Detection of pks Island mRNAs Using Toehold Sensors in Escherichia coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmid Construction and E. coli Strains
2.2. Cell Culture and Induction Condition
2.3. Fluorescence Measurements Using Flow Cytometry
2.4. Quantitative Reverse-Transcription PCR
2.5. In Silico Toehold Switch Sensor Design for clb ORFs
3. Results
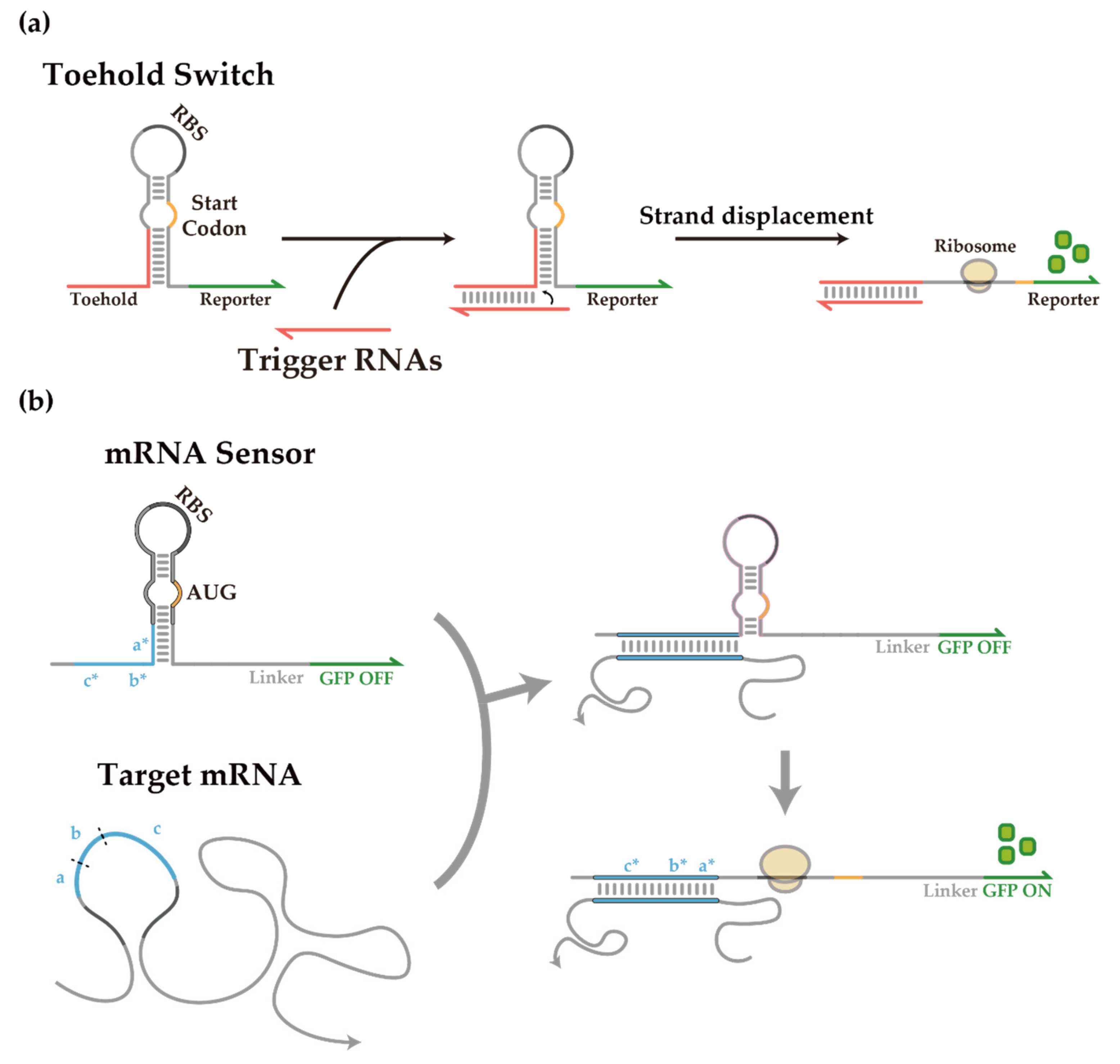
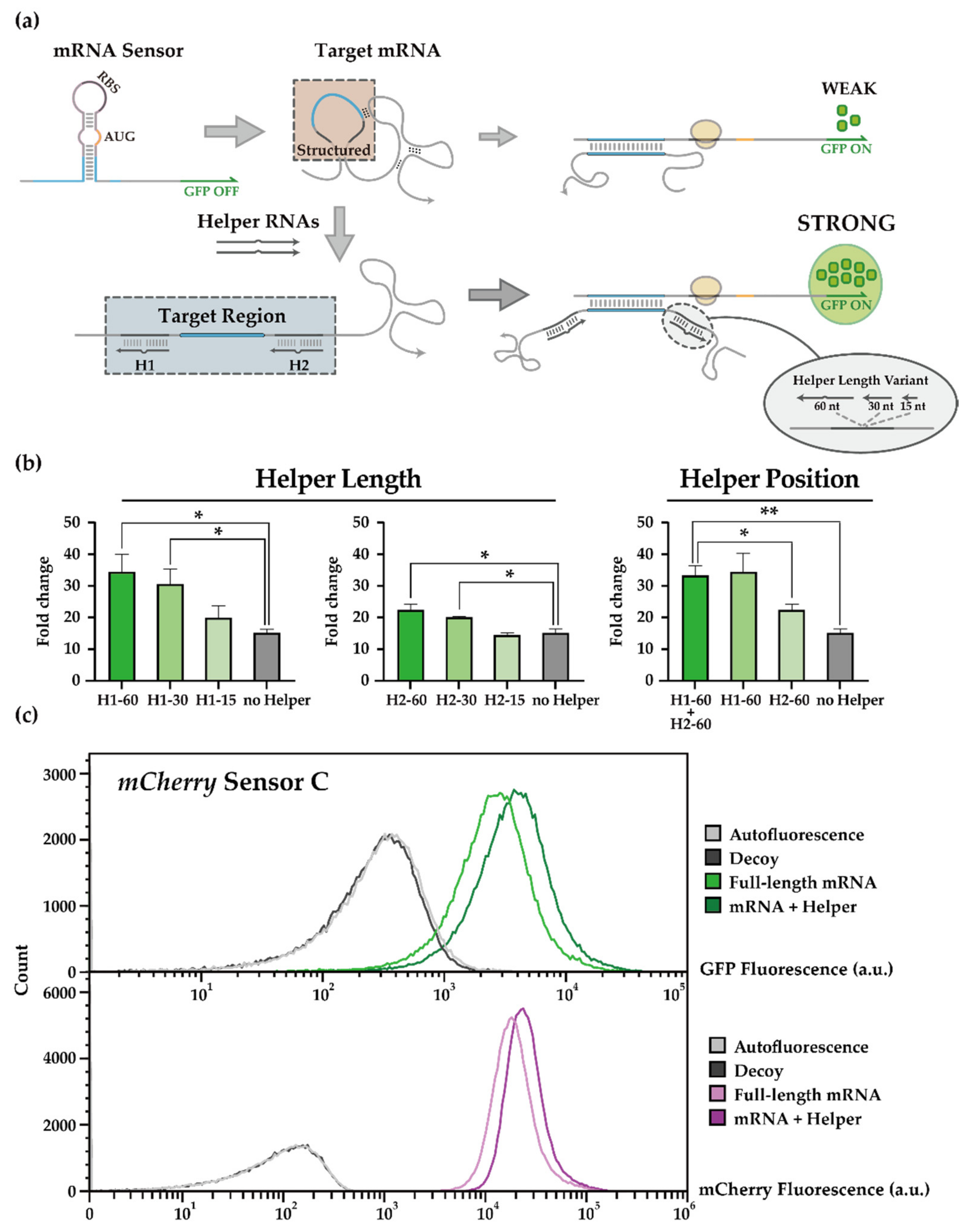
3.1. Helper-Assisted mRNA Sensing of Toehold Switch
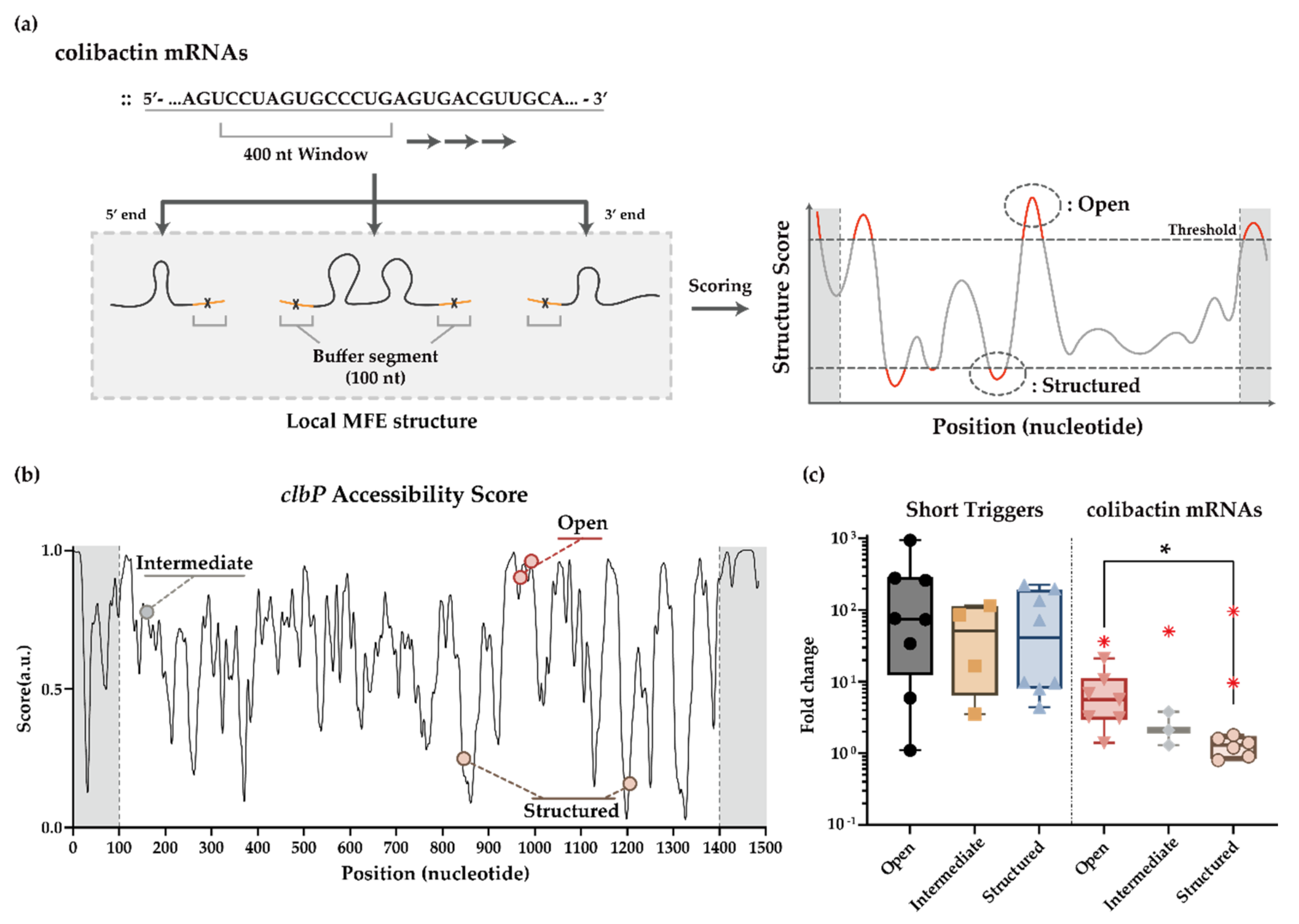
3.2. Automated Toehold Switch Design for clb ORFs in the pks Island
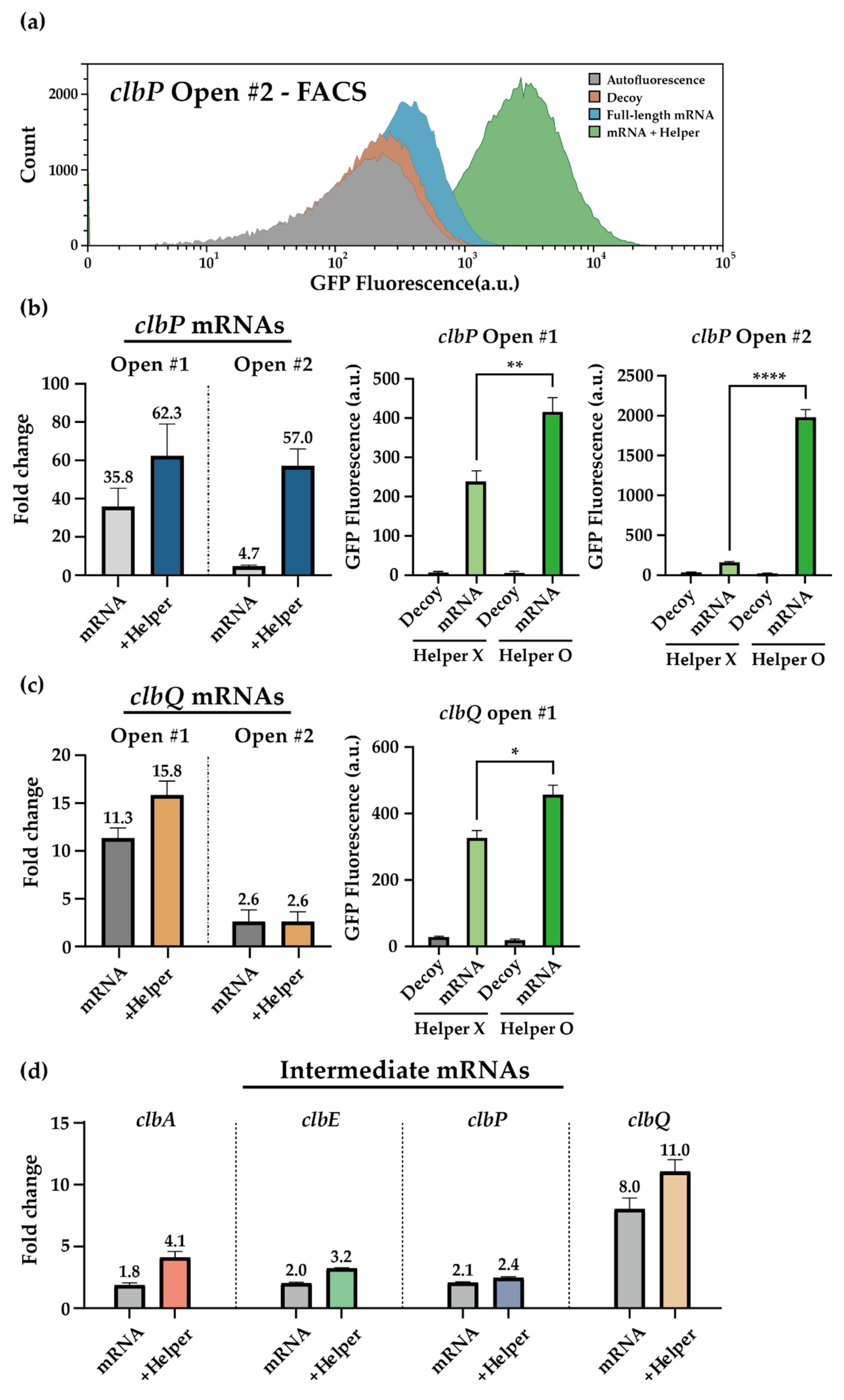
3.3. In Vivo Sensing of clb Genes Using Toehold Switch Sensors
3.4. HAM System for the clb ORFs Targeting Toehold Switch Sensors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khalil, A.S.; Collins, J.J. Synthetic biology: Applications come of age. Nat. Rev. Genet. 2010, 11, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Benner, S.A.; Sismour, A.M. Synthetic biology. Nat. Rev. Genet. 2005, 6, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Gardner, T.S.; Cantor, C.R.; Collins, J.J. Construction of a genetic toggle switch in Escherichia coli. Nature 2000, 403, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, M.R.; Savageau, M.A.; Myers, J.T.; Ninfa, A.J. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell 2003, 113, 597–607. [Google Scholar] [CrossRef]
- Elowitz, M.B.; Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 2000, 403, 335–338. [Google Scholar] [CrossRef]
- Friedland, A.E.; Lu, T.K.; Wang, X.; Shi, D.; Church, G.; Collins, J.J. Synthetic gene networks that count. Science 2009, 324, 1199–1202. [Google Scholar] [CrossRef]
- Ham, T.S.; Lee, S.K.; Keasling, J.D.; Arkin, A.P. Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS ONE 2008, 3, e2815. [Google Scholar] [CrossRef]
- Basu, S.; Mehreja, R.; Thiberge, S.; Chen, M.T.; Weiss, R. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl. Acad. Sci. USA 2004, 101, 6355–6360. [Google Scholar] [CrossRef]
- Hong, S.; Jeong, D.; Ryan, J.; Foo, M.; Tang, X.; Kim, J. Design and evaluation of synthetic RNA-based incoherent feed-forward loop circuits. Biomolecules 2021, 11, 1182. [Google Scholar] [CrossRef]
- Alnahhas, R.N.; Sadeghpour, M.; Chen, Y.; Frey, A.A.; Ott, W.; Josic, K.; Bennett, M.R. Majority sensing in synthetic microbial consortia. Nat. Commun. 2020, 11, 3659. [Google Scholar] [CrossRef]
- Wieland, M.; Fussenegger, M. Reprogrammed cell delivery for personalized medicine. Adv. Drug Deliv. Rev. 2012, 64, 1477–1487. [Google Scholar] [CrossRef]
- Xiang, S.; Fruehauf, J.; Li, C.J. Short hairpin RNA-expressing bacteria elicit RNA interference in mammals. Nat. Biotechnol. 2006, 24, 697–702. [Google Scholar] [CrossRef]
- Anderson, J.C.; Clarke, E.J.; Arkin, A.P.; Voigt, C.A. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol. 2006, 355, 619–627. [Google Scholar] [CrossRef]
- Porcar, M. Beyond directed evolution: Darwinian selection as a tool for synthetic biology. Syst. Synth. Biol. 2010, 4, 1–6. [Google Scholar] [CrossRef]
- Bereza-Malcolm, L.T.; Mann, G.; Franks, A.E. Environmental sensing of heavy metals through whole cell microbial biosensors: A synthetic biology approach. ACS Synth. Biol. 2015, 4, 535–546. [Google Scholar] [CrossRef]
- Callura, J.M.; Dwyer, D.J.; Isaacs, F.J.; Cantor, C.R.; Collins, J.J. Tracking, tuning, and terminating microbial physiology using synthetic riboregulators. Proc. Natl. Acad. Sci. USA 2010, 107, 15898–15903. [Google Scholar] [CrossRef]
- Callura, J.M.; Cantor, C.R.; Collins, J.J. Genetic switchboard for synthetic biology applications. Proc. Natl. Acad. Sci. USA 2012, 109, 5850–5855. [Google Scholar] [CrossRef]
- Chappell, J.; Takahashi, M.K.; Lucks, J.B. Creating small transcription activating RNAs. Nat. Chem. Biol. 2015, 11, 214–220. [Google Scholar] [CrossRef]
- Green, A.A.; Silver, P.A.; Collins, J.J.; Yin, P. Toehold switches: De-novo-designed regulators of gene expression. Cell 2014, 159, 925–939. [Google Scholar] [CrossRef] [PubMed]
- Isaacs, F.J.; Dwyer, D.J.; Ding, C.; Pervouchine, D.D.; Cantor, C.R.; Collins, J.J. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004, 22, 841–847. [Google Scholar] [CrossRef]
- Green, A.A.; Kim, J.; Ma, D.; Silver, P.A.; Collins, J.J.; Yin, P. Complex cellular logic computation using ribocomputing devices. Nature 2017, 548, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Zhou, Y.; Carlson, P.D.; Teichmann, M.; Chaudhary, S.; Simmel, F.C.; Silver, P.A.; Collins, J.J.; Lucks, J.B.; Yin, P.; et al. De novo-designed translation-repressing riboregulators for multi-input cellular logic. Nat. Chem. Biol. 2019, 15, 1173–1182. [Google Scholar] [CrossRef]
- Hong, S.; Kim, J.; Kim, J. Multilevel gene regulation using switchable transcription terminator and toehold switch in Escherichia coli. Appl. Sci. 2021, 11, 4532. [Google Scholar] [CrossRef]
- Yang, J.; Han, Y.H.; Im, J.; Seo, S.W. Synthetic protein quality control to enhance full-length translation in bacteria. Nat. Chem. Biol. 2021, 17, 421–427. [Google Scholar] [CrossRef]
- Hwang, Y.; Kim, S.G.; Jang, S.; Kim, J.; Jung, G.Y. Signal amplification and optimization of riboswitch-based hybrid inputs by modular and titratable toehold switches. J. Biol. Eng. 2021, 15, 11. [Google Scholar] [CrossRef]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef]
- Park, S.; Lee, J.W. Detection of Coronaviruses using RNA toehold switch sensors. Int. J. Mol. Sci. 2021, 22, 1772. [Google Scholar] [CrossRef]
- Takahashi, M.K.; Tan, X.; Dy, A.J.; Braff, D.; Akana, R.T.; Furuta, Y.; Donghia, N.; Ananthakrishnan, A.; Collins, J.J. A low-cost paper-based synthetic biology platform for analyzing gut microbiota and host biomarkers. Nat. Commun. 2018, 9, 3347. [Google Scholar] [CrossRef]
- Romano, J.W.; Williams, K.G.; Shurtliff, R.N.; Ginocchio, C.; Kaplan, M. NASBA technology: Isothermal RNA amplification in qualitative and quantitative diagnostics. Immunol. Investig. 1997, 26, 15–28. [Google Scholar] [CrossRef]
- Deiman, B.; van Aarle, P.; Sillekens, P. Characteristics and applications of nucleic acid sequence-based amplification (NASBA). Mol. Biotechnol. 2002, 20, 163–179. [Google Scholar] [CrossRef]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef]
- Akcakaya, P.; Bobbin, M.L.; Guo, J.A.; Malagon-Lopez, J.; Clement, K.; Garcia, S.P.; Fellows, M.D.; Porritt, M.J.; Firth, M.A.; Carreras, A.; et al. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 2018, 561, 416–419. [Google Scholar] [CrossRef]
- Kiga, K.; Tan, X.E.; Ibarra-Chavez, R.; Watanabe, S.; Aiba, Y.; Sato’o, Y.; Li, F.Y.; Sasahara, T.; Cui, B.; Kawauchi, M.; et al. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat. Commun. 2020, 11, 2934. [Google Scholar] [CrossRef]
- Sheth, R.U.; Cabral, V.; Chen, S.P.; Wang, H.H. Manipulating bacterial communities by in situ microbiome engineering. Trends Genet. 2016, 32, 189–200. [Google Scholar] [CrossRef]
- Hsu, B.B.; Plant, I.N.; Lyon, L.; Anastassacos, F.M.; Way, J.C.; Silver, P.A. In situ reprogramming of gut bacteria by oral delivery. Nat. Commun. 2020, 11, 5030. [Google Scholar] [CrossRef]
- Cho, S.; Choe, D.; Lee, E.; Kim, S.C.; Palsson, B.; Cho, B.K. High-level dCas9 expression induces abnormal cell morphology in Escherichia coli. ACS Synth. Biol. 2018, 7, 1085–1094. [Google Scholar] [CrossRef]
- Zhang, S.; Voigt, C.A. Engineered dCas9 with reduced toxicity in bacteria: Implications for genetic circuit design. Nucleic Acids Res. 2018, 46, 11115–11125. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.L.; Leroy, P.; Unoson, C.; Fange, D.; Curic, V.; Lawson, M.J.; Elf, J. Kinetics of dCas9 target search in Escherichia coli. Science 2017, 357, 1420–1424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.H.; Tee, L.Y.; Wang, X.G.; Huang, Q.S.; Yang, S.H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther. Nucleic Acids 2015, 4, e264. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Liu, W.; Shen, H.; Chen, S.; Liao, P.; Liu, Y. Molecular AND logic gate for multiple single-nucleotide mutations detection based on CRISPR/Cas9n system-trigged signal amplification. Anal. Chim. Acta 2020, 1112, 46–53. [Google Scholar] [CrossRef]
- Peng, L.; Zhou, J.; Yin, L.; Man, S.; Ma, L. Integration of logic gates to CRISPR/Cas12a system for rapid and sensitive detection of pathogenic bacterial genes. Anal. Chim. Acta 2020, 1125, 162–168. [Google Scholar] [CrossRef]
- Jin, M.; Garreau de Loubresse, N.; Kim, Y.; Kim, J.; Yin, P. Programmable CRISPR-Cas repression, activation, and computation with sequence-independent targets and triggers. ACS Synth. Biol. 2019, 8, 1583–1589. [Google Scholar] [CrossRef]
- Rusk, N. De novo-designed riboregulators. Nat. Methods 2014, 11, 1192–1193. [Google Scholar] [CrossRef]
- Peters, G.; Maertens, J.; Lammertyn, J.; De Mey, M. Exploring of the feature space of de novo developed post-transcriptional riboregulators. PLoS Comput. Biol. 2018, 14, e1006170. [Google Scholar] [CrossRef]
- Pardee, K.; Green, A.A.; Ferrante, T.; Cameron, D.E.; DaleyKeyser, A.; Yin, P.; Collins, J.J. Paper-based synthetic gene networks. Cell 2014, 159, 940–954. [Google Scholar] [CrossRef]
- Barken, K.B.; Gabig-Ciminska, M.; Holmgren, A.; Molin, S. Effect of unlabeled helper probes on detection of an RNA target by bead-based sandwich hybridization. Biotechniques 2004, 36, 124–132. [Google Scholar] [CrossRef]
- Lopez, R.; Wang, R.; Seelig, G. A molecular multi-gene classifier for disease diagnostics. Nat. Chem. 2018, 10, 746–754. [Google Scholar] [CrossRef]
- Fuchs, B.M.; Glockner, F.O.; Wulf, J.; Amann, R. Unlabeled helper oligonucleotides increase the in situ accessibility to 16S rRNA of fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 2000, 66, 3603–3607. [Google Scholar] [CrossRef]
- Hacker, J.; Knapp, S.; Goebel, W. Spontaneous deletions and flanking regions of the chromosomally inherited hemolysin determinant of an Escherichia coli O6 strain. J. Bacteriol. 1983, 154, 1145–1152. [Google Scholar] [CrossRef]
- Johnson, J.R.; Johnston, B.; Kuskowski, M.A.; Nougayrede, J.P.; Oswald, E. Molecular epidemiology and phylogenetic distribution of the Escherichia coli pks genomic island. J. Clin. Microbiol. 2008, 46, 3906–3911. [Google Scholar] [CrossRef]
- Raisch, J.; Buc, E.; Bonnet, M.; Sauvanet, P.; Vazeille, E.; de Vallee, A.; Dechelotte, P.; Darcha, C.; Pezet, D.; Bonnet, R.; et al. Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World J. Gastroenterol. 2014, 20, 6560–6572. [Google Scholar] [CrossRef]
- Nougayrede, J.P.; Homburg, S.; Taieb, F.; Boury, M.; Brzuszkiewicz, E.; Gottschalk, G.; Buchrieser, C.; Hacker, J.; Dobrindt, U.; Oswald, E. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 2006, 313, 848–851. [Google Scholar] [CrossRef]
- Dziubanska-Kusibab, P.J.; Berger, H.; Battistini, F.; Bouwman, B.A.M.; Iftekhar, A.; Katainen, R.; Cajuso, T.; Crosetto, N.; Orozco, M.; Aaltonen, L.A.; et al. Colibactin DNA-damage signature indicates mutational impact in colorectal cancer. Nat. Med. 2020, 26, 1063–1069. [Google Scholar] [CrossRef]
- Arthur, J.C. Microbiota and colorectal cancer: Colibactin makes its mark. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 317–318. [Google Scholar] [CrossRef]
- Pleguezuelos-Manzano, C.; Puschhof, J.; Rosendahl Huber, A.; van Hoeck, A.; Wood, H.M.; Nomburg, J.; Gurjao, C.; Manders, F.; Dalmasso, G.; Stege, P.B.; et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature 2020, 580, 269–273. [Google Scholar] [CrossRef]
- Prorok-Hamon, M.; Friswell, M.K.; Alswied, A.; Roberts, C.L.; Song, F.; Flanagan, P.K.; Knight, P.; Codling, C.; Marchesi, J.R.; Winstanley, C.; et al. Colonic mucosa-associated diffusely adherent afaC+ Escherichia coli expressing lpfA and pks are increased in inflammatory bowel disease and colon cancer. Gut 2014, 63, 761–770. [Google Scholar] [CrossRef]
- Arthur, J.C.; Perez-Chanona, E.; Muhlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Schultz, M. Clinical use of E. coli Nissle 1917 in inflammatory bowel disease. Inflamm. Bowel Dis. 2008, 14, 1012–1018. [Google Scholar] [CrossRef]
- Kruis, W.; Fric, P.; Pokrotnieks, J.; Lukas, M.; Fixa, B.; Kascak, M.; Kamm, M.A.; Weismueller, J.; Beglinger, C.; Stolte, M.; et al. Maintaining remission of ulcerative colitis with the probiotic Escherichia coli Nissle 1917 is as effective as with standard mesalazine. Gut 2004, 53, 1617–1623. [Google Scholar] [CrossRef]
- Malchow, H.A. Crohn’s disease and Escherichia coli. A new approach in therapy to maintain remission of colonic Crohn’s disease? J. Clin. Gastroenterol. 1997, 25, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Rembacken, B.J.; Snelling, A.M.; Hawkey, P.M.; Chalmers, D.M.; Axon, A.T. Non-pathogenic Escherichia coli versus mesalazine for the treatment of ulcerative colitis: A randomised trial. Lancet 1999, 354, 635–639. [Google Scholar] [CrossRef]
- Fais, T.; Delmas, J.; Barnich, N.; Bonnet, R.; Dalmasso, G. Colibactin: More than a new bacterial toxin. Toxins 2018, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Olier, M.; Marcq, I.; Salvador-Cartier, C.; Secher, T.; Dobrindt, U.; Boury, M.; Bacqui, V.; Penary, M.; Gaultier, E.; Nougayrede, J.P.; et al. Genotoxicity of Escherichia coli Nissle 1917 strain cannot be dissociated from its probiotic activity. Gut Microbes 2012, 3, 501–509. [Google Scholar] [CrossRef]
- Massip, C.; Branchu, P.; Bossuet-Grief, N.; Chagneau, C.V.; Gaillard, D.; Martin, P.; Boury, M.; Secher, T.; Dubois, D.; Nougayrede, J.P.; et al. Deciphering the interplay between the genotoxic and probiotic activities of Escherichia coli Nissle 1917. PLoS Pathog. 2019, 15, e1008029. [Google Scholar] [CrossRef]
- Fais, T.; Cougnoux, A.; Dalmasso, G.; Laurent, F.; Delmas, J.; Bonnet, R. Antibiotic activity of Escherichia coli against multiresistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2016, 60, 6986–6988. [Google Scholar] [CrossRef]
- Perez-Berezo, T.; Pujo, J.; Martin, P.; Le Faouder, P.; Galano, J.M.; Guy, A.; Knauf, C.; Tabet, J.C.; Tronnet, S.; Barreau, F.; et al. Identification of an analgesic lipopeptide produced by the probiotic Escherichia coli strain Nissle 1917. Nat. Commun. 2017, 8, 1314. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- Quan, J.; Tian, J. Circular polymerase extension cloning for high-throughput cloning of complex and combinatorial DNA libraries. Nat. Protoc. 2011, 6, 242–251. [Google Scholar] [CrossRef]
- Liu, H.; Naismith, J.H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 2008, 8, 91. [Google Scholar] [CrossRef]
- Fornace, M.E.; Porubsky, N.J.; Pierce, N.A. A unified dynamic programming framework for the analysis of interacting nucleic acid strands: Enhanced models, scalability, and speed. ACS Synth. Biol. 2020, 9, 2665–2678. [Google Scholar] [CrossRef]
- Dirks, R.M.; Bois, J.S.; Schaeffer, J.M.; Winfree, E.; Pierce, N.A. Thermodynamic analysis of interacting nucleic acid strands. SIAM Rev. 2007, 49, 65–88. [Google Scholar] [CrossRef]
- Dirks, R.M.; Pierce, N.A. An algorithm for computing nucleic acid base-pairing probabilities including pseudoknots. J. Comput. Chem. 2004, 25, 1295–1304. [Google Scholar] [CrossRef]
- Dirks, R.M.; Pierce, N.A. A partition function algorithm for nucleic acid secondary structure including pseudoknots. J. Comput. Chem. 2003, 24, 1664–1677. [Google Scholar] [CrossRef]
- Wolfe, B.R.; Pierce, N.A. Sequence design for a test tube of interacting nucleic acid strands. ACS Synth. Biol. 2015, 4, 1086–1100. [Google Scholar] [CrossRef]
- Reister, M.; Hoffmeier, K.; Krezdorn, N.; Rotter, B.; Liang, C.; Rund, S.; Dandekar, T.; Sonnenborn, U.; Oelschlaeger, T.A. Complete genome sequence of the gram-negative probiotic Escherichia coli strain Nissle 1917. J. Biotechnol. 2014, 187, 106–107. [Google Scholar] [CrossRef]
- Van Rossum, G. Python programming language. In Proceedings of the USENIX Annual Technical Conference, Santa Clara, CA, USA, 17–22 June 2007; p. 36. [Google Scholar]
- McKinney, W.; Van Der Walt, S.; Millman, J. Data structures for statistical computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010. [Google Scholar]
- Kaplinski, L.; Scheler, O.; Parkel, S.; Palta, P.; Toome, K.; Kurg, A.; Remm, M. Detection of tmRNA molecules on microarrays at low temperatures using helper oligonucleotides. BMC Biotechnol. 2010, 10, 34. [Google Scholar] [CrossRef]
- Dornenburg, J.E.; Devita, A.M.; Palumbo, M.J.; Wade, J.T. Widespread antisense transcription in Escherichia coli. mBio 2010, 1, 00010–e00024. [Google Scholar] [CrossRef]
- Eklof, V.; Lofgren-Burstrom, A.; Zingmark, C.; Edin, S.; Larsson, P.; Karling, P.; Alexeyev, O.; Rutegard, J.; Wikberg, M.L.; Palmqvist, R. Cancer-associated fecal microbial markers in colorectal cancer detection. Int. J. Cancer 2017, 141, 2528–2536. [Google Scholar] [CrossRef]
- Cougnoux, A.; Gibold, L.; Robin, F.; Dubois, D.; Pradel, N.; Darfeuille-Michaud, A.; Dalmasso, G.; Delmas, J.; Bonnet, R. Analysis of structure-function relationships in the colibactin-maturating enzyme ClbP. J. Mol. Biol. 2012, 424, 203–214. [Google Scholar] [CrossRef]
- Li, Z.R.; Li, J.; Gu, J.P.; Lai, J.Y.; Duggan, B.M.; Zhang, W.P.; Li, Z.L.; Li, Y.X.; Tong, R.B.; Xu, Y.; et al. Divergent biosynthesis yields a cytotoxic aminomalonate-containing precolibactin. Nat. Chem. Biol. 2016, 12, 773–775. [Google Scholar] [CrossRef]
- Jung, J.K.; Alam, K.K.; Verosloff, M.S.; Capdevila, D.A.; Desmau, M.; Clauer, P.R.; Lee, J.W.; Nguyen, P.Q.; Pasten, P.A.; Matiasek, S.J.; et al. Cell-free biosensors for rapid detection of water contaminants. Nat. Biotechnol. 2020, 38, 1451–1459. [Google Scholar] [CrossRef]
- Kim, J.; Yin, P.; Green, A.A. Ribocomputing: Cellular logic computation using RNA devices. Biochemistry 2018, 57, 883–885. [Google Scholar] [CrossRef]
- Wang, J.; Williams, B.; Chirasani, V.R.; Krokhotin, A.; Das, R.; Dokholyan, N.V. Limits in accuracy and a strategy of RNA structure prediction using experimental information. Nucleic Acids Res. 2019, 47, 5563–5572. [Google Scholar] [CrossRef]
- Mathews, D.H. Revolutions in RNA secondary structure prediction. J. Mol. Biol. 2006, 359, 526–532. [Google Scholar] [CrossRef]
- Lai, D.; Proctor, J.R.; Meyer, I.M. On the importance of cotranscriptional RNA structure formation. RNA 2013, 19, 1461–1473. [Google Scholar] [CrossRef]
- Yu, A.M.; Gasper, P.M.; Cheng, L.; Lai, L.B.; Kaur, S.; Gopalan, V.; Chen, A.A.; Lucks, J.B. Computationally reconstructing cotranscriptional RNA folding from experimental data reveals rearrangement of non-native folding intermediates. Mol. Cell 2021, 81, 870–883. [Google Scholar] [CrossRef]
- Watters, K.E.; Strobel, E.J.; Yu, A.M.; Lis, J.T.; Lucks, J.B. Cotranscriptional folding of a riboswitch at nucleotide resolution. Nat. Struct. Mol. Biol. 2016, 23, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Angenent-Mari, N.M.; Garruss, A.S.; Soenksen, L.R.; Church, G.; Collins, J.J. A deep learning approach to programmable RNA switches. Nat. Commun. 2020, 11, 5057. [Google Scholar] [CrossRef]
- Valeri, J.A.; Collins, K.M.; Ramesh, P.; Alcantar, M.A.; Lepe, B.A.; Lu, T.K.; Camacho, D.M. Sequence-to-function deep learning frameworks for engineered riboregulators. Nat. Commun. 2020, 11, 5058. [Google Scholar] [CrossRef] [PubMed]
- Woodson, S.A. Taming free energy landscapes with RNA chaperones. RNA Biol. 2010, 7, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Konermann, S.; Brideau, N.J.; Lotfy, P.; Wu, X.; Novick, S.J.; Strutzenberg, T.; Griffin, P.R.; Hsu, P.D.; Lyumkis, D. Structural basis for the RNA-guided ribonuclease activity of CRISPR-Cas13d. Cell 2018, 175, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 2018, 173, 665–676. [Google Scholar] [CrossRef] [PubMed]
- Charles, E.J.; Kim, S.E.; Knott, G.J.; Smock, D.; Doudna, J.; Savage, D.F. Engineering improved Cas13 effectors for targeted post-transcriptional regulation of gene expression. bioRxiv 2021, 5687. [Google Scholar] [CrossRef]
- Kharma, N.; Varin, L.; Abu-Baker, A.; Ouellet, J.; Najeh, S.; Ehdaeivand, M.R.; Belmonte, G.; Ambri, A.; Rouleau, G.; Perreault, J. Automated design of hammerhead ribozymes and validation by targeting the PABPN1 gene transcript. Nucleic Acids Res. 2016, 44, e39. [Google Scholar] [CrossRef]
- Carbonell, A.; Flores, R.; Gago, S. Trans-cleaving hammerhead ribozymes with tertiary stabilizing motifs: In vitro and in vivo activity against a structured viroid RNA. Nucleic Acids Res. 2011, 39, 2432–2444. [Google Scholar] [CrossRef]
- Choudhary, R.; Mahadevan, R. Toward a systematic design of smart probiotics. Curr. Opin. Biotechnol. 2020, 64, 199–209. [Google Scholar] [CrossRef]
- Rottinghaus, A.G.; Amrofell, M.B.; Moon, T.S. Biosensing in smart engineered probiotics. Biotechnol. J. 2020, 15, e1900319. [Google Scholar] [CrossRef]
- El Hage, R.; Hernandez-Sanabria, E.; Van de Wiele, T. Emerging trends in “Smart Probiotics”: Functional consideration for the development of novel health and industrial applications. Front. Microbiol. 2017, 8, 1889. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Heo, T.; Kang, H.; Choi, S.; Kim, J. Detection of pks Island mRNAs Using Toehold Sensors in Escherichia coli. Life 2021, 11, 1280. https://doi.org/10.3390/life11111280
Heo T, Kang H, Choi S, Kim J. Detection of pks Island mRNAs Using Toehold Sensors in Escherichia coli. Life. 2021; 11(11):1280. https://doi.org/10.3390/life11111280
Chicago/Turabian StyleHeo, Taeyang, Hansol Kang, Seungdo Choi, and Jongmin Kim. 2021. "Detection of pks Island mRNAs Using Toehold Sensors in Escherichia coli" Life 11, no. 11: 1280. https://doi.org/10.3390/life11111280
APA StyleHeo, T., Kang, H., Choi, S., & Kim, J. (2021). Detection of pks Island mRNAs Using Toehold Sensors in Escherichia coli. Life, 11(11), 1280. https://doi.org/10.3390/life11111280





