Advanced CRISPR-Cas Effector Enzyme-Based Diagnostics for Infectious Diseases, Including COVID-19
Abstract
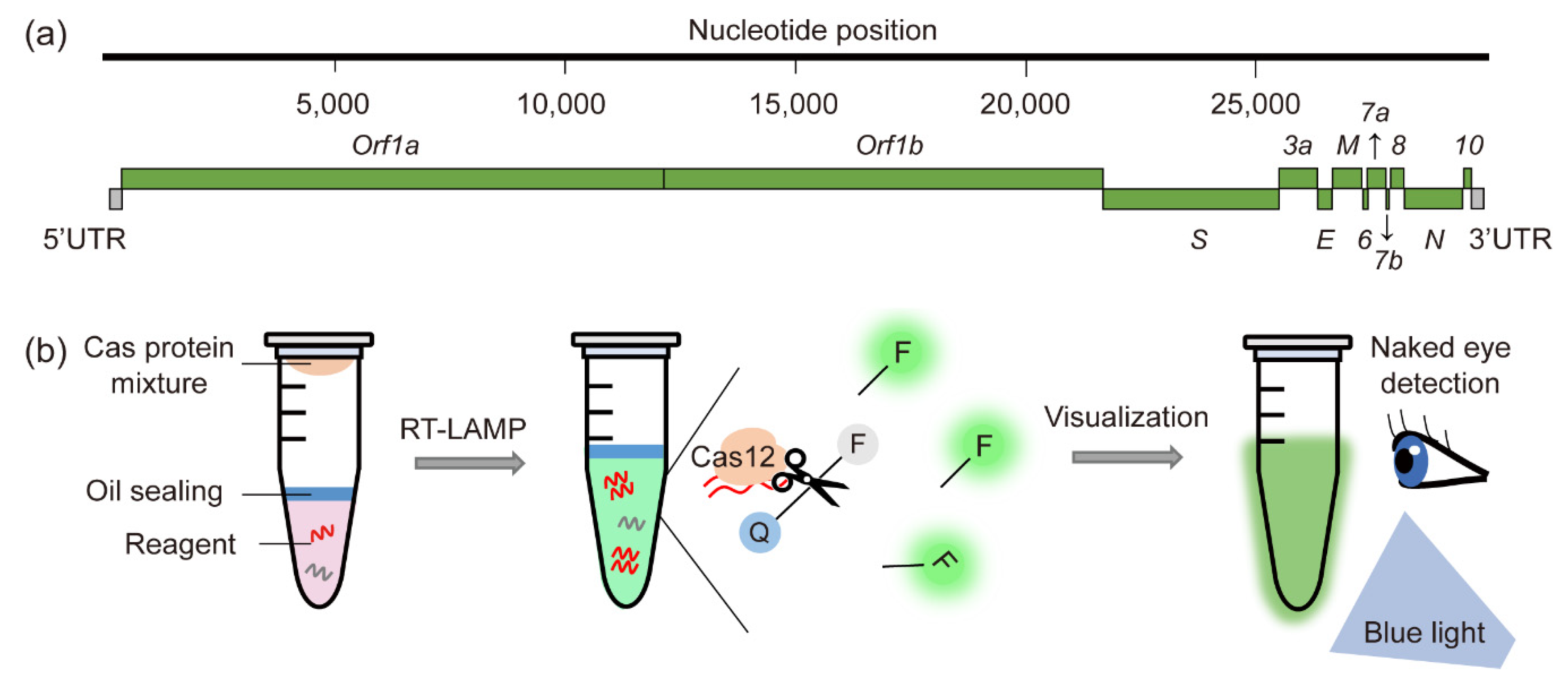
:1. Introduction
2. Molecular Diagnosis Based on the CRISPR Effectors Cas12 and Cas13
2.1. Cas12-Based Diagnostics
2.1.1. Fluorescence-Based Methods
2.1.2. Lateral Flow Readout
2.1.3. Electrochemistry-Based Methods
2.2. Cas13-Based Diagnostics
2.2.1. SHERLOCK-Based CRISPR-Dx
2.2.2. CARMEN-Cas13
3. Nucleic Acid Detection Platforms for SARS-CoV-2
3.1. Fluorescence Detection with Naked Eyes
3.2. Electrokinetic Microfluidic Chip
3.3. Divalent Ion Enhancement
4. Discussion and Future Perspectives
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Zhang, W.; Du, R.H.; Li, B.; Zheng, X.S.; Yang, X.L.; Hu, B.; Wang, Y.Y.; Xiao, G.F.; Yan, B.; Shi, Z.L.; et al. Molecular and serological investigation of 2019-nCoV infected patients: Implication of multiple shedding routes. Emerg. Microbes Infect. 2020, 9, 386–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharyya, R.P.; Thakku, S.G.; Hung, D.T. Harnessing CRISPR Effectors for Infectious Disease Diagnostics. ACS Infect. Dis. 2018, 4, 1278–1282. [Google Scholar] [CrossRef] [PubMed]
- Benda, A.; Zerajic, L.; Ankita, A.; Cleary, E.; Park, Y.; Pandey, S. COVID-19 Testing and Diagnostics: A Review of Commercialized Technologies for Cost, Convenience and Quality of Tests. Sensors 2021, 21, 6581. [Google Scholar] [CrossRef] [PubMed]
- Mpoza, E.; Mukaremera, L.; Kundura, D.A.; Akampurira, A.; Luggya, T.; Tadeo, K.K.; Pastick, K.A.; Bridge, S.C.; Tugume, L.; Kiggundu, R.; et al. Evaluation of a point-of-care immunoassay test kit ‘StrongStep’ for cryptococcal antigen detection. PLoS ONE 2018, 13, e0190652. [Google Scholar] [CrossRef] [Green Version]
- Torres, I.; Poujois, S.; Albert, E.; Colomina, J.; Navarro, D. Evaluation of a rapid antigen test (Panbio COVID-19 Ag rapid test device) for SARS-CoV-2 detection in asymptomatic close contacts of COVID-19 patients. Clin. Microbiol. Infect. 2021, 27, 636.e631–636.e634. [Google Scholar] [CrossRef]
- Larese Filon, F.; Purpuri, A.; Camata, D.; Bovenzi, M.; Rui, F.; Ronchese, F.; De Michieli, P.; Marcello, A.; Poggianella, M.; Confalonieri, M.; et al. Low sensitivity of rapid tests detecting anti-CoV-2 IgG and IgM in health care workers’ serum for COVID-19 screening. Med. Lav. 2021, 112, 331–339. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Rothman, R.E. PCR-based diagnostics for infectious diseases: Uses, limitations, and future applications in acute-care settings. Lancet Infect. Dis. 2004, 4, 337–348. [Google Scholar] [CrossRef]
- Chen, J.S.; Ma, E.; Harrington, L.B.; Da Costa, M.; Tian, X.; Palefsky, J.M.; Doudna, J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 2018, 360, 436–439. [Google Scholar] [CrossRef] [Green Version]
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017, 356, 438–442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef] [Green Version]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [Green Version]
- Knott, G.J.; Doudna, J.A. CRISPR-Cas guides the future of genetic engineering. Science 2018, 361, 866–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haurwitz, R.E.; Jinek, M.; Wiedenheft, B.; Zhou, K.; Doudna, J.A. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 2010, 329, 1355–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef] [Green Version]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017, 15, 169–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef]
- Jiang, F.; Doudna, J.A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; La Russa, M.; Qi, L.S. CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 2016, 85, 227–264. [Google Scholar] [CrossRef] [Green Version]
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M.; et al. Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, M.; Zhou, X.; Wang, H.; Xing, D. Clustered Regularly Interspaced Short Palindromic Repeats/Cas9 Triggered Isothermal Amplification for Site-Specific Nucleic Acid Detection. Anal. Chem. 2018, 90, 2193–2200. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Hu, L.; Ying, L.; Zhao, Z.; Chu, P.K.; Yu, X.F. A CRISPR-Cas9-triggered strand displacement amplification method for ultrasensitive DNA detection. Nat. Commun. 2018, 9, 5012. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uppada, V.; Gokara, M.; Rasineni, G.K. Diagnosis and therapy with CRISPR advanced CRISPR based tools for point of care diagnostics and early therapies. Gene 2018, 656, 22–29. [Google Scholar] [CrossRef]
- Li, S.Y.; Cheng, Q.X.; Liu, J.K.; Nie, X.Q.; Zhao, G.P.; Wang, J. CRISPR-Cas12a has both cis- and trans-cleavage activities on single-stranded DNA. Cell Res. 2018, 28, 491–493. [Google Scholar] [CrossRef] [PubMed]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [Green Version]
- Abudayyeh, O.O.; Gootenberg, J.S.; Essletzbichler, P.; Han, S.; Joung, J.; Belanto, J.J.; Verdine, V.; Cox, D.B.T.; Kellner, M.J.; Regev, A.; et al. RNA targeting with CRISPR-Cas13. Nature 2017, 550, 280–284. [Google Scholar] [CrossRef] [Green Version]
- Teng, F.; Guo, L.; Cui, T.; Wang, X.G.; Xu, K.; Gao, Q.; Zhou, Q.; Li, W. CDetection: CRISPR-Cas12b-based DNA detection with sub-attomolar sensitivity and single-base specificity. Genome Biol. 2019, 20, 132. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.N.; Nakagawa, R.; Okazaki, S.; Hirano, H.; Kobayashi, K.; Kusakizako, T.; Nishizawa, T.; Yamashita, K.; Nishimasu, H.; Nureki, O. Structure of the miniature type V-F CRISPR-Cas effector enzyme. Mol. Cell 2021, 81, 558–570.e553. [Google Scholar] [CrossRef]
- Karvelis, T.; Bigelyte, G.; Young, J.K.; Hou, Z.; Zedaveinyte, R.; Budre, K.; Paulraj, S.; Djukanovic, V.; Gasior, S.; Silanskas, A.; et al. PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res. 2020, 48, 5016–5023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, W.X.; Hunnewell, P.; Alfonse, L.E.; Carte, J.M.; Keston-Smith, E.; Sothiselvam, S.; Garrity, A.J.; Chong, S.; Makarova, K.S.; Koonin, E.V.; et al. Functionally diverse type V CRISPR-Cas systems. Science 2019, 363, 88–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef]
- Li, S.Y.; Cheng, Q.X.; Wang, J.M.; Li, X.Y.; Zhang, Z.L.; Gao, S.; Cao, R.B.; Zhao, G.P.; Wang, J. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018, 4, 20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Li, S.; Wu, N.; Wu, J.; Wang, G.; Zhao, G.; Wang, J. HOLMESv2: A CRISPR-Cas12b-Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation. ACS Synth. Biol. 2019, 8, 2228–2237. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.W.; Zhou, X.; Xu, T.; Yang, M.; Chen, Y.; He, G.Q.; Pan, N.; Cai, Y.; Li, Y.; Wang, X.; et al. CRISPR-based rapid and ultra-sensitive diagnostic test for Mycobacterium tuberculosis. Emerg. Microbes Infect. 2019, 8, 1361–1369. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Ji, P.; Fan, H.; Dang, L.; Wan, W.; Liu, S.; Li, Y.; Yu, W.; Li, X.; Ma, X.; et al. CRISPR/Cas12a technology combined with immunochromatographic strips for portable detection of African swine fever virus. Commun. Biol. 2020, 3, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukama, O.; Wu, J.; Li, Z.; Liang, Q.; Yi, Z.; Lu, X.; Liu, Y.; Liu, Y.; Hussain, M.; Makafe, G.G.; et al. An ultrasensitive and specific point-of-care CRISPR/Cas12 based lateral flow biosensor for the rapid detection of nucleic acids. Biosens. Bioelectron. 2020, 159, 112143. [Google Scholar] [CrossRef]
- Tsou, J.H.; Leng, Q.; Jiang, F. A CRISPR Test for Detection of Circulating Nuclei Acids. Transl. Oncol. 2019, 12, 1566–1573. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Lin, H.; Li, H.; Zhou, Y.; Liu, J.; Zhong, G.; Wu, L.; Jiang, W.; Du, H.; Yang, J.; et al. Cas12a-Based On-Site and Rapid Nucleic Acid Detection of African Swine Fever. Front. Microbiol. 2019, 10, 2830. [Google Scholar] [CrossRef] [Green Version]
- Dai, Y.; Somoza, R.A.; Wang, L.; Welter, J.F.; Li, Y.; Caplan, A.I.; Liu, C.C. Exploring the Trans-Cleavage Activity of CRISPR-Cas12a (cpf1) for the Development of a Universal Electrochemical Biosensor. Angew. Chem. Int. Ed. Engl. 2019, 58, 17399–17405. [Google Scholar] [CrossRef] [PubMed]
- Shao, N.; Han, X.; Song, Y.; Zhang, P.; Qin, L. CRISPR-Cas12a Coupled with Platinum Nanoreporter for Visual Quantification of SNVs on a Volumetric Bar-Chart Chip. Anal. Chem. 2019, 91, 12384–12391. [Google Scholar] [CrossRef]
- Gootenberg, J.S.; Abudayyeh, O.O.; Kellner, M.J.; Joung, J.; Collins, J.J.; Zhang, F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 2018, 360, 439–444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myhrvold, C.; Freije, C.A.; Gootenberg, J.S.; Abudayyeh, O.O.; Metsky, H.C.; Durbin, A.F.; Kellner, M.J.; Tan, A.L.; Paul, L.M.; Parham, L.A.; et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 2018, 360, 444–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ackerman, C.M.; Myhrvold, C.; Thakku, S.G.; Freije, C.A.; Metsky, H.C.; Yang, D.K.; Ye, S.H.; Boehm, C.K.; Kosoko-Thoroddsen, T.F.; Kehe, J.; et al. Massively multiplexed nucleic acid detection with Cas13. Nature 2020, 582, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Augustine, R.; Hasan, A.; Das, S.; Ahmed, R.; Mori, Y.; Notomi, T.; Kevadiya, B.D.; Thakor, A.S. Loop-Mediated Isothermal Amplification (LAMP): A Rapid, Sensitive, Specific, and Cost-Effective Point-of-Care Test for Coronaviruses in the Context of COVID-19 Pandemic. Biology 2020, 9, 182. [Google Scholar] [CrossRef] [PubMed]
- Chavez, A.; Pruitt, B.W.; Tuttle, M.; Shapiro, R.S.; Cecchi, R.J.; Winston, J.; Turczyk, B.M.; Tung, M.; Collins, J.J.; Church, G.M. Precise Cas9 targeting enables genomic mutation prevention. Proc. Natl. Acad. Sci. USA 2018, 115, 3669–3673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Lubin, A.A.; Plaxco, K.W. Folding-based electrochemical biosensors: The case for responsive nucleic acid architectures. Acc. Chem. Res. 2010, 43, 496–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smargon, A.A.; Cox, D.B.T.; Pyzocha, N.K.; Zheng, K.; Slaymaker, I.M.; Gootenberg, J.S.; Abudayyeh, O.A.; Essletzbichler, P.; Shmakov, S.; Makarova, K.S.; et al. Cas13b Is a Type VI-B CRISPR-Associated RNA-Guided RNase Differentially Regulated by Accessory Proteins Csx27 and Csx28. Mol. Cell 2017, 65, 618–630.e617. [Google Scholar] [CrossRef] [Green Version]
- Niewoehner, O.; Jinek, M. Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA 2016, 22, 318–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- East-Seletsky, A.; O’Connell, M.R.; Knight, S.C.; Burstein, D.; Cate, J.H.; Tjian, R.; Doudna, J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 2016, 538, 270–273. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef] [Green Version]
- Scherer, L.C.; Sperhacke, R.D.; Ruffino-Netto, A.; Rossetti, M.L.; Vater, C.; Klatser, P.; Kritski, A.L. Cost-effectiveness analysis of PCR for the rapid diagnosis of pulmonary tuberculosis. BMC Infect. Dis. 2009, 9, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.H.; Yeh, I.J.; Phan, N.N.; Yen, M.C.; Hung, J.H.; Chiao, C.C.; Chen, C.F.; Sun, Z.; Hsu, H.P.; Wang, C.Y.; et al. Gene signatures and potential therapeutic targets of Middle East respiratory syndrome coronavirus (MERS-CoV)-infected human lung adenocarcinoma epithelial cells. J. Microbiol. Immunol. Infect. 2021, 54, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.L.; Yeh, I.J.; Phan, N.N.; Wu, Y.H.; Yen, M.C.; Hung, J.H.; Chiao, C.C.; Chen, C.F.; Sun, Z.; Jiang, J.Z.; et al. Gene signatures of SARS-CoV/SARS-CoV-2-infected ferret lungs in short- and long-term models. Infect. Genet. Evol. 2020, 85, 104438. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Peng, J.; Xiong, Q.; Liu, Z.; Lin, H.; Tan, X.; Kang, M.; Yuan, R.; Zeng, L.; Zhou, P.; et al. Clinical, immunological and virological characterization of COVID-19 patients that test re-positive for SARS-CoV-2 by RT-PCR. EBioMedicine 2020, 59, 102960. [Google Scholar] [CrossRef]
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef] [Green Version]
- Chu, D.K.W.; Pan, Y.; Cheng, S.M.S.; Hui, K.P.Y.; Krishnan, P.; Liu, Y.; Ng, D.Y.M.; Wan, C.K.C.; Yang, P.; Wang, Q.; et al. Molecular Diagnosis of a Novel Coronavirus (2019-nCoV) Causing an Outbreak of Pneumonia. Clin. Chem. 2020, 66, 549–555. [Google Scholar] [CrossRef] [Green Version]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brunink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Shi, Y.; Chen, Y.; Yang, Z.; Wu, H.; Zhou, Z.; Li, J.; Ping, J.; He, L.; Shen, H.; et al. Contamination-free visual detection of SARS-CoV-2 with CRISPR/Cas12a: A promising method in the point-of-care detection. Biosens. Bioelectron. 2020, 169, 112642. [Google Scholar] [CrossRef]
- Wang, X.; Zhong, M.; Liu, Y.; Ma, P.; Dang, L.; Meng, Q.; Wan, W.; Ma, X.; Liu, J.; Yang, G.; et al. Rapid and sensitive detection of COVID-19 using CRISPR/Cas12a-based detection with naked eye readout, CRISPR/Cas12a-NER. Sci. Bull. 2020, 65, 1436–1439. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Sun, X.; Wang, X.; Liang, C.; Jiang, H.; Gao, Q.; Dai, M.; Qu, B.; Fang, S.; Mao, Y.; et al. SARS-CoV-2 detection with CRISPR diagnostics. Cell Discov. 2020, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Meng, Q.; Sun, B.; Zhao, B.; Dang, L.; Zhong, M.; Liu, S.; Xu, H.; Mei, H.; Liu, J.; et al. MeCas12a, a Highly Sensitive and Specific System for COVID-19 Detection. Adv. Sci. 2020, 7, 2001300. [Google Scholar] [CrossRef] [PubMed]
- Ding, X.; Yin, K.; Li, Z.; Lalla, R.V.; Ballesteros, E.; Sfeir, M.M.; Liu, C. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat. Commun. 2020, 11, 4711. [Google Scholar] [CrossRef] [PubMed]
- Mayuramart, O.; Nimsamer, P.; Rattanaburi, S.; Chantaravisoot, N.; Khongnomnan, K.; Chansaenroj, J.; Puenpa, J.; Suntronwong, N.; Vichaiwattana, P.; Poovorawan, Y.; et al. Detection of severe acute respiratory syndrome coronavirus 2 and influenza viruses based on CRISPR-Cas12a. Exp. Biol. Med. 2021, 246, 400–405. [Google Scholar] [CrossRef] [PubMed]
- Pang, B.; Xu, J.; Liu, Y.; Peng, H.; Feng, W.; Cao, Y.; Wu, J.; Xiao, H.; Pabbaraju, K.; Tipples, G.; et al. Isothermal Amplification and Ambient Visualization in a Single Tube for the Detection of SARS-CoV-2 Using Loop-Mediated Amplification and CRISPR Technology. Anal. Chem. 2020, 92, 16204–16212. [Google Scholar] [CrossRef]
- Wang, R.; Qian, C.; Pang, Y.; Li, M.; Yang, Y.; Ma, H.; Zhao, M.; Qian, F.; Yu, H.; Liu, Z.; et al. opvCRISPR: One-pot visual RT-LAMP-CRISPR platform for SARS-cov-2 detection. Biosens. Bioelectron. 2021, 172, 112766. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Tian, D.; Liu, Y.; Lin, Z.; Lyon, C.J.; Lai, W.; Fusco, D.; Drouin, A.; Yin, X.; Hu, T.; et al. Ultra-sensitive and high-throughput CRISPR-p owered COVID-19 diagnosis. Biosens. Bioelectron. 2020, 164, 112316. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Smith, B.M.; Jain, P.K. Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection. Nat. Commun. 2020, 11, 4906. [Google Scholar] [CrossRef] [PubMed]
- Broughton, J.P.; Deng, X.; Yu, G.; Fasching, C.L.; Servellita, V.; Singh, J.; Miao, X.; Streithorst, J.A.; Granados, A.; Sotomayor-Gonzalez, A.; et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat. Biotechnol. 2020, 38, 870–874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joung, J.; Ladha, A.; Saito, M.; Kim, N.G.; Woolley, A.E.; Segel, M.; Barretto, R.P.J.; Ranu, A.; Macrae, R.K.; Faure, G.; et al. Detection of SARS-CoV-2 with SHERLOCK One-Pot Testing. N. Engl. J. Med. 2020, 383, 1492–1494. [Google Scholar] [CrossRef]
- Ramachandran, A.; Huyke, D.A.; Sharma, E.; Sahoo, M.K.; Huang, C.; Banaei, N.; Pinsky, B.A.; Santiago, J.G. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 29518–29525. [Google Scholar] [CrossRef] [PubMed]
- Arizti-Sanz, J.; Freije, C.A.; Stanton, A.C.; Petros, B.A.; Boehm, C.K.; Siddiqui, S.; Shaw, B.M.; Adams, G.; Kosoko-Thoroddsen, T.F.; Kemball, M.E.; et al. Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat. Commun. 2020, 11, 5921. [Google Scholar] [CrossRef]
- Hou, T.; Zeng, W.; Yang, M.; Chen, W.; Ren, L.; Ai, J.; Wu, J.; Liao, Y.; Gou, X.; Li, Y.; et al. Development and evaluation of a rapid CRISPR-based diagnostic for COVID-19. PLoS Pathog. 2020, 16, e1008705. [Google Scholar] [CrossRef] [PubMed]
- Sundaresan, R.; Parameshwaran, H.P.; Yogesha, S.D.; Keilbarth, M.W.; Rajan, R. RNA-Independent DNA Cleavage Activities of Cas9 and Cas12a. Cell Rep. 2017, 21, 3728–3739. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Cas9 | Cas12 | Cas13 | |
|---|---|---|---|
| Class | 2 | ||
| Type | II | V | VI |
| Target | dsDNA | dsDNA, ssDNA | ssRNA |
| Trans-cleavage activity | N | Y | Y |
| Target motif | G-rich PAM | T-rich PAM | Non-G PFS |
| Cleavage pattern | Blunt end | Staggered end | Near U |
| Effect or Enzyme | Reporter System | Diagnostic Platform | Nucleic Acid Extraction | Sample Amplification | Single-Base Resolution | Sensitivity | Turnaround Time | Pathogenic Application | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Cas12 | Fluorescence | DETECTR | N | RPA | N | aM | ~2 h | HPV16, HPV18 | [8] |
| HOLMES | Y | PCR | Y | aM | ~3 h | PRV, JEV | [34] | ||
| HOLMESv2 | Y | LAMP | Y | nM | ~3 h | JEV | [35] | ||
| CDetection | Y | RPA | Y | aM | ~2 h | HPV | [29] | ||
| CRISPR-MTB | N | RPA | N | 5 copies | ~2 h | Mycobacterium tuberculosis | [36] | ||
| Cas14-DETECTR | N | PCR | N | - | ~3 h | - | [10] | ||
| Lateral flow | CRISPR-Cas12a-LFD | N | RAA | N | 20 copies | ~1 h | ASFV | [37] | |
| CIA | N | LAMP | N | aM | ~1.5 h | Pseudomonas aeruginosa | [38] | ||
| Fluorescence and lateral flow | Cas12-based POC | N | RPA | N | fM | ~3 h | HPV16, HPV18 | [39] | |
| CORDS | Y | RAA | N | aM | ~2.5 h | ASFV | [40] | ||
| Electrochemistry | E-CRISPR | N | N/A | Y | pM | ~1 h | HPV16, PB-19 | [41] | |
| MAV-chip | N | N/A | Y | - | ~1 h | - | [42] | ||
| Cas13 | Fluorescence | SHERLOCK | Y | RPA | Y | aM | ~4 h | ZIKV, DENV | [9] |
| Multiplexed fluorescence and lateral flow | SHERLOCKv2 | Y | RPA | Y | aM | ~3.5 h | ZIKV, DENV | [43] | |
| Fluorescence and lateral flow | HUDSON +SHERLOCK | N | RPA | Y | aM | ~2 h | ZIKV, DENV, WNV, YFV | [44] | |
| Multiplexed fluorescence | CARMEN-Cas13 | Y | PCR/RPA | Y | aM | ~3 h | 169 Human associated virus | [45] |
| Effect or Enzyme | Detection Method | Diagnostic Platform | Nucleic Acid Extraction | Sample Amplification | Single-Base Resolution | Sensitivity | Turnaround Time | Target Gene | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Cas12 | Naked eye | Contamination-free visual Cas12a | Y | RT-LAMP | N | 20 copies | ~1 h 45 min | ORF | [61] |
| CRISPR-Cas12a-NER | Y | RT-RAA | N | 10 copies | ~1 h 45 min | E | [62] | ||
| CASdetec | Y | RT-RAA | N | 10 copies | ~2 h | RdRp | [63] | ||
| MeCas12a | Y | RT-RAA | Y | 5 copies | ~1 h 45 min | E | [64] | ||
| AIOD-CRISPR | Y | NA | N | 5 copies | ~1 h 40 min | N | [65] | ||
| RT-RPA-CRISPR-Cas12a assay | Y | RT-RPA | N | 10 copies | ~2 h | S | [66] | ||
| RT-LAMP-Cas12a assay | Y | RT-LAMP | N | 30 copies | ~1 h 40 min | E, N | [67] | ||
| opvCRISPR | Y | RT-LAMP | N | 5 copies | ~1 h 45 min | S | [68] | ||
| Fluorescence | CRISPR-FDS | Y | RT-RPA | N | 2 copies | ~1 h 40 min | ORF1ab, N | [69] | |
| Lateral flow strip | ENHANCE | Y | RT-LAMP | N | 3~300 copies | ~1 h 50 min | N | [70] | |
| SARS-CoV-2 DETECTR | Y | RT-LAMP | N | 10 copies | ~1 h 40 min | E, N | [71] | ||
| Fluorescence or lateral flow strip | STOPCovid.v2 | N | RT-LAMP | N | 100 copies | ~2 h | N | [72] | |
| Electrokinetic chip | Microfluidic ITP-CRISPR-based assay | N | RT-LAMP | N | 10 copies | ~40 min | E, N | [73] | |
| Cas13 | Naked eye and later flow strip | SHINE | N | NA | N | >1000 copies | ~55 min | ORF1a | [74] |
| Fluorescence | CRISPR-COVID | Y | RT-RPA | N | 7.5 copies | ~1 h 40 min | ORF1ab | [75] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, S.; Shin, H.Y. Advanced CRISPR-Cas Effector Enzyme-Based Diagnostics for Infectious Diseases, Including COVID-19. Life 2021, 11, 1356. https://doi.org/10.3390/life11121356
Kwon S, Shin HY. Advanced CRISPR-Cas Effector Enzyme-Based Diagnostics for Infectious Diseases, Including COVID-19. Life. 2021; 11(12):1356. https://doi.org/10.3390/life11121356
Chicago/Turabian StyleKwon, Sangha, and Ha Youn Shin. 2021. "Advanced CRISPR-Cas Effector Enzyme-Based Diagnostics for Infectious Diseases, Including COVID-19" Life 11, no. 12: 1356. https://doi.org/10.3390/life11121356






