Activation of Blood Vessel Development in Endometrial Stromal Cells In Vitro Cocultured with Human Peri-Implantation Embryos Revealed by Single-Cell RNA-Seq
Abstract
1. Introduction
2. Materials and Methods
2.1. Coculture Model and Sample Collection
2.2. Transcriptome Data Processing and Validation
2.3. Weighted Gene Coexpression Analysis (WGCNA)
2.4. Gene Ontology (GO) and Gene Set Enrichment Analysis (GSEA)
3. Results
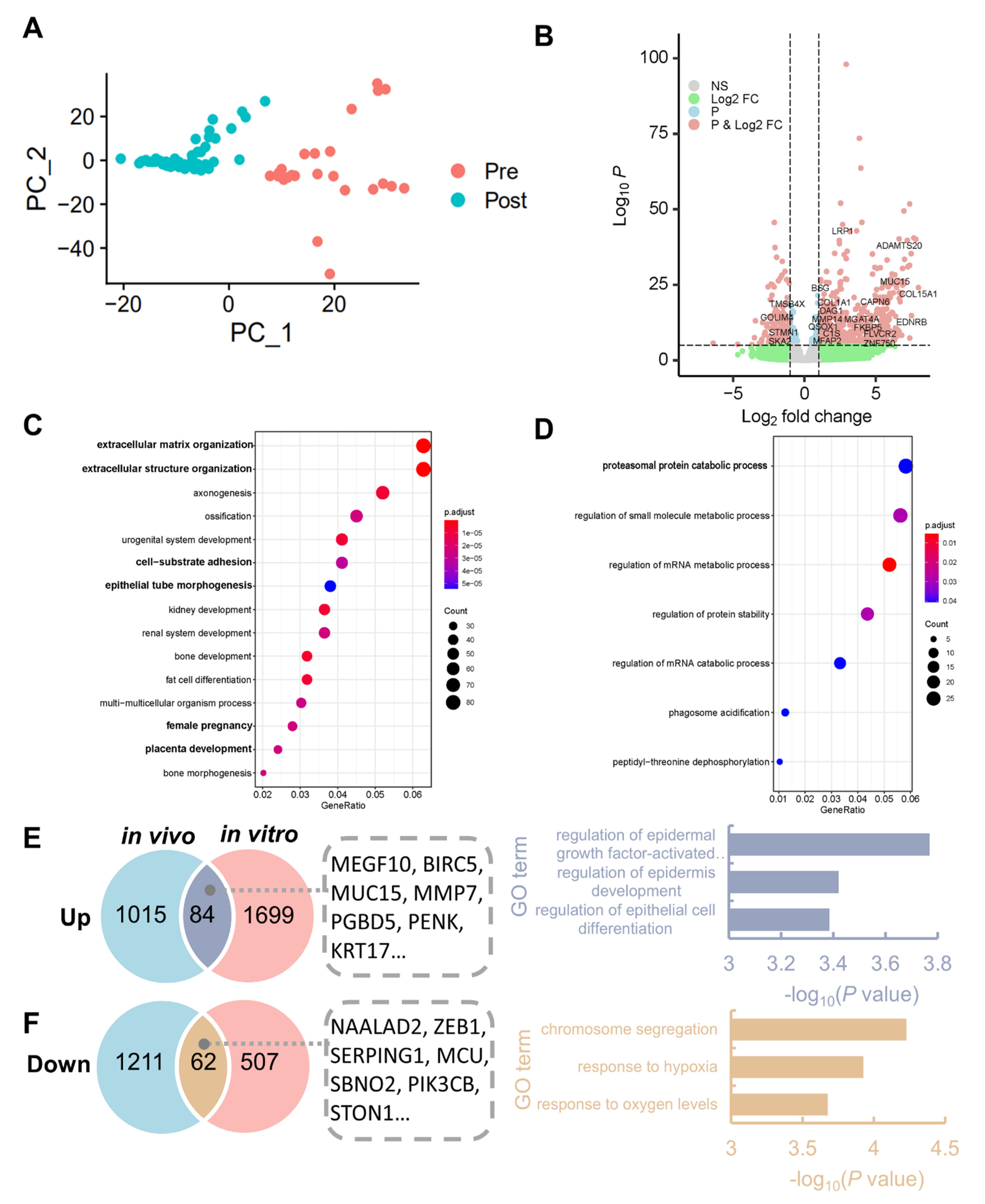
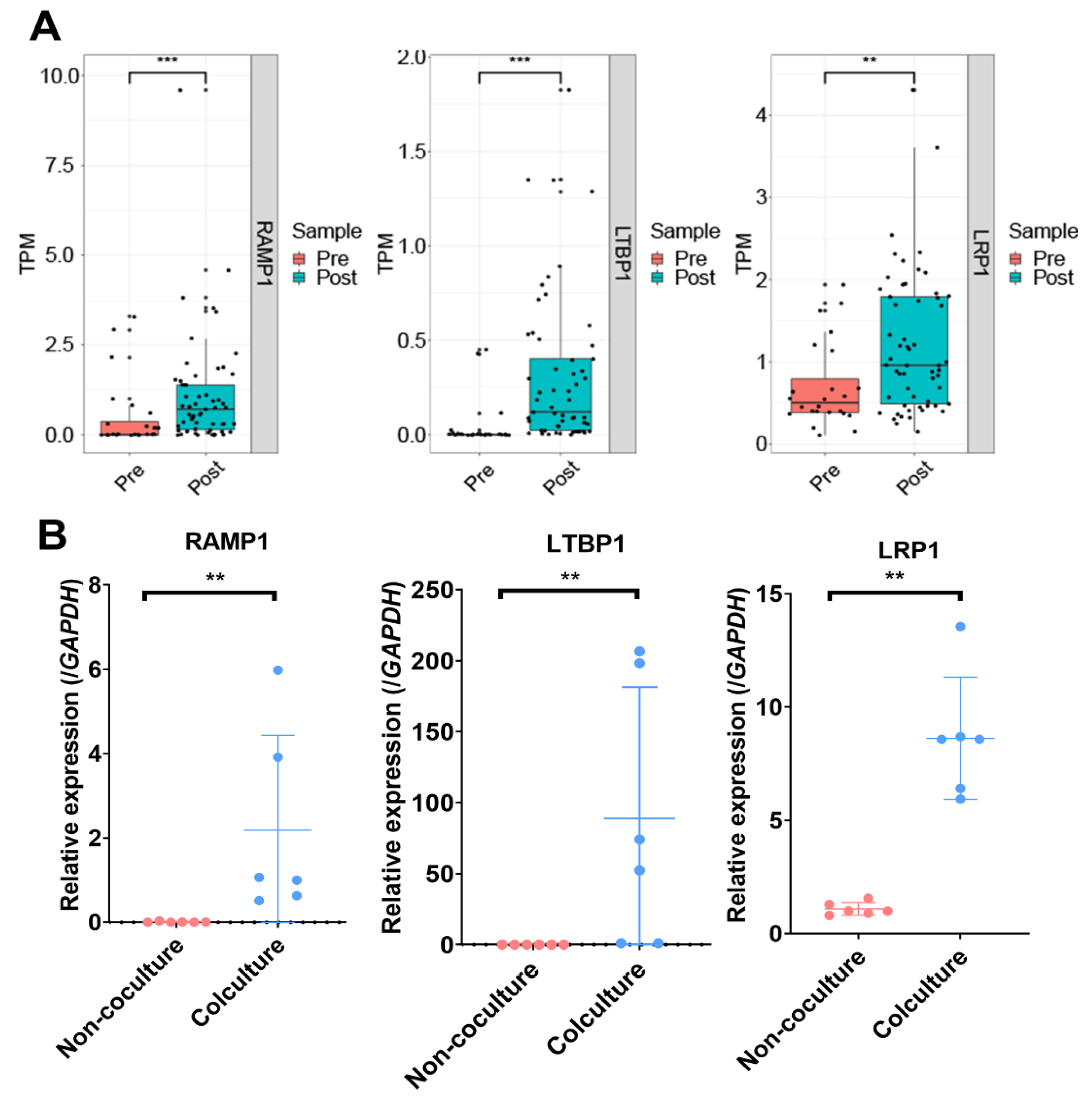
3.1. Identification of DEGs and Enriched Pathways after Implantation
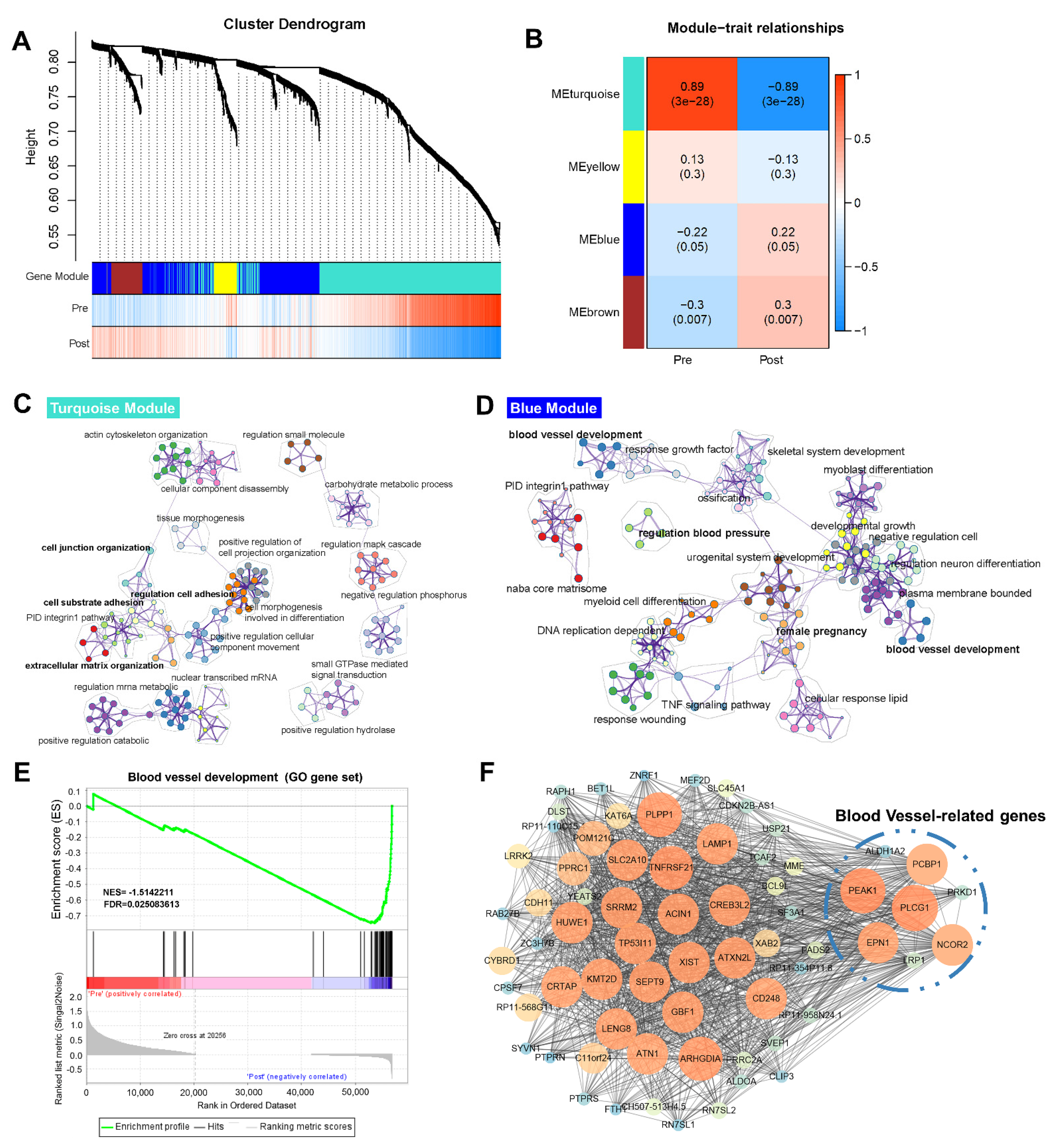
3.2. Genetic Programs of Receptivity Development in EmSCs Revealed by WGCNA
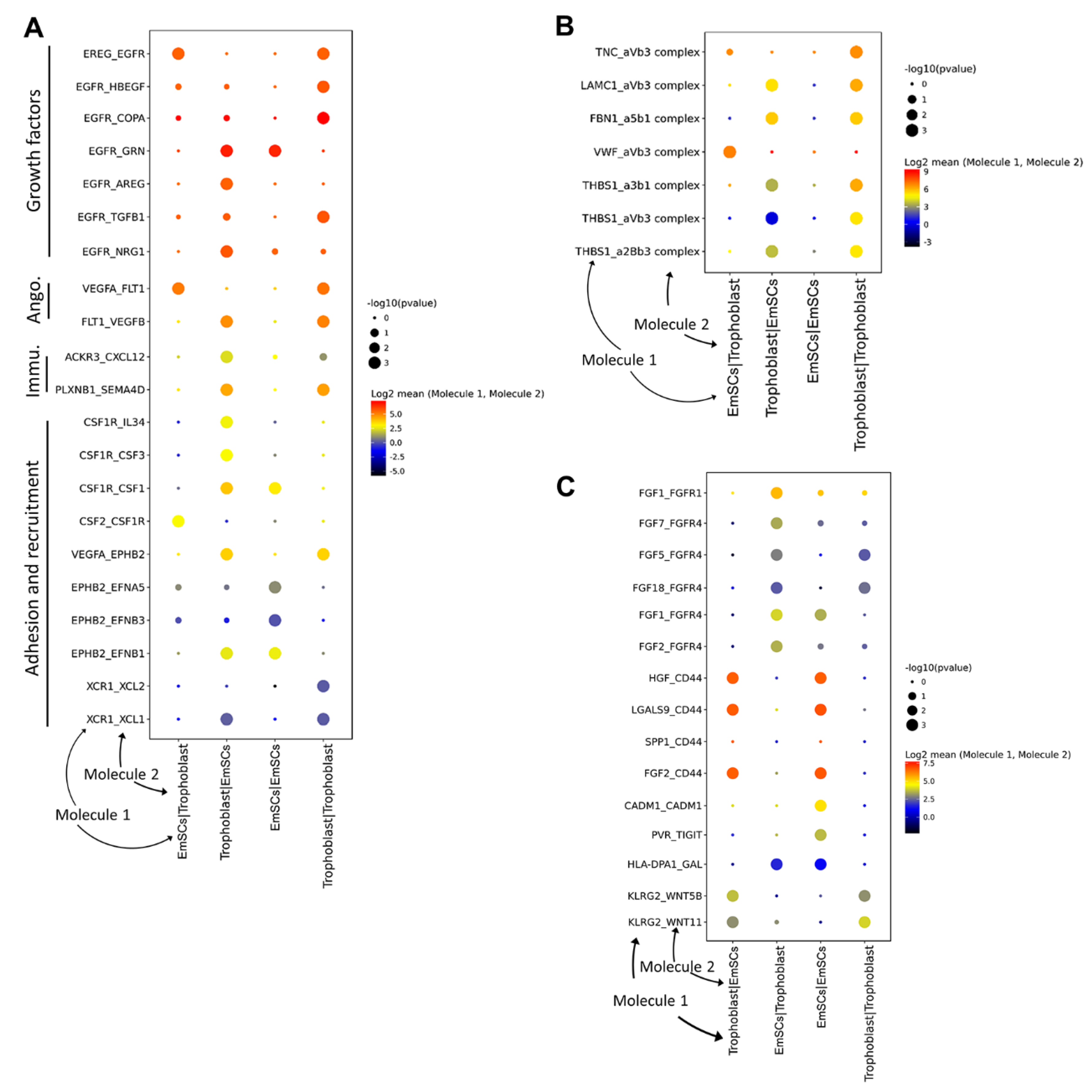
3.3. Crosstalk between Endometrium and Trophoblast Cells during Implantation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Zhang, S.; Lin, H.; Kong, S.; Wang, S.; Wang, H.; Wang, H.; Armant, D.R. Physiological and molecular determinants of embryo implantation. Mol. Asp. Med. 2013, 34, 939–980. [Google Scholar] [CrossRef]
- Valdes, C.T.; Schutt, A.; Simon, C. Implantation failure of endometrial origin: It is not pathology, but our failure to synchronize the developing embryo with a receptive endometrium. Fertil. Steril. 2017, 108, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Karizbodagh, M.P.; Rashidi, B.; Sahebkar, A.; Masoudifar, A.; Mirzaei, H. Implantation window and angiogenesis. J. Cell. Biochem. 2017, 118, 4141–4151. [Google Scholar] [CrossRef]
- Koot, Y.E.; Van Hooff, S.R.; Boomsma, C.M.; Van Leenen, D.; Koerkamp, M.J.G.; Goddijn, M.; Eijkemans, M.J.; Fauser, B.C.; Holstege, F.C.; Macklon, N.S. An endometrial gene expression signature accurately predicts recurrent implantation failure after IVF. Sci. Rep. 2016, 6, 19411. [Google Scholar] [CrossRef]
- Demir, R.; Yaba, A.; Huppertz, B. Vasculogenesis and angiogenesis in the endometrium during menstrual cycle and implantation. Acta Histochem. 2010, 112, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Weimar, C.H.; Uiterweer, E.D.P.; Teklenburg, G.; Heijnen, C.J.; Macklon, N.S. In-vitro model systems for the study of human embryo–endometrium interactions. Reprod. Biomed. Online 2013, 27, 461–476. [Google Scholar] [CrossRef] [PubMed]
- Shahbazi, M.N.; Jedrusik, A.; Vuoristo, S.; Recher, G.; Hupalowska, A.; Bolton, V.; Fogarty, N.M.; Campbell, A.; Devito, L.G.; Ilic, D. Self-organization of the human embryo in the absence of maternal tissues. Nat. Cell Biol. 2016, 18, 700. [Google Scholar] [CrossRef]
- Deglincerti, A.; Croft, G.F.; Pietila, L.N.; Zernicka-Goetz, M.; Siggia, E.D.; Brivanlou, A.H. Self-organization of the in vitro attached human embryo. Nature 2016, 533, 251. [Google Scholar] [CrossRef]
- Aberkane, A.; Essahib, W.; Spits, C.; De Paepe, C.; Sermon, K.; Adriaenssens, T.; Mackens, S.; Tournaye, H.; Brosens, J.; Van de Velde, H. Expression of adhesion and extracellular matrix genes in human blastocysts upon attachment in a 2D co-culture system. MHR Basic Sci. Reprod. Med. 2018, 24, 375–387. [Google Scholar] [CrossRef]
- Lv, B.; An, Q.; Zeng, Q.; Zhang, X.; Lu, P.; Wang, Y.; Zhu, X.; Ji, Y.; Fan, G.; Xue, Z. Single-cell RNA sequencing reveals regulatory mechanism for trophoblast cell-fate divergence in human peri-implantation conceptuses. PLoS Biol. 2019, 17, e3000187. [Google Scholar] [CrossRef]
- Zhou, F.; Wang, R.; Yuan, P.; Ren, Y.; Mao, Y.; Li, R.; Lian, Y.; Li, J.; Wen, L.; Yan, L.; et al. Reconstituting the transcriptome and DNA methylome landscapes of human implantation. Nature 2019, 572, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Huang, K.; Cai, C.; Cai, L.; Jiang, C.-y.; Feng, Y.; Liu, Z.; Zeng, Q.; Cheng, L.; Sun, Y.E. Genetic programs in human and mouse early embryos revealed by single-cell RNA sequencing. Nature 2013, 500, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Picelli, S.; Faridani, O.R.; Bjorklund, A.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014, 9, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Petropoulos, S.; Edsgärd, D.; Reinius, B.; Deng, Q.; Panula, S.P.; Codeluppi, S.; Reyes, A.P.; Linnarsson, S.; Sandberg, R.; Lanner, F. Single-cell RNA-seq reveals lineage and X chromosome dynamics in human preimplantation embryos. Cell 2016, 165, 1012–1026. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Hu, S.; Yao, G.; Wang, Y.; Xu, H.; Ji, X.; He, Y.; Zhu, Q.; Chen, Z.; Sun, Y. Transcriptomic changes during the pre-receptive to receptive transition in human endometrium detected by RNA-Seq. J. Clin. Endocrinol. Metab. 2014, 99, E2744–E2753. [Google Scholar] [CrossRef]
- Vento-Tormo, R.; Efremova, M.; Botting, R.A.; Turco, M.Y.; Vento-Tormo, M.; Meyer, K.B.; Park, J.E.; Stephenson, E.; Polanski, K.; Goncalves, A.; et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature 2018, 563, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Ginestet, C. ggplot2: Elegant graphics for data analysis. J. R. Stat. Soc. Ser. A (Stat. Soc.) 2011, 174, 245–246. [Google Scholar] [CrossRef]
- Li, F.; Devi, Y.S.; Bao, L.; Mao, J.; Gibori, G. Involvement of cyclin D3, CDKN1A (p21), and BIRC5 (Survivin) in interleukin 11 stimulation of decidualization in mice. Biol. Reprod. 2008, 78, 127–133. [Google Scholar] [CrossRef]
- Hamutoğlu, R.; Bulut, H.E.; Kaloğlu, C.; Önder, O.; Dağdeviren, T.; Aydemir, M.N.; Korkmaz, E.M. The regulation of trophoblast invasion and decidual reaction by matrix metalloproteinase-2, metalloproteinase-7, and metalloproteinase-9 expressions in the rat endometrium. Reprod. Med. Biol. 2020, 19, 385–397. [Google Scholar] [CrossRef]
- Shyu, M.-K.; Lin, M.-C.; Shih, J.-C.; Lee, C.-N.; Huang, J.; Liao, C.-H.; Huang, I.-F.; Chen, H.-Y.; Huang, M.-C.; Hsieh, F.-J. Mucin 15 is expressed in human placenta and suppresses invasion of trophoblast-like cells in vitro. Hum. Reprod. 2007, 22, 2723–2732. [Google Scholar] [CrossRef]
- Suzuki, E.; Nakayama, M. The mammalian Ced-1 ortholog MEGF10/KIAA1780 displays a novel adhesion pattern. Exp. Cell Res. 2007, 313, 2451–2464. [Google Scholar] [CrossRef] [PubMed]
- Sheikh, B.N.; Phipson, B.; El-Saafin, F.; Vanyai, H.K.; Downer, N.L.; Bird, M.J.; Kueh, A.J.; May, R.E.; Smyth, G.K.; Voss, A.K.; et al. MOZ (MYST3, KAT6A) inhibits senescence via the INK4A-ARF pathway. Oncogene 2015, 34, 5807–5820. [Google Scholar] [CrossRef]
- Froimchuk, E.; Jang, Y.; Ge, K. Histone H3 lysine 4 methyltransferase KMT2D. Gene 2017, 627, 337–342. [Google Scholar] [CrossRef] [PubMed]
- D’Hooghe, T.M.; Nugent, N.P.; Cuneo, S.; Chai, D.C.; Deer, F.; Debrock, S.; Kyama, C.M.; Mihalyi, A.; Mwenda, J.M. Recombinant human TNFRSF1A (r-hTBP1) inhibits the development of endometriosis in baboons: A prospective, randomized, placebo- and drug-controlled study. Biol. Reprod. 2006, 74, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Yue, Y.; Xiong, S. POM121 inhibits the macrophage inflammatory response by impacting NF-kappaB P65 nuclear accumulation. Exp. Cell Res. 2019, 377, 17–23. [Google Scholar] [CrossRef]
- Kim, N.H.; Choi, S.H.; Lee, T.R.; Lee, C.H.; Lee, A.Y. Cadherin 11, a miR-675 target, induces N-cadherin expression and epithelial-mesenchymal transition in melasma. J. Investig. Derm. 2014, 134, 2967–2976. [Google Scholar] [CrossRef]
- Ferri-Lagneau, K.F.; Haider, J.; Sang, S.; Leung, T. Rescue of hematopoietic stem/progenitor cells formation in plcg1 zebrafish mutant. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Wei, Y.; Ma, D.; Gao, Y.; Zhang, C.; Wang, L.; Liu, F. Ncor2 is required for hematopoietic stem cell emergence by inhibiting Fos signaling in zebrafish. Blood 2014, 124, 1578–1585. [Google Scholar] [CrossRef]
- Pasula, S.; Cai, X.; Dong, Y.; Messa, M.; McManus, J.; Chang, B.; Liu, X.; Zhu, H.; Mansat, R.S.; Yoon, S.-J. Endothelial epsin deficiency decreases tumor growth by enhancing VEGF signaling. J. Clin. Investig. 2012, 122, 4424–4438. [Google Scholar] [CrossRef] [PubMed]
- Klemke, R.L. A PEAK1/GATA2 signaling axis controls VEGFR2 expression to mediate angiogenesis. In Proceedings of the AACR Annual Meeting, Washington, DC, USA, 1–5 April 2017. [Google Scholar]
- Wang, H.; Vardy, L.A.; Tan, C.P.; Loo, J.M.; Guo, K.; Li, J.; Lim, S.G.; Zhou, J.; Chng, W.J.; Ng, S.B. PCBP1 suppresses the translation of metastasis-associated PRL-3 phosphatase. Cancer Cell 2010, 18, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Lessey, B.A.; Castelbaum, A.J. Integrins and implantation in the human. Rev. Endocr. Metab. Disord. 2002, 3, 107–117. [Google Scholar] [CrossRef]
- Smith, S.D.; Choudhury, R.H.; Matos, P.; Horn, J.A.; Lye, S.J.; Dunk, C.E.; Aplin, J.D.; Jones, R.L.; Harris, L.K. Changes in vascular extracellular matrix composition during decidual spiral arteriole remodeling in early human pregnancy. Histol. Histopathol. 2016, 31, 557–571. [Google Scholar]
- Wang, W.; Vilella, F.; Alama, P.; Moreno, I.; Mignardi, M.; Isakova, A.; Pan, W.; Simon, C.; Quake, S.R. Single-cell transcriptomic atlas of the human endometrium during the menstrual cycle. Nat. Med. 2020, 26, 1644–1653. [Google Scholar] [CrossRef]
- Egashira, M.; Hirota, Y. Uterine receptivity and embryo–uterine interactions in embryo implantation: Lessons from mice. Reprod. Med. Biol. 2013, 12, 127–132. [Google Scholar] [CrossRef]
- Stanietsky, N.; Simic, H.; Arapovic, J.; Toporik, A.; Levy, O.; Novik, A.; Levine, Z.; Beiman, M.; Dassa, L.; Achdout, H.; et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 17858–17863. [Google Scholar] [CrossRef]
- Zhang, Q.; Yan, J. Update of Wnt signaling in implantation and decidualization. Reprod. Med. Biol. 2016, 15, 95–105. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lv, B.; Xu, X.; Zhang, X.; Qi, L.; He, W.; Wang, L.; Chen, X.; Peng, L.; Xue, J.; Ji, Y.; et al. Activation of Blood Vessel Development in Endometrial Stromal Cells In Vitro Cocultured with Human Peri-Implantation Embryos Revealed by Single-Cell RNA-Seq. Life 2021, 11, 367. https://doi.org/10.3390/life11050367
Lv B, Xu X, Zhang X, Qi L, He W, Wang L, Chen X, Peng L, Xue J, Ji Y, et al. Activation of Blood Vessel Development in Endometrial Stromal Cells In Vitro Cocultured with Human Peri-Implantation Embryos Revealed by Single-Cell RNA-Seq. Life. 2021; 11(5):367. https://doi.org/10.3390/life11050367
Chicago/Turabian StyleLv, Bo, Xiaoyu Xu, Xunyi Zhang, Lingbin Qi, Wen He, Lu Wang, Xian Chen, Luying Peng, Jinfeng Xue, Yazhong Ji, and et al. 2021. "Activation of Blood Vessel Development in Endometrial Stromal Cells In Vitro Cocultured with Human Peri-Implantation Embryos Revealed by Single-Cell RNA-Seq" Life 11, no. 5: 367. https://doi.org/10.3390/life11050367
APA StyleLv, B., Xu, X., Zhang, X., Qi, L., He, W., Wang, L., Chen, X., Peng, L., Xue, J., Ji, Y., & Xue, Z. (2021). Activation of Blood Vessel Development in Endometrial Stromal Cells In Vitro Cocultured with Human Peri-Implantation Embryos Revealed by Single-Cell RNA-Seq. Life, 11(5), 367. https://doi.org/10.3390/life11050367





