evolution of glucose transporters (GLUT) with implications for diabetes. Evol. Bioinform. 2007, 2, 317–331. [Google Scholar] [CrossRef]
- Dwyer, D.S. Molecular model of interleukin 12 that highlights amino acid sequence homologies with adhesion domains and gastrointestinal peptides. J. Mol. Graphics 1996, 14, 148–157. [Google Scholar] [CrossRef]
- Sadanandam, A.; Varney, M.L.; Singh, R.K. Identification of semaphorin 5A interacting protein by applying a priori knowledge and peptide complementarity related to protein evolution and structure. Genom. Proteom. Bioinform. 2008, 6, 163–174. [Google Scholar] [CrossRef]
- Root-Bernstein, R.S.; Dillon, P.F. Molecular complementarity I: The complementarity theory of the origin and evolution of life. J. Theor. Biol. 1997, 188, 447–479. [Google Scholar] [CrossRef]
- Hunding, A.; Keeps, F.; Lancet, D.; Minsky, A.; Norris, V.; Raine, D.; Sriram, K.; Root-Bernstein, R. Compositional complementarity and prebiotic ecology in the origin of life. Bioessays 2006, 28, 399–412. [Google Scholar] [CrossRef]
- Root-Bernstein, R. A modular hierarchy-based theory of the chemical origins of life based on molecular complementarity. Acc. Chem. Res. 2012, 45, 2169–2177. [Google Scholar] [CrossRef]
- Root-Bernstein, R.S.; Dillon, P.F. Small molecule complementarity as a source of novel pharmaceutical agents and combination therapies. Curr. Pharm. Des. 2008, 14, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Root-Bernstein, R.; Podufaly, A.; Dillon, P.F. Estradiol binds to insulin and insulin receptor decreasing insulin binding in vitro. Front. Endocrinol. 2014, 5, 118. [Google Scholar] [CrossRef] [PubMed]
- Štambuk, N.; Manojlović, Z.; Turčić, P.; Martinić, R.; Konjevoda, P.; Weitner, T.; Wardega, P.; Gabričević, M. A simple three-step method for design and affinity testing of new antisense peptides: An example of erythropoietin. Int. J. Mol. Sci. 2014, 15, 9209–9223. [Google Scholar] [CrossRef]
- Root-Bernstein, R.S. Catecholamines bind to enkephalins, morphiceptin, and morphine. Brain Res. Bull. 1987, 18, 509–532. [Google Scholar] [CrossRef]
- Dillon, P.F.; Root-Bernstein, R.S.; Lieder, C.M. Molecular shielding of electric field complex dissociation. Biophys. J. 2006, 90, 1432–1438. [Google Scholar] [CrossRef][Green Version]
- Root-Bernstein, R.; Churchill, B.; Turke, M.; Subhramanyam, U.K.T.; Labahn, J. Mutual enhancement of opioid and adrenergic receptors by combinations of opioids and adrenergic ligands is reflected in molecular complementarity of ligands: Drug development possibilities. Int. J. Mol. Sci. 2019, 20, 4137. [Google Scholar] [CrossRef]
- Madhusudanan, K.P.; Katti, S.B.; Haq, W.; Misra, P.K. Antisense peptide interactions studied by electrospray ionization mass spectrometry. J. Mass Spectrom. 2000, 35, 237–241. [Google Scholar] [CrossRef]
- Higashijima, T.; Kobayashi, J.; Nagai, U.; Miyazawa, T. Nuclear-magnetic-resonance study on Met-enkephalin and Met-enkephalinamide. Molecular association and conformation. Eur. J. Biochem. 1979, 97, 43–57. [Google Scholar] [CrossRef]
- Khaled, M.A.; Long, M.M.; Thompson, W.D.; Bradley, R.J.; Brown, G.B.; Urry, D.W. Conformational states of enkephalins in solution. Biochem. Biophys. Res. Commun. 1976, 76, 224–231. [Google Scholar] [CrossRef]
- Griffin, J.F.; Langs, D.A.; Smith, G.D.; Blundell, T.L.; Tickle, I.J.; Bedarkar, S. The crystal structures of [Met5]enkephalin and a third form of [Leu5]enkephalin: Observations of a novel pleated beta-sheet. Proc. Natl. Acad. Sci. USA 1986, 83, 3272–3276. [Google Scholar] [CrossRef] [PubMed]
- Bleiholder, C.; Dupuis, N.F.; Bowers, M.T. Dimerization of chirally mutated Enkephalin neurotransmitters: Implications for peptide and protein aggregation mechanisms. J. Phys. Chem. B 2013, 117, 1770–1779. [Google Scholar] [CrossRef]
- Do, T.D.; LaPointe, N.E.; Sangwan, S.; Teplow, D.B.; Feinstein, S.C.; Sawaya, M.R.; Eisenberg, D.S.; Bowers, M.T. Factors that drive peptide assembly from native to amyloid structures: Experimental and theoretical analysis of [leu-5]-enkephalin mutants. J. Phys. Chem. B 2014, 118, 7247–7256. [Google Scholar] [CrossRef] [PubMed]
- Carlacci, L. Conformational analysis of [Met5]-enkephalin: Solvation and ionization considerations. J. Comput. Aided Mol. Des. 1998, 12, 195–213. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, S.; Singh, P.K.; Singh, U.; Singru, P.S.; Padinhateeri, R.; Maji, S. K Molecular interpretation of ACTH-β-endorphin coaggregation: Relevance to secretory granule biogenesis. PLoS ONE 2012, 7, e31924. [Google Scholar] [CrossRef]
- Nespovitaya, N.; Mahou, P.; Laine, R.; Kaminskim, S.G.; Kaminski, C. Heparin acts as a structural component of β-endorphin amyloid fibrils rather than a simple aggregation promoter. Chem. Commun. 2017, 53, 1273–1276. [Google Scholar] [CrossRef] [PubMed]
- Nespovitaya, N.; Gath, J.; Barylyuk, K.; Seuring, C.; Meier, B.H.; Riek, R. Dynamic assembly and disassembly of functional β-endorphin amyloid fibrils. J. Am. Chem. Soc. 2016, 138, 846–856. [Google Scholar] [CrossRef] [PubMed]
- Seuring, C.; Verasdonck, J.; Gath, J.; Ghosh, D.; Nespovitaya, N.; Wälti, M.A.; Maji, S.K.; Cadalbert, R.; Güntert, P.; Meier, B.H.; et al. The three-dimensional structure of human β-endorphin amyloid fibrils. Nat. Struct. Mol. Biol. 2020, 27, 1178–1184. [Google Scholar] [CrossRef]
- Seuring, C.; Gath, J.; Verasdonck, J.; Cadalbert, R.; Rivier, J.; Böckmann, A.; Meier, B.H.; Riek, R. Solid-state NMR sequential assignment of the β-endorphin peptide in its amyloid form. Biomol. NMR Assign. 2016, 10, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.K.; Joyce, P.B.M.; Gorr, S.-U. Aggregation chaperones enhance aggregation and storage of secretory proteins in endocrine cell. J. Biol. Chem. 2000, 275, 27032–27036. [Google Scholar] [CrossRef]
- Blank, M.S.; Diez, J.A.; Roberts, D.L. Monoaminergic antagonists which block naloxone-induced release of luteinizing hormone bind selectively to hypothalamic opiate receptors. Brain Res. 1983, 279, 153–158. [Google Scholar] [CrossRef]
- Jacobson, W.; Wilkinson, M.; Gibson, C.J. Direct effects of the adrenergic neurotoxin DSP4 on central opiate receptors: Implications for neuroendocrine studies. Exp. Brain Res. 1985, 59, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.; Jacobson, W.; Wilkinson, D.A. The noradrenergic neurotoxins DSP4 and xylamine bind to opiate receptors. Brain Res. Bull. 1985, 14, 493–495. [Google Scholar] [CrossRef]
- Takayama, H.; Ogawa, N.; Asanuma, M.; Hirata, H.; Ogura, T.; Ota, Z. Effects of beta-adrenergic blocking agents on specific binding of [3H]D-Ala2-Met5-enkephalinamide and [3H]naloxone. Acta Med. Okayama 1991, 45, 295–299. [Google Scholar] [CrossRef]
- Cicero, T.J.; Wilcox, C.E.; Meyer, E.R. Effect of alpha-adrenergic blockers on naloxone-binding in brain. Biochem. Pharmacol. 1974, 23, 2349–2352. [Google Scholar] [CrossRef]
- Spiehler, V.; Fairhurst, A.S.; Randall, L.O. The interaction of phenoxybenzamine with the mouse brain opiate receptor. Mol. Pharmacol. 1978, 14, 587–595. [Google Scholar]
- Tampier, L.; Alpers, H.S.; Davis, V.E. Influence of catecholamine-derived alkaloids and beta-adrenergic blocking agents on stereospecific binding of 3H-naloxone. Res. Commun. Chem. Pathol. Pharmacol. 1977, 17, 731–734. [Google Scholar] [PubMed]
- Root-Bernstein, R.; Churchill, B.; Turke, M. Glutathione and glutathione-like sequences of opioid and aminergic receptors bind ascorbic acid: Adrenergic and opioid drugs mediating antioxidant function: Relevance for anesthesia and abuse. Int. J. Mol. Sci. 2020, 21, 6230. [Google Scholar] [CrossRef] [PubMed]
- Meneghini, V.; Cuccurazzu, B.; Bortolotto, V.; Ramazzotti, V.; Ubezio, F.; Tzschentke, T.M.; Canonico, P.L.; Grilli, M. The noradrenergic component in tapentadol action counteracts μ-opioid receptor-mediated adverse effects on adult neurogenesis. Mol. Pharmacol. 2014, 85, 658–670. [Google Scholar] [CrossRef]
- Li, C.; Chen, S.Q.; Chen, B.X.; Huang, W.Q.; Liu, K.X. The antinociceptive effect of intrathecal tramadol in rats: The role of alpha 2-adrenoceptors in the spinal cord. J. Anesth. 2012, 26, 230–235. [Google Scholar] [CrossRef]
- Kayser, V.; Besson, J.M.; Guilbaud, G. Evidence for a noradrenergic component in the antinociceptive effect of the analgesic agent tramadol in an animal model of clinical pain.; the arthritic rat. Eur. J. Pharmacol. 1992, 224, 83–88. [Google Scholar] [CrossRef]
- Wu, X.; Bartel, D.P. kpLogo: Positional k-mer analysis reveals hidden specificity in biological sequences. Nucleic Acids Res. 2017, 45, W534–W538. [Google Scholar] [CrossRef]
- Rubenstein, L.A.; Lanzara, R.G. Activation of G protein-coupled receptors entails cysteine modulation of agonist binding. J. Molec. Struc. 1998, 430, 57–71. [Google Scholar] [CrossRef]
- Gouldson, P.R.; Snell, C.R.; Bywater, R.P.; Higgs, C.; Reynolds, C.A. Domain swapping in G-protein coupled receptor dimers. Protein Eng. 1998, 11, 1181–1193. [Google Scholar] [CrossRef] [PubMed]
- Hebert, T.E.; Moffett, S.; Morello, J.P.; Loisel, T.P.; Bichet, D.G.; Barret, C.; Bouvier, M. A peptide derived from a beta2-adrenergic receptor transmembrane domain inhibits both receptor dimerization and activation. J. Biol. Chem. 1996, 271, 16384–16392. [Google Scholar] [CrossRef]
- Stanasila, L.; Perez, J.B.; Vogel, H.; Cotecchia, S. Oligomerization of the alpha1a- and alpha1b-adrenergic receptor subytpes: Potential implications in receptor internalization. J. Biol. Chem. 2003, 278, 40239–40251. [Google Scholar] [CrossRef]
- Cotecchia, S.; Stanasila, L.; Diviani, D. Protein-protein interactions at the adrenergic receptors. Curr. Drug Targets 2012, 13, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Parmar, V.K.; Grinde, E.; Mazurkiewicz, J.E.; Herrick-Davis, K. Beta2-adrenergic receptor homodimers: Role of transmembrane domain 1 and helix 8 in dimerization and cell surface expression. Biochim. Biophys. Acta Biomembr. 2017, 1859, 1445–1455. [Google Scholar] [CrossRef]
- Huang, J.; Chen, S.; Zhang, J.J.; Huang, X.Y. Crystal structure of oligomeric β1-adrenergic G protein-coupled receptors in ligand-free basal state. Nat. Struct. Mol. Biol. 2013, 20, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, J.J.; López-Giménez, J.F.; Milligan, G. Multiple interactions between transmembrane helices generate the oligomeric alpha1b-adrenoceptor. Mol. Pharmacol. 2004, 66, 1123–1137. [Google Scholar] [CrossRef]
- Lee, S.P.; O’Dowd, B.F.; Rajaram, R.D.; Nguyen, T.; George, S.R. D2 dopamine receptor homodimerization is mediated by multiple sites of interaction: Including an intermolecular interaction involving transmembrane domain 4. Biochemistry 2003, 42, 11023–11031. [Google Scholar] [CrossRef]
- Johnston, J.M.; Filizola, M. Differential stability of the crystallographic interfaces of mu- and kappa-opioid receptors. PLoS ONE 2014, 9, e90694. [Google Scholar] [CrossRef]
- Zhang, T.; Xu, J.; Pan, Y.X. A truncated six transmembrane splice variant MOR-1G enhances expression of the full-length seven transmembrane μ-opioid receptor through heterodimerization. Mol. Pharmacol. 2020, 98, 518–527. [Google Scholar] [CrossRef]
- Samoshkin, A.; Convertino, M.; Viet, C.T.; Wieskopf, J.S.; Kambur, O.; Marcovitz, J.; Patel, P.; Stone, L.S.; Kalso, E.; Mogil, J.S.; et al. Structural and functional interactions between six-transmembrane μ-opioid receptors and β2-adrenoreceptors modulate opioid signaling. Sci. Rep. 2015, 5, 18198. [Google Scholar] [CrossRef]
- Johnston, J.M.; Aburi, M.; Provasi, D.; Bortolato, A.; Urizar, E.; Lambert, N.A.; Javitch, J.A.; Filizola, M. Making structural sense of dimerization interfaces of delta opioid receptor homodimers. Biochemistry 2011, 50, 1682–1690. [Google Scholar] [CrossRef]
- Manglik, A.; Kruse, A.C.; Kobilka, T.S.; Thian, F.S.; Mathiesen, J.M.; Sunahara, R.K.; Pardo, L.; Weis, W.I.; Kobilka, B.K.; Granier, S. Crystal structure of the µ-opioid receptor bound to a morphinan antagonist. Nature 2012, 485, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Provasi, D.; Boz, M.B.; Johnston, J.M.; Filizola, M. Preferred supramolecular organization and dimer interfaces of opioid receptors from simulated self-association. PLoS Comput. Biol. 2015, 11, e1004148. [Google Scholar] [CrossRef]
- Wang, L.; Yuan, Y.; Chen, X.; Chen, J.; Guo, Y.; Li, M.; Li, C.; Pu, X. Probing the cooperative mechanism of the μ–δ opioid receptor heterodimer by multiscale simulation. Phys. Chem. Chem. Phys. 2018, 20, 29969–29982. [Google Scholar] [CrossRef]
- Waterman, M.S.; Eggert, M.; Lander, E. Parametric sequence comparisons. Proc. Natl. Acad. Sci. USA 1992, 89, 6090–6093. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Grünewald, S. Sequence, structure and ligand binding evolution of rhodopsin-like g protein-coupled receptors: A crystal structure-based phylogenetic analysis. PLoS ONE 2015, 10, e0123533. [Google Scholar] [CrossRef] [PubMed]
- Larhammar, D.; Dreborg, S.; Larsson, T.A.; Sundström, G. Early duplications of opioid receptor and peptide genes in vertebrate evolution. Ann. N. Y. Acad. Sci. 2009, 1163, 451–453. [Google Scholar] [CrossRef] [PubMed]
- Larhammar, D.; Bergqvist, C.; Sundström, G. Ancestral vertebrate complexity of the opioid system. Vitam. Horm. 2015, 97, 95–122. [Google Scholar] [CrossRef] [PubMed]
- Stevens, R.C.; Cherezov, V.; Katritch, V.; Abagyan, R.; Kuhn, P.; Rosen, H.; Wüthrich, K. The GPCR network: A large-scale collaboration to determine human GPCR structure and function. Nat. Rev. Drug Discov. 2013, 12, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.M.; Mai, T.L.; Chen, C.M. Visualizing the GPCR network: Classification and evolution. Sci. Rep. 2017, 7, 15495. [Google Scholar] [CrossRef] [PubMed]
- van der Horst, E.; Peironcely, J.E.; Ijzerman, A.P.; Beukers, M.W.; Lane, J.R.; van Vlijmen, H.W.; Emmerich, M.T.; Okuno, Y.; Bender, A. A novel chemogenomics analysis of G protein-coupled receptors (GPCRs) and their ligands: A potential strategy for receptor de-orphanization. BMC Bioinform. 2010, 11, 316. [Google Scholar] [CrossRef]
- Krishnan, A.; Mustafa, A.; Almén, M.S.; Fredriksson, R.; Williams, M.J.; Schiöth, H.B. Evolutionary hierarchy of vertebrate-like heterotrimeric G protein families. Mol. Phylogenet. Evol. 2015, 91, 27–40. [Google Scholar] [CrossRef]
- Pfeiffer, M.; Koch, T.; Schröder, H.; Laugsch, M.; Höllt, V.; Schulz, S. Heterodimerization of somatostatin and opioid receptors cross-modulates phosphorylation, internalization, and desensitization. J. Biol. Chem. 2002, 277, 19762–19772. [Google Scholar] [CrossRef]
- Somvanshi, R.K.; Kumar, U. δ-opioid receptor and somatostatin receptor-4 heterodimerization: Possible implications in modulation of pain associated signaling. PLoS ONE 2014, 9, e85193. [Google Scholar] [CrossRef]
- Cussac, D.; Rauly-Lestienne, I.; Heusler, P.; Finana, F.; Cathala, C.; Bernois, S.; De Vries, L. μ-Opioid and 5-HT1A receptors heterodimerize and show signalling crosstalk via G protein and MAP-kinase pathways. Cell Signal. 2012, 24, 1648–1657. [Google Scholar] [CrossRef]
- Ugur, M.; Derouiche, L.; Massotte, D. Heteromerization modulates mu opioid receptor functional properties in vivo. Front Pharmacol. 2018, 9, 1240. [Google Scholar] [CrossRef]
- Rocheville, M.; Lange, D.C.; Kumar, U.; Patel, S.C.; Patel, R.C.; Patel, Y.C. Receptors for dopamine and somatostatin: Formation of hetero-oligomers with enhanced functional activity. Science 2000, 288, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Somvanshi, R.K.; Chaudhari, N.; Qiu, X.; Kumar, U. Heterodimerization of β2 adrenergic receptor and somatostatin receptor 5: Implications in modulation of signaling pathway. J. Mol. Signal. 2011, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Somvanshi, R.K.; War, S.A.; Chaudhari, N.; Qiu, X.; Kumar, U. Receptor specific crosstalk and modulation of signaling upon heterodimerization between β1-adrenergic receptor and somatostatin receptor-5. Cell Signal. 2011, 23, 794–811. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekera, P.C.; Wan, T.C.; Gizewski, E.T.; Auchampach, J.A.; Lasley, R.D. Adenosine A1 receptors heterodimerize with β1- and β2-adrenergic receptors creating novel receptor complexes with altered G protein coupling and signaling. Cell Signal. 2013, 25, 736–742. [Google Scholar] [CrossRef] [PubMed]
- Dillon, P.F.; Root-Bernstein, R.S.; Lieder, C.M. Ascorbate enhancement of H1 histamine receptor sensitivity coincides with ascorbate oxidation inhibition by histamine receptors. Am. J. Physiol. Cell. Physiol. 2006, 291, C977–C984. [Google Scholar] [CrossRef]
- Forman, H.J.; Zhang, H.; Rinna, A. Glutathione: Overview of its protective roles, measurement, and biosynthesis. Mol. Aspects Med. 2009, 30, 1–12. [Google Scholar] [CrossRef]
- Roston, S. Studies of the epinephrine-glutathione reaction in aqueous solution and human blood. Arch. Biochem. Biophys. 1965, 109, 41–48. [Google Scholar] [CrossRef]
- Heacock, R.A.; Mattok, G.L. The reaction of adrenochrome with glutathione. Arch. Biochem. Biophys. 1964, 107, 352–353. [Google Scholar] [CrossRef]
- Powell, W.S.; Heacock, R.A. Adrenochrome-thiol addition products. Experientia 1972, 28, 124–125. [Google Scholar] [CrossRef]
- Nappi, A.J.; Vass, E. The effects of glutathione and ascorbic acid on the oxidations of 6-hydroxydopa and 6-hydroxydopamine. Biochim. Biophys. Acta. 1994, 1201, 498–504. [Google Scholar] [CrossRef]
- Aizenman, E.; Boeckman, F.A.; Rosenberg, P.A. Glutathione prevents 2,4,5-trihydroxyphenylalanine excitotoxicity by maintaining it in a reduced, non-active form. Neurosci. Lett. 1992, 144, 233–236. [Google Scholar] [CrossRef]
- Drukarch, B.; Jongenelen, C.A.; Schepens, E.; Langeveld, C.H.; Stoof, J.C. Glutathione is involved in the granular storage of dopamine in rat PC 12 pheochromocytoma cells: Implications for the pathogenesis of Parkinson’s disease. J. Neurosci. 1996, 16, 6038–6045. [Google Scholar] [CrossRef] [PubMed]
- Remião, F.; Carmo, H.; Carvalho, F.D.; Bastos, M.L. Inhibition of glutathione reductase by isoproterenol oxidation products. J. Enz. Inhib. 1999, 15, 47–61. [Google Scholar] [CrossRef]
- Costa, V.M.; Silva, R.; Ferreira, L.M.; Branco, P.S.; Carvalho, F.; Bastos, M.L.; Carvalho, R.A.; Carvalho, M.; Remião, F. Oxidation process of adrenaline in freshly isolated rat cardiomyocytes: Formation of adrenochrome, quinoproteins, and GSH adduct. Chem. Res. Toxicol. 2007, 20, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, M.; Milhazes, N.; Remião, F.; Borges, F.; Fernandes, E.; Amado, F.; Monks, T.J.; Carvalho, F.; Bastos, M.L. Hepatotoxicity of 3,4-methylenedioxyamphetamine and alpha-methyldopamine in isolated rat hepatocytes: Formation of glutathione conjugates. Arch. Toxicol. 2004, 78, 16–24. [Google Scholar] [CrossRef]
- Correia, M.A.; Krowech, G.; Caldera-Munoz, P.; Yee, S.L.; Straub, K.; Castagnoli, N., Jr. Morphine metabolism revisited. II. Isolation and chemical characterization of a glutathionylmorphine adduct from rat liver microsomal preparations. Chem. Biol. Interact. 1984, 51, 13–24. [Google Scholar] [CrossRef]
- Ishida, T.; Kumagai, Y.; Ikeda, Y.; Ito, K.; Yano, M.; Tok, I.S.; Mihashi, K.; Fujioka, T.; Iwase, Y.; Hachiyama, S. (8S)-(glutathion-S-yl)dihydromorphinone: Novel metabolite of morphine from guinea pig bile. Drug Metab. Dispos. 1989, 17, 77–81. [Google Scholar]
- Kumagai, Y.; Todaka, T.; Toki, S. A new metabolic pathway of morphine: In vivo and in vitro formation of morphinone and morphine-glutathione adduct in guinea pig. J. Pharmacol. Exp. Ther. 1990, 255, 504–510. [Google Scholar]
- Armstrong, S.C.; Cozza, K.L. Pharmacokinetic drug interactions of morphine, codeine, and their derivatives: Theory and clinical reality, Part I. Psychosomatics 2003, 44, 167–171. [Google Scholar] [CrossRef]
- Nagamatsu, K.; Inoue, K.; Terao, T.; Toki, S. Effects of glutathione and phenobarbital on the toxicity of codeinone. Biochem. Pharmacol. 1986, 35, 1675–1678. [Google Scholar] [CrossRef]
- Ishida, T.; Yano, M.; Toki, S. In vivo formation of codeinone-glutathione adduct: Isolation and identification of a new metabolite in the bile of codeine-treated guinea pig. J. Anal. Toxicol. 1998, 22, 567–572. [Google Scholar] [CrossRef]
- Costa, V.M.; Ferreira, L.M.; Branco, P.S.; Carvalho, F.; Bastos, M.L.; Carvalho, R.A.; Carvalho, M.; Remio, F. Cross-functioning between the extraneuronal monoamine transporter and multidrug resistance protein 1 in the uptake of adrenaline and export of 5-(glutathion-S-yl)adrenaline in rat cardiomyocytes. Chem. Res. Toxicol. 2009, 22, 129–135. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Root-Bernstein, R. Adrenergic agonists and the mu opioid receptor. In The Neurobiology, Physiology, and Behavior of Pain; Rajendram, R., Patel, V.B., Preedy, V.R., Eds.; Elsevier Science: Amsterdam, The Netherlands; New York, NY, USA, 2022; pp. 1–22. ISBN 9780128206089. [Google Scholar]
- Dwyer, D.S. Assembly of exons from unitary transposable genetic elements: Implications for the evolution of protein-protein interactions. J. Theor. Biol. 1998, 194, 11–27. [Google Scholar] [CrossRef] [PubMed]
- Delihas, N. Enterobacterial small mobile sequences carry open reading frames and are found intragenically--evolutionary implications for formation of new peptides. Gene Regul. Syst. Biol. 2007, 1, 191–205. [Google Scholar] [CrossRef]
- 156 Fattash, I.; Rooke, R.; Wong, A.; Hui, C.; Luu, T.; Bhardwaj, P.; Yang, G. Miniature inverted-repeat transposable elements: Discovery, distribution, and activity. Genome 2013, 56, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, C.Y.; Clark, K.B. Modularity in the design of complex engineering systems. In Complex Engineered Systems. Understanding Complex Systems; Braha, D., Minai, A., Bar-Yam, Y., Eds.; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar] [CrossRef]
- Andrianantoandro, E.; Basu, S.; Karig, D.K.; Weiss, R. Synthetic biology: New engineering rules for an emerging discipline. Mol. Syst. Biol. 2006, 2, 2006-0028. [Google Scholar] [CrossRef] [PubMed]
- Hurley, J.H. Membrane binding domains. Biochim. Biophys. Acta 2006, 1761, 805–811. [Google Scholar] [CrossRef] [PubMed]
- Rooklin, D.; Wang, C.; Katigbak, J.; Arora, P.S.; Zhang, Y. AlphaSpace: Fragment-centric topographical mapping to target protein-protein interaction interfaces. J. Chem. Inf. Model. 2015, 55, 1585–1599. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Boyken, S.E.; Jia, M.; Busch, F.; Flores-Solis, D.; Bick, M.J.; Lu, P.; Van Aernum, Z.L.; Sahasrabuddhe, A.; Langan, R.A.; et al. Programmable design of orthogonal protein heterodimers. Nature 2019, 565, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Kočar, V.; Božič, A.S.; Doles, T.; Bašić, N.; Gradišar, H.; Pisanski, T.; Jerala, R. TOPOFOLD, the designed modular biomolecular folds: Polypeptide-based molecular origami nanostructures following the footsteps of DNA. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2015, 7, 218–237. [Google Scholar] [CrossRef]
- Lapenta, F.; Aupič, J.; Strmšek, Ž.; Jerala, R. Coiled coil protein origami: From modular design principles towards biotechnological applications. Chem. Soc. Rev. 2018, 47, 3530–3542. [Google Scholar] [CrossRef]
- Källberg, M.; Bhardwaj, N.; Langlois, R.; Lu, H. A structure-based protocol for learning the family-specific mechanisms of membrane-binding domains. Bioinformatics 2012, 28, i431–i437. [Google Scholar] [CrossRef][Green Version]
- Zhang, S.; Tao, F.; Qing, R.; Tang, H.; Skuhersky, M.; Corin, K.; Tegler, L.; Wassie, A.; Wassie, B.; Kwon, Y.; et al. QTY code enables design of detergent-free chemokine receptors that retain ligand-binding activities. Proc. Natl. Acad. Sci. USA 2018, 115, E8652–E8659. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Holmes, T.; Lockshin, C.; Rich, A. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc. Natl. Acad. Sci. USA 1993, 90, 3334–3338. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S. Fabrication of novel materials through molecular self-assembly. Nat. Biotechnol. 2003, 21, 1171–1178. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Holmes, T.C.; DiPersio, C.M.; Hynes, R.O.; Su, X.; Rich, A. Self-complementary oligopeptide matrices support mammalian cell attachment. Biomaterials 1995, 16, 1385–1393. [Google Scholar] [CrossRef]
- Lu, J.; Wang, X. Biomimetic self-assembling peptide hydrogels for tissue engineering applications. Adv. Exp. Med. Biol. 2018, 1064, 297–312. [Google Scholar] [CrossRef]
- Zhang, S. Beyond the petri dish. Nat. Biotechnol. 2004, 22, 151–152. [Google Scholar] [CrossRef]
- Ardejani, M.S.; Orner, B.P. Obey the peptide assembly rules. Science 2013, 340, 561–562. [Google Scholar] [CrossRef]
- Dinca, V.; Kasotakis, E.; Catherine, J.; Mourka, A.; Ranella, A.; Ovsianikov, A.; Chichkov, B.N.; Farsari, M.; Mitraki, A.; Fotakis, C. Directed three-dimensional patterning of self-assembled peptide fibrils. Nano Lett. 2008, 8, 538–543. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, S. Designer self-assembling peptide materials. Macromol. Biosci. 2007, 7, 13–22. [Google Scholar] [CrossRef]
- Qing, R.; Tao, F.; Chatterjee, P.; Yang, G.; Han, Q.; Chung, H.; Ni, J.; Suter, B.P.; Kubicek, J.; Maertens, B.; et al. Non-full-length water-soluble CXCR4QTY and CCR5QTY chemokine receptors: Implication for overlooked truncated but functional membrane receptors. Iscience 2020, 23, 101670. [Google Scholar] [CrossRef] [PubMed]














| Kd (µM) U. V. @ 200 nm | M-Enk | Endomorphin | Morph |
|---|---|---|---|
| Epinephrine HCl | 5.8 * | 0.3 | 8.0 @ 0.5 * |
| Norepinephrine HCL | 5.3 * | 0.4 | 0.4 * |
| Dopamine | 30 * | 0.5 | 0.6 * |
| L-DOPA | 70 * | >1000 * | |
| Amphetamine | 80 * | 0.1 * | |
| Propranolol | 25 | 45 | |
| Salbutamol | 30 | 0.3 | |
| Isoproterenol | 40 | 0.1 | |
| Phenylephrine | 30 | 0.13 | |
| Tyramine | 12 | 50 | |
| Octopamine | 80 * | 0.35 | 3.2 * |
| Homovanillic Acid | 80 * | >1000 * | |
| Tyrosine | >1000 * | >1000 * | |
| Phenylalanine | >1000 * | >1000 * | |
| Serotonin | 45 * | 0.2 | 0.7 * |
| Melatonin | 130 | 1.2 | 300 |
| Histamine | >1000 * | 0.17 | >1000 * |
| Acetylcholine | 80 * | >1000 | >1000 * |
| Ascorbic Acid | 600 | >1000 | >1000 |
| Kd (μM) @ 200 nm | MOR | NALOX | MENK | Epi | NorEpi | tyro | phenyl |
|---|---|---|---|---|---|---|---|
| Mu OPR 38–51, EC1 | 35 | 0.5/35 | 0.15/55 | 1.2/35 | 1.4/45 | 85 | 50 |
| Mu OPR 111–122, TM2 | 50 | 0.5/38 | 0.33/80 | 1.3/40 | 1.3/40 | 700 | >1000 |
| Mu OPR 121–131, TM2 | 900 | >1000 | 3.5/90 | >1000 | >1000 | >1000 | >1000 |
| Mu OPR 132–143, EC2 | 35 | 0.5/42 | 0.4/70 | 1.4/35 | 1.4/40 | 60 | 80 |
| Mu OPR 211–226, EC3 | 30 | 1.0/45 | 1.0/65 | 1.2/40 | 1.3/45 | 160 | 200 |
| B2AR 97–103, EC2 | 1 | 6 | 130 | 120 | 600 | >1000 | >1000 |
| B2AR 103–113, TM5 | 40 | 50 | >1000 | >1000 | |||
| B2AR 105–108, EC2/TM5 | 30 | 30 | 30 | 35 | 35 | 50 | 55 |
| B2AR 175–188, EC3 | 50 | 40 | 700 | 900 | 1000 | >1000 | >1000 |
| B2AR 183–185, EC3 | >1000 | >1000 | >1000 | 25 | 12 | 35 | 30 |
| SP | EC 1 | EC 2 | EC 3 | EC 4 | TM 1 | TM 2 | TM 3 | TM 4 | TM 5 | TM 6 | TM 7 | IC 1 | IC 2 | IC 3 | IC 4 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ZEB | 21/53 | 14/19 | 13/26 | 6/12 | 20/29 | 13/19 | 13/21 | 12/21 | 15/25 | 19/20 | 15/23 | 9/10 | 13/20 | 14/51 | 32/79 |
| STK | 13/52 | 11/18 | 8/20 | 4/11 | 18/29 | 21/25 | 17/22 | 14/20 | 13/25 | 22/28 | 14/22 | 9/12 | 8/14 | 11/51 | 32/64 |
| TO | 15/45 | 6/8 | 8/24 | 7/19 | 17/25 | 21/31 | 13/22 | 14/24 | 15/23 | 23/28 | 12/25 | 6/11 | 11/19 | 22/60 | 6/42 |
| MAL | 3/26 | 12/18 | 8/24 | 8/11 | 15/27 | 15/18 | 13/23 | 10/21 | 15/26 | 19/28 | 14/24 | 7/11 | 16/27 | 19/62 | 17/68 |
| MS | 17/56 | 9/13 | 12/26 | 3/15 | 19/29 | 17/25 | 14/22 | 16/23 | 14/25 | 17/26 | 15/23 | 10/11 | 14/21 | 21/54 | 25/90 |
| HUM | 19/68 | 10/13 | 12/19 | 7/12 | 14/24 | 20/28 | 12/21 | 13/27 | 14/28 | 16/24 | 14/23 | 4/11 | 10/14 | 18/67 | 41/137 |
| HUK | 18/63 | 9/13 | 16/26 | 4/15 | 17/26 | 16/24 | 14/22 | 15/24 | 14/25 | 19/24 | 15/23 | 9/11 | 13/21 | 18/54 | 21/64 |
| Avg. Cons. | 29% | 70% | 47% | 41% | 64% | 72% | 63% | 60% | 57% | 76% | 61% | 70% | 63% | 31% | 32% |
| HUMAN | Homologous to: | Homologous to: | Homologous to: |
|---|---|---|---|
| Location | Extracellular 2 loop | Transmembrane helix 2 | Transmembrane helix 6 |
| Sequence | LMGSWPFGRVLCK | FIVNLAVADLLLTSTVLPFSA | VVAVFVLCWTPIFI |
| Receptors | Adenosine | ||
| Adrenergic, Alpha | Adrenergic, Alpha | Adrenergic, Alpha | |
| Adrenergic, Beta | Adrenergic, Beta | Adrenergic, Beta | |
| Angiotensin II | Angiotensin II | ||
| Cannabinoid | |||
| C3a anaphylatoxin | C3a anaphylatoxin | ||
| C-X-C chemokine | C-X-C chemokine | ||
| Cholecystokinin | Cholecystokinin | ||
| Dopamine | Dopamine | ||
| fMet-Leu-Phe | |||
| Histamine | Histamine | Histamine | |
| Melanocortin | Melanocortin | ||
| Melatonin | |||
| Neuropeptide FF | |||
| Neuropeptide Y | Neuropeptide Y | ||
| Neuropeptides B/W | Neuropeptides B/W | ||
| Nociceptin | Nociceptin | ||
| Opioid, Delta | Opioid, Delta | Opioid, Delta | |
| Opioid, Kappa | Opioid, Kappa | Opioid, Kappa | |
| Opioid, Mu | Opioid, Mu | Opioid, Mu | |
| Orexin | Orexin | ||
| P2Y purinoceptor | |||
| Relaxin | Relaxin | ||
| Serotonin | Serotonin | Serotonin | |
| Somatostatin | Somatostatin | Somatostatin | |
| Thyrotropin-releasing hormone | |||
| Urotensin II | |||
| Vasopressin |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Root-Bernstein, R.; Churchill, B. Co-Evolution of Opioid and Adrenergic Ligands and Receptors: Shared, Complementary Modules Explain Evolution of Functional Interactions and Suggest Novel Engineering Possibilities. Life 2021, 11, 1217. https://doi.org/10.3390/life11111217
Root-Bernstein R, Churchill B. Co-Evolution of Opioid and Adrenergic Ligands and Receptors: Shared, Complementary Modules Explain Evolution of Functional Interactions and Suggest Novel Engineering Possibilities. Life. 2021; 11(11):1217. https://doi.org/10.3390/life11111217
Chicago/Turabian StyleRoot-Bernstein, Robert, and Beth Churchill. 2021. "Co-Evolution of Opioid and Adrenergic Ligands and Receptors: Shared, Complementary Modules Explain Evolution of Functional Interactions and Suggest Novel Engineering Possibilities" Life 11, no. 11: 1217. https://doi.org/10.3390/life11111217
APA StyleRoot-Bernstein, R., & Churchill, B. (2021). Co-Evolution of Opioid and Adrenergic Ligands and Receptors: Shared, Complementary Modules Explain Evolution of Functional Interactions and Suggest Novel Engineering Possibilities. Life, 11(11), 1217. https://doi.org/10.3390/life11111217







