Gene Expression Meta-Analysis of Cerebellum Samples Supports the FKBP5 Gene-Environment Interaction Model for Schizophrenia
Abstract
:1. Introduction
2. Results
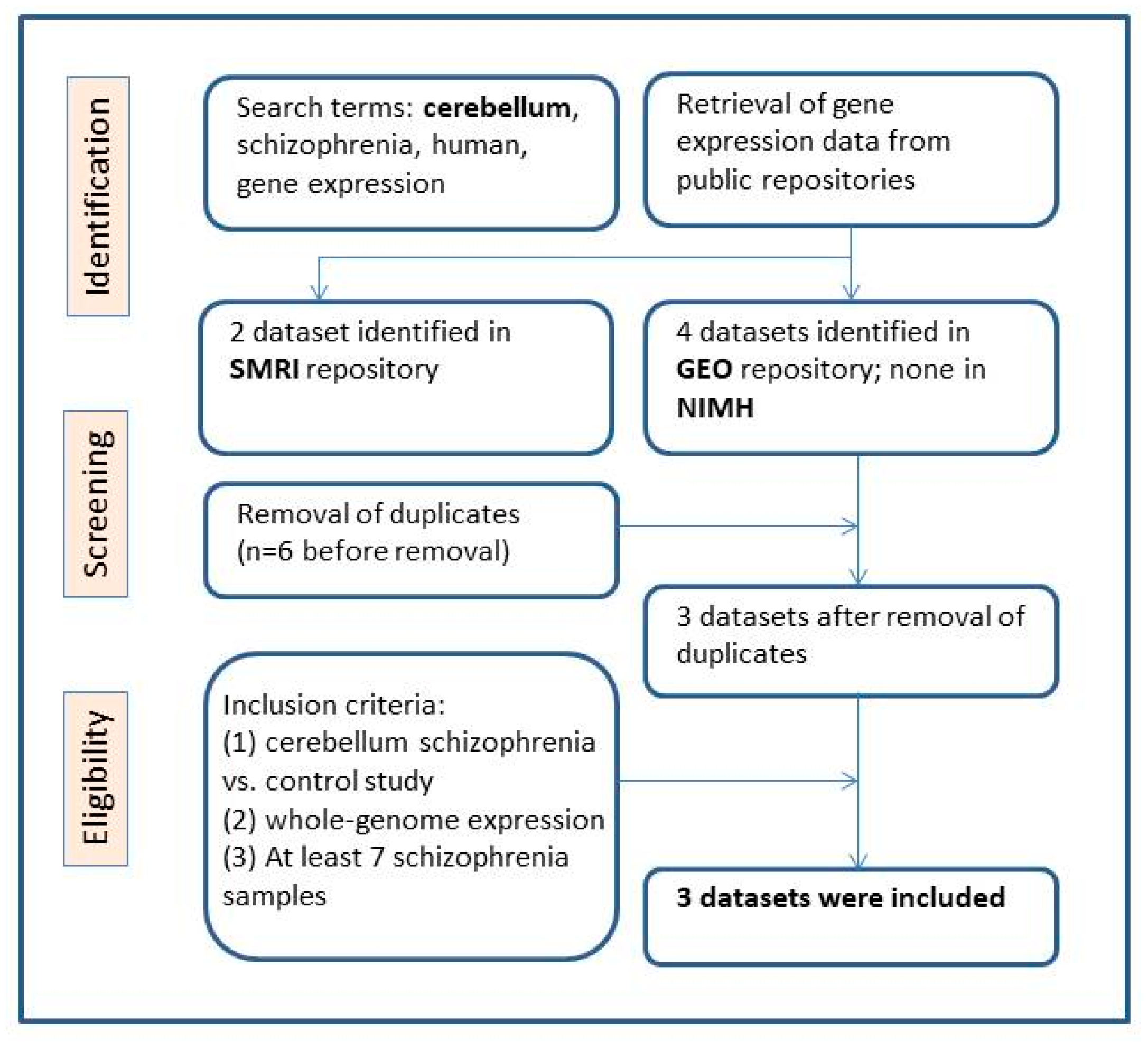
2.1. Selection of Eligible Gene Expression Datasets
2.2. The Cerebellum Gene Expression Datasets are Comparable
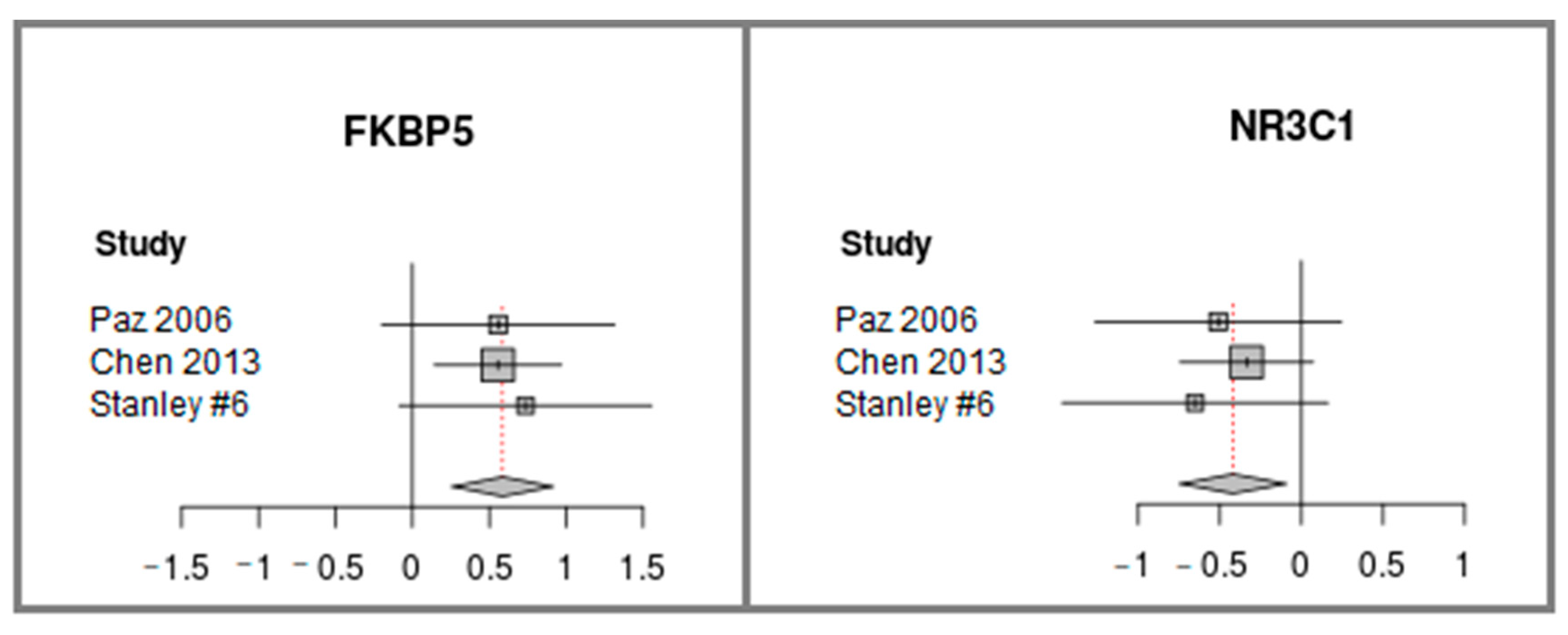
2.3. FKBP5 is Up-regulated and NR3C1, Encoding GR, is Down-regulated in Cerebellum Samples of Individuals with Schizophrenia
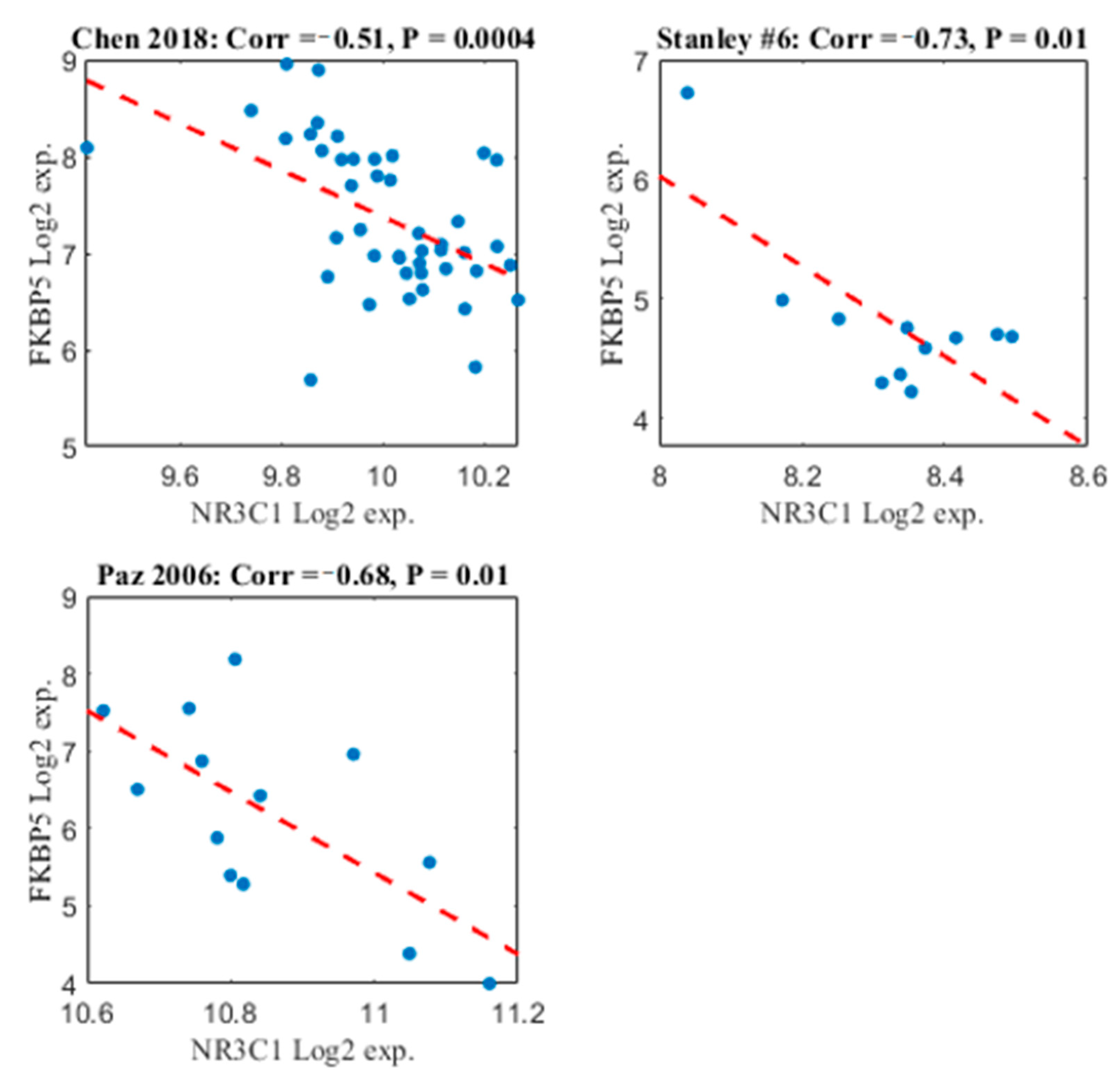
2.4. FKBP5 and NR3C1 Expression Patterns are Negatively Correlated
2.5. Examination of Potential Confounding Factors
2.6. FKBP5 and NR3C1 are not Differentially Expressed in Blood Samples of Individuals with Schizophrenia Versus Healthy Controls
2.7. FKBP5 and IL6 Expression Patterns are Positively Correlated
3. Discussion
4. Materials and Methods
4.1. Eligible Gene Expression Datasets Selection for Meta-analysis
4.2. Gene Expression Meta-analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Daskalakis, N.P.; Binder, E.B. Schizophrenia in the Spectrum of Gene-Stress Interactions: The FKBP5 Example. Schizophr. Bull. 2015, 41, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Varese, F.; Smeets, F.; Drukker, M.; Lieverse, R.; Lataster, T.; Viechtbauer, W.; Read, J.; Van Os, J.; Bentall, R.P. Childhood Adversities Increase the Risk of Psychosis: A Meta-analysis of Patient-Control, Prospective- and Cross-sectional Cohort Studies. Schizophr. Bull. 2012, 38, 661–671. [Google Scholar] [CrossRef] [PubMed]
- Smoller, J.W.; Andreassen, O.A.; Edenberg, H.J.; Faraone, S.V.; Glatt, S.J.; Kendler, K.S. Psychiatric genetics and the structure of psychopathology. Mol. Psychiatry 2019, 24, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Ellis, B.J.; Boyce, W.T.; Belsky, J.; Bakermans-Kranenburg, M.J.; Van Ijzendoorn, M.H. Differential susceptibility to the environment: An evolutionary–neurodevelopmental theory. Dev. Psychopathol. 2011, 23, 7–28. [Google Scholar] [CrossRef] [Green Version]
- Grad, I.; Picard, D. The glucocorticoid responses are shaped by molecular chaperones. Mol. Cell. Endocrinol. 2007, 275, 2–12. [Google Scholar] [CrossRef]
- Matosin, N.; Halldorsdottir, T.; Binder, E.B. Understanding the Molecular Mechanisms Underpinning Gene by Environment Interactions in Psychiatric Disorders: The FKBP5 Model. Biol. Psychiatry 2018, 83, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Binder, E.B. The role of FKBP5, a co-chaperone of the glucocorticoid receptor in the pathogenesis and therapy of affective and anxiety disorders. Psychoneuroendocrinology 2009, 34 (Suppl. S1), S186–S195. [Google Scholar] [CrossRef]
- Klengel, T.; Mehta, D.; Anacker, C.; Rex-Haffner, M.; Pruessner, J.C.; Pariante, C.M.; Pace, T.W.W.; Mercer, K.B.; Mayberg, H.S.; Bradley, B.; et al. Allele-specific FKBP5 DNA demethylation mediates gene–childhood trauma interactions. Nat. Neurosci. 2013, 16, 33–41. [Google Scholar] [CrossRef] [Green Version]
- Buchmann, A.F.; Holz, N.; Boecker, R.; Blomeyer, D.; Rietschel, M.; Witt, S.H.; Schmidt, M.H.; Esser, G.; Banaschewski, T.; Brandeis, D.; et al. Moderating role of FKBP5 genotype in the impact of childhood adversity on cortisol stress response during adulthood. Eur. Neuropsychopharmacol. 2014, 24, 837–845. [Google Scholar] [CrossRef] [Green Version]
- Klengel, T.; Binder, E.B. Allele-specific epigenetic modification: A molecular mechanism for gene–environment interactions in stress-related psychiatric disorders? Epigenomics 2013, 5, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Young, K.A.; Thompson, P.M.; Cruz, D.A.; Williamson, D.E.; Selemon, L.D. BA11 FKBP5 expression levels correlate with dendritic spine density in postmortem PTSD and controls. Neurobiol. Stress 2015, 2, 67–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albu, S.; Romanowski, C.P.N.; Curzi, M.L.; Jakubcakova, V.; Flachskamm, C.; Gassen, N.C.; Hartmann, J.; Schmidt, M.V.; Schmidt, U.; Rein, T.; et al. Deficiency of FK506-binding protein (FKBP) 51 alters sleep architecture and recovery sleep responses to stress in mice. J. Sleep Res. 2013, 23, 176–185. [Google Scholar] [CrossRef] [Green Version]
- Galatzer-Levy, I.R.; Andero, R.; Sawamura, T.; Jovanovic, T.; Papini, S.; Ressler, K.J.; Norrholm, S.D. A cross species study of heterogeneity in fear extinction learning in relation to FKBP5 variation and expression: Implications for the acute treatment of posttraumatic stress disorder. Neuropharmacology 2017, 116, 188–195. [Google Scholar] [CrossRef]
- Hartmann, J.; Wagner, K.V.; Gaali, S.; Kirschner, A.; Kozany, C.; Rühter, G.; Dedic, N.; Häusl, A.S.; Hoeijmakers, L.; Westerholz, S.; et al. Pharmacological Inhibition of the Psychiatric Risk Factor FKBP51 Has Anxiolytic Properties. J. Neurosci. 2015, 35, 9007–9016. [Google Scholar] [CrossRef] [Green Version]
- Touma, C.; Gassen, N.C.; Herrmann, L.; Cheung-Flynn, J.; Büll, D.R.; Ionescu, I.A.; Heinzmann, J.-M.; Knapman, A.; Siebertz, A.; Depping, A.-M.; et al. FK506 Binding Protein 5 Shapes Stress Responsiveness: Modulation of Neuroendocrine Reactivity and Coping Behavior. Biol. Psychiatry 2011, 70, 928–936. [Google Scholar] [CrossRef]
- Holtzman, C.W.; Trotman, H.D.; Goulding, S.M.; Ryan, A.T.; Macdonald, A.N.; Shapiro, D.I.; Brasfield, J.L.; Walker, E.F. Stress and neurodevelopmental processes in the emergence of psychosis. Neuroscience 2013, 249, 172–191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, L.J.; McGorry, P.D.; Garner, B.; Thompson, K.N.; Pantelis, C.; Wood, S.J.; Berger, G. Stress, the Hippocampus and the Hypothalamic-Pituitary-Adrenal Axis: Implications for the Development of Psychotic Disorders. Aust. N. Z. J. Psychiatry 2006, 40, 725–741. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.J.; Knable, M.B.; O’Grady, J.; Orthmann, J.; Weickert, C.S. Regional specificity of brain glucocorticoid receptor mRNA alterations in subjects with schizophrenia and mood disorders. Mol. Psychiatry 2002, 7, 985–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perlman, W.R.; Webster, M.J.; Kleinman, J.E.; Weickert, C.S. Reduced glucocorticoid and estrogen receptor alpha messenger ribonucleic acid levels in the amygdala of patients with major mental illness. Biol. Psychiatry 2004, 56, 844–852. [Google Scholar] [CrossRef]
- Patel, P.D.; Katz, M.; Karssen, A.M.; Lyons, D.M. Stress-induced changes in corticosteroid receptor expression in primate hippocampus and prefrontal cortex. Psychoneuroendocrinology 2008, 33, 360–367. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Wu, J.; Qing, L.; Li, J.; Yang, H.; Ji, A.; Yan, M.; Hu, L.; Nie, S. DNA Methylation Analysis of the NR3C1 Gene in Patients with Schizophrenia. J. Mol. Neurosci. 2020, 70, 1177–1185. [Google Scholar] [CrossRef]
- Ajnakina, O.; Borges, S.; Di Forti, M.; Patel, Y.; Xu, X.; Green, P.; Stilo, S.A.; Kolliakou, A.; Sood, P.; Marques, T.R.; et al. Role of Environmental Confounding in the Association between FKBP5 and First-Episode Psychosis. Front. Psychiatry 2014, 5, 84. [Google Scholar] [CrossRef] [Green Version]
- Collip, D.; Myin-Germeys, I.; Wichers, M.; Jacobs, N.; Derom, C.; Thiery, E.; Lataster, T.; Simons, C.; Delespaul, P.; Marcelis, M.; et al. FKBP5 as a possible moderator of the psychosis-inducing effects of childhood trauma. Br. J. Psychiatry 2013, 202, 261–268. [Google Scholar] [CrossRef] [Green Version]
- Darby, M.M.; Yolken, R.H.; Sabunciyan, S. Consistently altered expression of gene sets in postmortem brains of individuals with major psychiatric disorders. Transl. Psychiatry 2016, 6, e890. [Google Scholar] [CrossRef]
- Gandal, M.J.; Haney, J.R.; Parikshak, N.N.; Leppa, V.; Ramaswami, G.; Hartl, C.; Schork, A.J.; Appadurai, V.; Buil, A.; Werge, T.M.; et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 2018, 359, 693–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, W.; Xu, C.; Rao, V. Brain Tissue Matters in the Study of FKBP5 Gene Expression Activities in Schizophrenia – A Meta-Analysis. J. Psy. and Brain Sci. 2017, 2, 1. [Google Scholar] [CrossRef] [Green Version]
- Fromer, M.; Roussos, P.; Sieberts, S.K.; Johnson, J.S.; Kavanagh, D.H.; Perumal, T.M.; Ruderfer, D.M.; Oh, E.C.; Topol, A.; Shah, H.R.; et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 2016, 19, 1442–1453. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Li, X.; Liu, J.; Luo, X.-J.; Yao, Y.-G. SZDB2.0: An updated comprehensive resource for schizophrenia research. Qual. Life Res. 2020, 139, 1285–1297. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yao, Y.-G.; Luo, X.-J. SZDB: A Database for Schizophrenia Genetic Research. Schizophr. Bull. 2016, 43, 459–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinclair, D.; Fillman, S.G.; Webster, M.J.; Weickert, C.S. Dysregulation of glucocorticoid receptor co-factors FKBP5, BAG1 and PTGES3 in prefrontal cortex in psychotic illness. Sci. Rep. 2013, 3, 3539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirnics, K.; Levitt, P.; Lewis, D.A. Critical Appraisal of DNA Microarrays in Psychiatric Genomics. Biol. Psychiatry 2006, 60, 163–176. [Google Scholar] [CrossRef]
- Ahmed, A.O.; Strauss, G.P.; Buchanan, R.W.; Kirkpatrick, B.; Carpenter, W.T. Schizophrenia heterogeneity revisited: Clinical, cognitive, and psychosocial correlates of statistically-derived negative symptoms subgroups. J. Psychiatr. Res. 2018, 97, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Khaitovich, P.; Muetzel, B.; She, X.; Lachmann, M.; Hellmann, I.; Dietzsch, J.; Steigele, S.; Do, H.-H.; Weiss, G.; Enard, W.; et al. Regional Patterns of Gene Expression in Human and Chimpanzee Brains. Genome Res. 2004, 14, 1462–1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; Prisma Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cierpka, M.; Wolf, N.D.; Kubera, K.M.; Schmitgen, M.M.; Vasic, N.; Frasch, K.; Wolf, R.C. Cerebellar Contributions to Persistent Auditory Verbal Hallucinations in Patients with Schizophrenia. Cerebellum 2017, 16, 964–972. [Google Scholar] [CrossRef]
- He, H.; Luo, C.; Luo, Y.; Duan, M.; Yi, Q.; Biswal, B.B.; Yao, D. Reduction in gray matter of cerebellum in schizophrenia and its influence on static and dynamic connectivity. Hum. Brain Mapp. 2019, 40, 517–528. [Google Scholar] [CrossRef] [Green Version]
- Lein, E.S.; Hawrylycz, M.J.; Ao, N.; Ayres, M.; Bensinger, A.; Bernard, A.; Boe, A.F.; Boguski, M.S.; Brockway, K.S.; Byrnes, E.J.; et al. Genome-wide atlas of gene expression in the adult mouse brain. Nat. Cell Biol. 2006, 445, 168–176. [Google Scholar] [CrossRef]
- Fleiss, J.L. Review papers: The statistical basis of meta-analysis. Stat. Methods Med. Res. 1993, 2, 121–145. [Google Scholar] [CrossRef]
- Paz, R.D.; Andreasen, N.C.; Daoud, S.Z.; Conley, R.; Roberts, R.; Bustillo, J.; Perrone-Bizzozero, N.I. Increased Expression of Activity-Dependent Genes in Cerebellar Glutamatergic Neurons of Patients With Schizophrenia. Am. J. Psychiatry 2006, 163, 1829–1831. [Google Scholar] [CrossRef]
- Chen, C.; Members of the Bipolar Disorder Genome Study (BiGS) Consortium; Cheng, L.; Grennan, K.; Pibiri, F.; Zhang, C.; Badner, J.A.; Gershon, E.S.; Liu, C. Two gene co-expression modules differentiate psychotics and controls. Mol. Psychiatry 2013, 18, 1308–1314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duyckaerts, C.; Godefroy, G. Voronoi tessellation to study the numerical density and the spatial distribution of neurones. J. Chem. Neuroanat. 2000, 20, 83–92. [Google Scholar] [CrossRef]
- Schwarzer, G. Meta: An R package for meta-analysis. R News 2007, 7, 40–45. [Google Scholar]
- Bousman, C.A.; Chana, G.; Glatt, S.J.; Chandler, S.D.; Lucero, G.R.; Tatro, E.; May, T.; Lohr, J.B.; Kremen, W.S.; Tsuang, M.T.; et al. Preliminary evidence of ubiquitin proteasome system dysregulation in schizophrenia and bipolar disorder: Convergent pathway analysis findings from two independent samples. Am. J. Med Genet. Part B Neuropsychiatr. Genet. 2009, 153, 494–502. [Google Scholar] [CrossRef] [Green Version]
- Janusek, L.W.; Tell, D.; Gaylord-Harden, N.; Mathews, H.L. Relationship of childhood adversity and neighborhood violence to a proinflammatory phenotype in emerging adult African American men: An epigenetic link. Brain, Behav. Immun. 2017, 60, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Tomassi, S.; Tosato, S. Epigenetics and gene expression profile in first-episode psychosis: The role of childhood trauma. Neurosci. Biobehav. Rev. 2017, 83, 226–237. [Google Scholar] [CrossRef] [PubMed]
- Andreasen, N.C.; Pierson, R. The Role of the Cerebellum in Schizophrenia. Biol. Psychiatry 2008, 64, 81–88. [Google Scholar] [CrossRef] [Green Version]
- Moberget, T.; Doan, N.T.; Alnæs, D.; Kaufmann, T.; Córdova-Palomera, A.; Lagerberg, T.V.; Diedrichsen, J.; Schwarz, E.; Zink, M.; Eisenacher, S.; et al. Cerebellar volume and cerebellocerebral structural covariance in schizophrenia: A multisite mega-analysis of 983 patients and 1349 healthy controls. Mol. Psychiatry 2018, 23, 1512–1520. [Google Scholar] [CrossRef]
- Lee, R.S.; Mahon, P.B.; Zandi, P.P.; McCaul, M.E.; Yang, X.; Bali, U.; Wand, G.S. DNA methylation and sex-specific expression of FKBP5 as correlates of one-month bedtime cortisol levels in healthy individuals. Psychoneuroendocrinology 2018, 97, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Szabo, C.; Kelemen, O.; Kéri, S. Changes in FKBP5 expression and memory functions during cognitive–behavioral therapy in posttraumatic stress disorder: A preliminary study. Neurosci. Lett. 2014, 569, 116–120. [Google Scholar] [CrossRef] [Green Version]
- Ising, M.; Maccarrone, G.; Brückl, T.; Scheuer, S.; Hennings, J.; Holsboer, F.; Turck, C.W.; Uhr, M.; Lucae, S. FKBP5 Gene Expression Predicts Antidepressant Treatment Outcome in Depression. Int. J. Mol. Sci. 2019, 20, 485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaali, S.; Gopalakrishnan, R.; Wang, Y.; Kozany, C.; Hausch, F. The chemical biology of immunophilin ligands. Curr. Med. Chem. 2011, 18, 5355–5379. [Google Scholar] [CrossRef]
- Tatro, E.T.; Everall, I.P.; Masliah, E.; Hult, B.J.; Lucero, G.; Chana, G.; Soontornniyomkij, V.; Achim, C.L. Differential Expression of Immunophilins FKBP51 and FKBP52 in the Frontal Cortex of HIV-Infected Patients with Major Depressive Disorder. J. Neuroimmune Pharmacol. 2009, 4, 218–226. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Wang, N.; Zhao, X.; Ross, C.A.; O’Shea, K.S.; McInnis, M.G. Gene expression alterations in bipolar disorder postmortem brains. Bipolar Disord. 2013, 15, 177–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gogtay, N.; Vyas, N.S.; Testa, R.; Wood, S.J.; Pantelis, C. Age of Onset of Schizophrenia: Perspectives From Structural Neuroimaging Studies. Schizophr. Bull. 2011, 37, 504–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoffman, G.E.; Bendl, J.; Voloudakis, G.; Montgomery, K.S.; Sloofman, L.; Wang, Y.-C.; Shah, H.R.; Hauberg, M.E.; Johnson, J.S.; Girdhar, K.; et al. CommonMind Consortium provides transcriptomic and epigenomic data for Schizophrenia and Bipolar Disorder. Sci. Data 2019, 6, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gandal, M.J.; Zhang, P.; Hadjimichael, E.; Walker, R.L.; Chen, C.; Liu, S.; Won, H.; Van Bakel, H.; Varghese, M.; Wang, Y.; et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 2018, 362, eaat8127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedges, L.V. Distribution Theory for Glass’s Estimator of Effect Size and Related Estimators. J. Educ. Stat. 1981, 6, 107. [Google Scholar] [CrossRef]


| Study | # Subjects with Schizophrenia and Controls per Region and Cohort | Method | FKBP5 Differential Expression |
|---|---|---|---|
| (Darby et al., 2016) [24] | Hippocampus: SZ (35), CNT (32); | RNA sequencing; | Up-regulation; |
| Orbitofrontal cortex: SZ (13), CNT (15); | RNA sequencing; | No differential expression | |
| (Sinclair, Fillman, Webster, & Weickert, 2013) [30] | Prefrontal cortex: SZ (20), CNT (20); | RNA sequencing; | Up-regulation; |
| Prefrontal cortex: SZ (35), CNT (35) | qPCR | Up-regulation | |
| (Gandal et al. 2018) Meta-analysis | BA46: SZ (35), CNT (34); BA46: SZ (30), CNT (29); BA46: SZ (15), CNT (19); BA10: SZ (28), CNT (23); Parietal cortex: SZ(51), CNT (50); Total: SZ (159), CNT (155) | Microarray | Up-regulation (union of the datasets) |
| (Jiang et al. 2017 Meta-analysis [26] | BA46: SZ (35), CNT (34); | Microarray | No differential expression (union of the datasets) |
| BA46: SZ (30), CNT (29); | |||
| BA46: SZ (15), CNT (19); | |||
| BA10: SZ (28), CNT (23); BA10: SZ (13), CNT (15); Hippocampus: SZ (15), CNT (18); Striatum: SZ (18), CNT (18); Entorhinal cortex: SZ (10), CNT (10); STG: SZ (23), CNT (19); STG: SZ (9), CNT (9); STG: SZ (8), CNT (8); DLPFC: SZ (65), CNT (72); Total: SZ(196), CNT (72) | |||
| SZDB2.0 database (Wu et al., 2020, 2017), using (Fromer et al., 2016) CMC data [27,28,29] | DLPFC: SZ (258), CNT (279); | RNA sequencing | No differential expression |
| Study | Accession | # SZ | # CNT | Platform | Mean Age (Standard Deviation | Mean PMI (Standard Deviation | Mean pH (Standard Deviation) |
|---|---|---|---|---|---|---|---|
| Paz 2006 [39] | GDS1917 | 14,14M:0F | 14,14M:0F | U133 Plus 2.0 Array | SZ: 45 (12) | SZ: 15.6 (6) | Not provided |
| CNT: 43 (10) | CNT: 12.2 (5) | ||||||
| p = 0.57 | p = 0.11 | ||||||
| Chen 2013 [40] | GSE35978 | 44,33M:12F | 50,31M:19F | Gene 1.0 ST Array | SZ: 46 (9) | SZ: 33 (15) | SZ: 6.5 (0.3) |
| CNT: 43 (9) p = 0.2 | CNT: 28 (11) p = 0.042 | CNT: 6.4 (0.3) p = 0.44 | |||||
| Stanley#6 | 11, 8M:3F | 14,9M:5F | U95 Av2 Array | SZ: 47 (9) | SZ: 25 (10) | SZ: 6.3 (0.3) | |
| CNT: 46 (14) | CNT: 34 (14) | CNT: 6.2 (0.3) | |||||
| p = 0.83 | p = 0.071 | p = 0.82 | |||||
| Total: 69 | Total: 78 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hertzberg, L.; Zohar, A.H.; Yitzhaky, A. Gene Expression Meta-Analysis of Cerebellum Samples Supports the FKBP5 Gene-Environment Interaction Model for Schizophrenia. Life 2021, 11, 190. https://doi.org/10.3390/life11030190
Hertzberg L, Zohar AH, Yitzhaky A. Gene Expression Meta-Analysis of Cerebellum Samples Supports the FKBP5 Gene-Environment Interaction Model for Schizophrenia. Life. 2021; 11(3):190. https://doi.org/10.3390/life11030190
Chicago/Turabian StyleHertzberg, Libi, Ada H. Zohar, and Assif Yitzhaky. 2021. "Gene Expression Meta-Analysis of Cerebellum Samples Supports the FKBP5 Gene-Environment Interaction Model for Schizophrenia" Life 11, no. 3: 190. https://doi.org/10.3390/life11030190
APA StyleHertzberg, L., Zohar, A. H., & Yitzhaky, A. (2021). Gene Expression Meta-Analysis of Cerebellum Samples Supports the FKBP5 Gene-Environment Interaction Model for Schizophrenia. Life, 11(3), 190. https://doi.org/10.3390/life11030190







