Molecular Interactions between Two LMP2A PY Motifs of EBV and WW Domains of E3 Ubiquitin Ligase AIP4
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Isothermal Titration Calorimetry
2.3. NMR Experiments and WW2 Structure Calculation
2.4. NMR Titrations of WW2 with Various LMP2A Constructs
2.5. Protein–Peptide Docking
3. Results
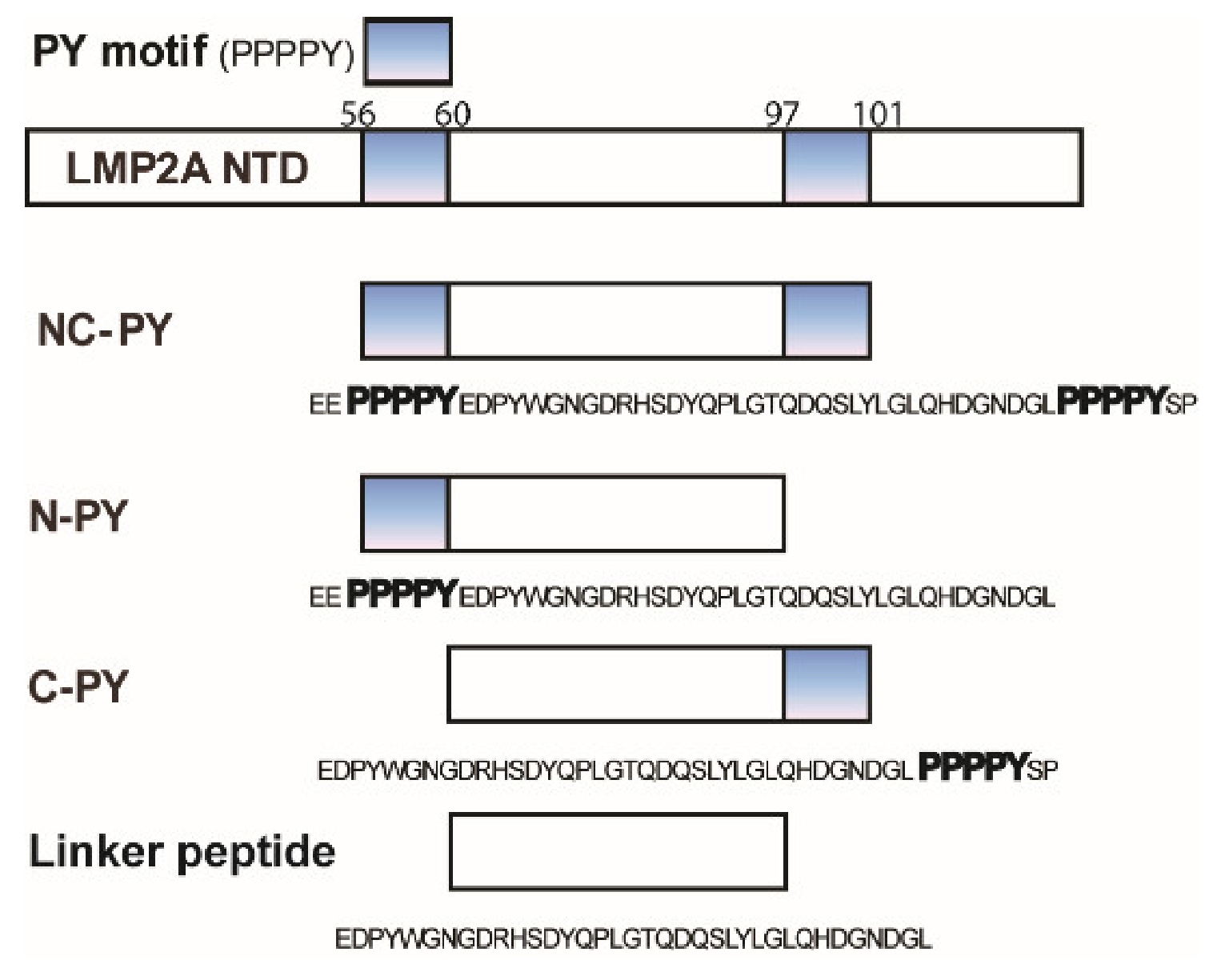
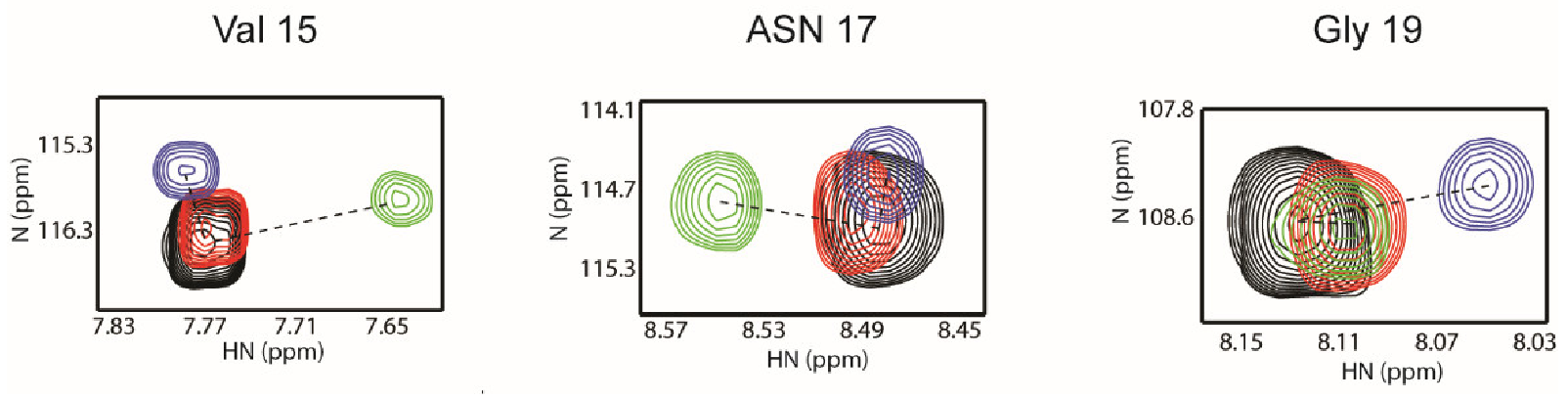
3.1. WW2 Domain Binding Properties of Two LMP2A PY Motifs
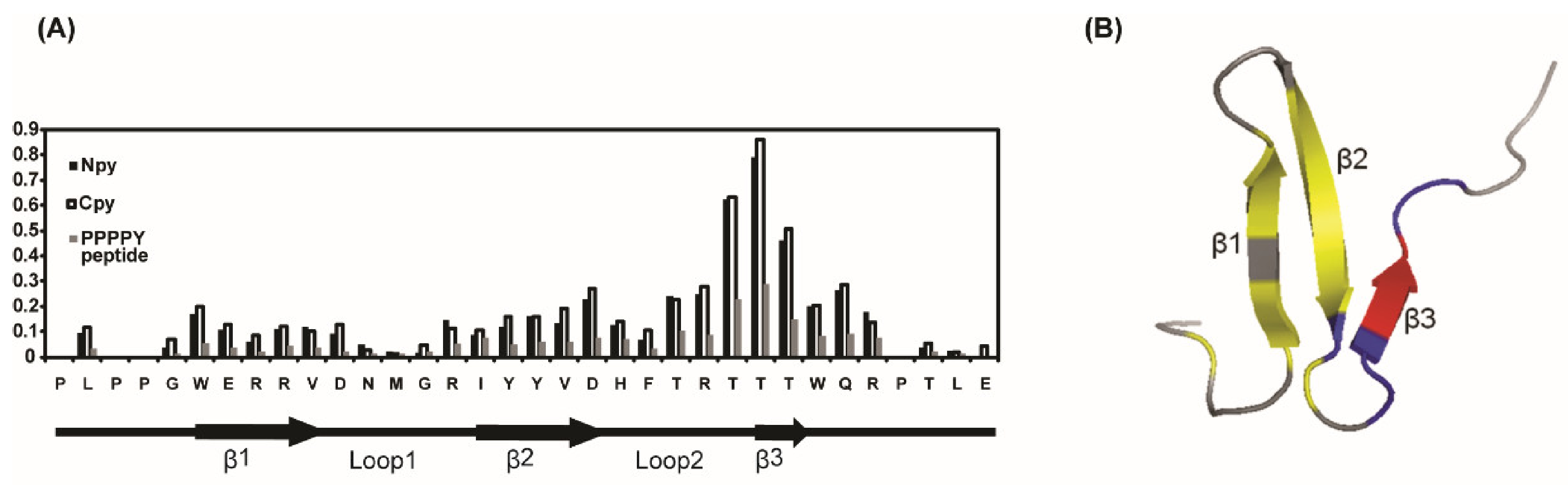
3.2. Solution Structure of the WW2 Domain
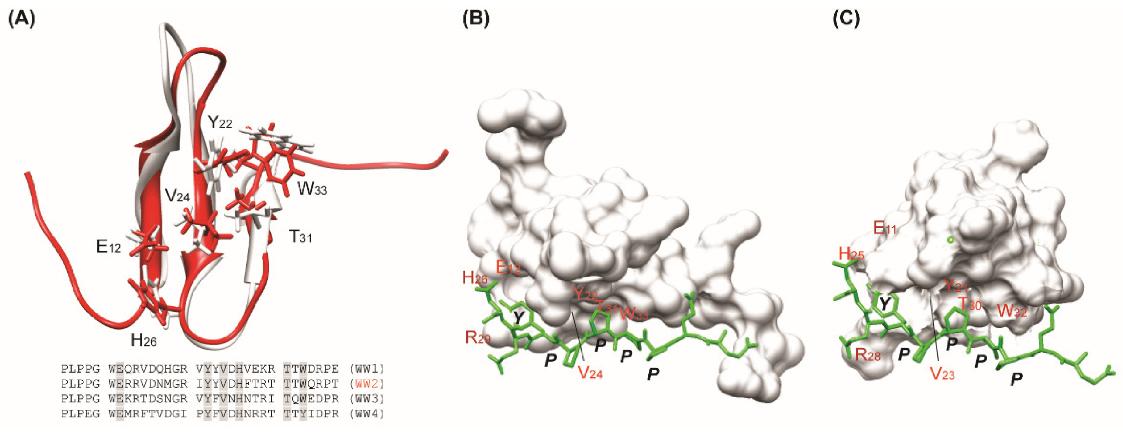
3.3. Modeled Structure of WW2 Domain–PY Motif Complex
3.4. Differences Between Two Tailed PY Motifs Upon Binding
4. Discussion
4.1. Importance of Linker Region Outside of PY Motif
4.2. Models of AIP4 and LMP2A Interaction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Henle, W.; Henle, G. Seroepidemiology of the Virus. In The Epstein-Barr Virus, 2nd ed.; Epstein, M.A., Achong, B.G., Eds.; Springer: Berlin, Germany, 1979; pp. 61–78. [Google Scholar]
- Kieff, E. Epstein-Barr virus and its replication. In Fields Virology, 2nd ed.; Fields, B.N., Knipe, D.M., Howley, P.M., Eds.; Lippincott-Raven: Philadelphia, PA, USA, 1996; pp. 1109–1162. [Google Scholar]
- Longnecker, R. Molecular biology of Epstein-Barr virus. In Human Tumor Viruses, 2nd ed.; McCance, D., Ed.; American Society for Microbiology: Washington, DC, USA, 1998; pp. 133–172. [Google Scholar]
- Rickinson, A.B.; Kieff, E. Epstein-Barr virus. In Fields Virology, 2nd ed.; Fields, B.N., Knipe, D.M., Howley, P.M., Eds.; Lippincott-Raven: Philadelphia, PA, USA, 1996; pp. 2379–2446. [Google Scholar]
- Hooykaas, M.J.G.; Van Gent, M.; Soppe, J.A.; Kruse, E.; Boer, I.G.J.; Van Leenen, D.; Groot Koerkamp, M.J.A.; Holstege, F.C.P.; Ressing, M.E.; Wiertz, E.J.H.J.; et al. EBV microrna BART16 suppresses type I IFN signaling. J. Immunol. 2017, 198, 4062–4073. [Google Scholar] [CrossRef] [PubMed]
- Jin, Q.E.; Su, J.R.; Yan, D.H.; Wu, S.N. Epstein-Barr Virus Infection and Increased Sporadic Breast Carcinoma Risk: A Meta-Analysis. Med. Princ. Pract. 2019, 29, 2. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.L.; Longnecker, R.; Kieff, E. Epstein-Barr virus latent membrane protein 2A blocks calcium mobilization in B lymphocytes. J. Virol. 1993, 67, 3087–3094. [Google Scholar] [CrossRef] [PubMed]
- Miller, C.L.; Lee, J.H.; Kieff, E.; Burkhardt, A.L.; Bolen, J.B.; Longnecker, R. Epstein-Barr virus protein LMP2A regulates reactivation from latency by negatively regulating tyrosine kinases involved in sIg-mediated signal transduction. Infect. Agents Dis. 1994, 3, 128–136. [Google Scholar]
- Miller, C.L.; Lee, J.H.; Kieff, E.; Longnecker, R. An integral membrane protein (LMP2) blocks reactivation of Epstein-Barr virus from latency following surface immunoglobulin crosslinking. Proc. Natl. Acad. Sci. USA 1994, 91, 772–776. [Google Scholar] [CrossRef]
- Miller, C.L.; Burkhardt, A.L.; Lee, J.H.; Stealey, B.; Longnecker, R.; Bolen, J.B.; Kieff, E. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity 1995, 2, 155–166. [Google Scholar] [CrossRef]
- Fish, K.; Comoglio, F.; Shaffer, A.L.; Ji, Y.; Pan, K.T.; Scheich, S.; Oellerich, A.; Doebele, C.; Ikeda, M.; Schaller, S.J.; et al. Rewiring of B cell receptor signaling by Epstein-Barr virus LMP2A. Proc. Natl. Acad. Sci. USA 2020, 117, 26318–26327. [Google Scholar] [CrossRef]
- Chen, Y.; Fachko, D.; Ivanov, N.S.; Skinner, C.M.; Skalsky, R.L. Epstein-Barr virus microRNAs regulate B cell receptor signal transduction and lytic reactivation. PLoS Pathog. 2019, 15, e1007535. [Google Scholar] [CrossRef]
- Longnecker, R.; Druker, B.; Roberts, T.M.; Kieff, E. An Epstein-Barr virus protein associated with cell growth transformation interacts with a tyrosine kinase. J. Virol. 1991, 65, 3681–3692. [Google Scholar] [CrossRef]
- Incrocci, R.; Hussain, S.; Stone, A.; Bieging, K.; Alt, L.A.; Fay, M.J.; Swanson-Mungerson, M. Epstein-Barr virus Latent Membrane Protein 2A (LMP2A)-mediated changes in Fas expression and Fas-dependent apoptosis: Role of Lyn/Syk activation. Cell Immunol. 2015, 297, 108–119. [Google Scholar] [CrossRef]
- Dergai, O.; Dergai, M.; Skrypkina, I.; Matskova, L.; Tsyba, L.; Gudkova, D.; Rynditch, A. The LMP2A protein of Epstein-Barr virus regulates phosphorylation of ITSN1 and Shb adaptors by tyrosine kinases. Cell Signal. 2013, 25, 33–40. [Google Scholar] [CrossRef]
- Caldwell, R.G.; Wilson, J.B.; Anderson, S.J.; Longnecker, R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 1998, 9, 405–411. [Google Scholar] [CrossRef]
- Incrocci, R.; McAloon, J.; Montesano, M.; Bardahl, J.; Vagvala, S.; Stone, A.; Swanson-Mungerson, M. Epstein-Barr virus LMP2A utilizes Syk and PI3K to activate NF-κB in B-cell lymphomas to increase MIP-1α production. J. Med. Virol. 2019, 91, 845–855. [Google Scholar] [CrossRef]
- Stewart, S.; Dawson, C.W.; Takada, K.; Curnow, J.; Moody, C.A.; Sixbey, J.W.; Young, L.S. Epstein-Barr virus-encoded LMP2A regulates viral and cellular gene expression by modulation of the NF-kappaB transcription factor pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 15730–15735. [Google Scholar] [CrossRef]
- Hino, R.; Uozaki, H.; Inoue, Y.; Shintani, Y.; Ushiku, T.; Sakatani, T.; Takada, K.; Fukayama, M. Survival advantage of EBV-associated gastric carcinoma: Survivin up-regulation by viral latent membrane protein 2A. Cancer Res. 2008, 68, 1427–1435. [Google Scholar] [CrossRef]
- McLaren, J.E.; Zuo, J.; Grimstead, J.; Poghosyan, Z.; Bell, A.I.; Rowe, M.; Brennan, P. STAT1 contributes to the maintenance of the latency III viral programme observed in Epstein-Barr virus-transformed B cells and their recognition by CD8+ T cells. J. Gen. Virol. 2009, 90, 2239–2250. [Google Scholar] [CrossRef]
- Nagel, S.; Uphoff, C.C.; Dirks, W.G.; Pommerenke, C.; Meyer, C.; Drexler, H.G. Epstein-Barr virus (EBV) activates NKL homeobox gene HLX in DLBCL. PLoS ONE 2019, 14, e0216898. [Google Scholar] [CrossRef]
- Sora, R.P.; Ikeda, M.; Longnecker, R. Two Pathways of p27Kip1 Degradation Are Required for Murine Lymphoma Driven by Myc and EBV Latent Membrane Protein 2A. mBio 2019, 10, e00548-19. [Google Scholar] [CrossRef]
- Fotheringham, J.A.; Mazzucca, S.; Raab-Traub, N. Epstein barr-virus latent membrane protein-2A-induced DeltaNp63alpha expression is associated with impaired epithelial-cell differentiation. Oncogene 2010, 29, 4287–4296. [Google Scholar] [CrossRef]
- Bieging, K.T.; Swanson-Mungerson, M.; Amick, A.C.; Longnecker, R. Epstein-Barr virus in Burkitt’s lymphoma: A role for latent membrane protein 2A. Cell Cycle 2010, 9, 901–908. [Google Scholar] [CrossRef]
- Ingham, R.J.; Raaijmakers, J.; Lim, C.S.; Mbamalu, G.; Gish, G.; Chen, F.; Matskova, L.; Ernberg, I.; Winberg, G.; Pawson, T. The Epstein-Barr virus protein, latent membrane protein 2A, co-opts tyrosine kinases used by the T cell receptor. J. Biol. Chem. 2005, 280, 34133–34142. [Google Scholar] [CrossRef] [PubMed]
- Huibregtse, J.; Scheffner, M.; Beaudenon, S.; Howley, P. A family of proteins structurally and functionally related to the E6-AP ubiquitinprotein ligase. Proc. Natl. Acad. Sci. USA 1995, 92, 2563–2567. [Google Scholar] [CrossRef] [PubMed]
- Ingham, R.J.; Gish, G.; Pawson, T. The Nedd4 family of E3 ubiquitin ligases: Functional diversity within a common modular architecture. Oncogene 2004, 23, 1972–1984. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Ma, X.; Yuan, M.; Yi, Y.; Liu, G.; Wen, M.; Jiang, W.; Ji, R.; Zhu, L.; Tang, Z.; et al. E3 ligase Nedd4l promotes antiviral innate immunity by catalyzing K29-linked cysteine ubiquitination of TRAF3. Nat. Commun. 2021, 12, 1194. [Google Scholar] [CrossRef]
- Harvey, K.F.; Kumar, S. Nedd4-like proteins: An emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends Cell Biol. 1999, 9, 166–169. [Google Scholar] [CrossRef]
- Macias, M.J.; Hyvonen, M.; Baraldi, E.; Schultz, J.; Sudol, M.; Saraste, M.; Oschkinat, H. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature 1996, 382, 646–649. [Google Scholar] [CrossRef]
- Sudol, M. Structure and function of the WW domain. Prog. Biophys. Mol. Biol. 1996, 65, 113–132. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Z.; Chen, X.; Li, J.; Yao, W.; Huang, S.; Gu, A.; Lei, Q.Y.; Mao, Y.; Wen, W. A multi-lock inhibitory mechanism for fine-tuning enzyme activities of the HECT family E3 ligases. Nat. Commun. 2019, 10, 3162. [Google Scholar] [CrossRef]
- Meiyappan, M.; Birrane, G.; Ladias, J.A.A. Structural basis for polyproline recognition by the FE65 WW domain. J. Mol. Biol. 2007, 372, 970–980. [Google Scholar] [CrossRef]
- Lin, Z.; Yang, Z.; Xie, R.; Ji, Z.; Guan, K.; Zhang, M. Decoding WW domain tandem-mediated target recognitions in tissue growth and cell polarity. Elife 2019, 8, e49439. [Google Scholar] [CrossRef]
- Staub, O.; Dho, S.; Henry, P.; Correa, J.; Ishikawa, T.; McGlade, J.; Rotin, D. WW domains of Nedd4 bind to the proline-rich PY motifs in the epithelial Na+ channel deleted in Liddle’s syndrome. EMBO J. 1996, 15, 2371–2380. [Google Scholar] [CrossRef]
- Bork, P.; Sudol, M. The WW domain, A signaling site in dystrophin. Trends Biochem. Sci. 1994, 19, 531–553. [Google Scholar] [CrossRef]
- Wang, Y.; Du, Y.; Luo, L.; Hu, P.; Yang, G.; Li, T.; Han, X.; Ma, A.; Wang, T. Alterations of Nedd4-2-binding capacity in PY-motif of NaV 1.5 channel underlie long QT syndrome and Brugada syndrome. Acta Physiol. 2020, 229, e13438. [Google Scholar] [CrossRef]
- Vargas, R.E.; Duong, V.T.; Han, H.; Ta, A.P.; Chen, Y.; Zhao, S.; Yang, B.; Seo, G.; Chuc, K.; Oh, S.; et al. Elucidation of WW domain ligand binding specificities in the Hippo pathway reveals STXBP4 as YAP inhibitor. EMBO J. 2020, 39, e102406. [Google Scholar] [CrossRef]
- Hansson, J.H.; Nelson-Williams, C.; Suzuki, H.; Schild, L.; Shimkets, R.; Lu, Y.; Canessa, C.; Iwasaki, T.; Rossier, B.; Lifton, R.P. Hypertension caused by a truncated epithelial sodium channel gamma subunit: Genetic heterogeneity of Liddle syndrome. Nat. Genet. 1995, 11, 76–82. [Google Scholar] [CrossRef]
- Hansson, J.H.; Schild, L.; Lu, Y.; Wilson, T.A.; Gautschi, I.; Shimkets, R.; Nelson-Williams, C.; Rossier, B.C.; Lifton, R.P. A de novo missense mutation of the beta subunit of the epithelial sodium channel causes hypertension and Liddle syndrome, identifying a proline-rich segment critical for regulation of channel activity. Proc. Natl. Acad. Sci. USA 1995, 92, 11495–11499. [Google Scholar] [CrossRef] [PubMed]
- Inoue, J.; Iwaoka, T.; Tokunaga, H.; Takamune, K.; Naomi, S.; Araki, M.; Takahama, K.; Yamaguchi, K.; Tomita, K. A family with Liddle’s syndrome caused by a new missense mutation in the beta subunit of the epithelial sodium channel. J. Clin. Endocrinol. Metab. 1998, 83, 2210–2213. [Google Scholar] [CrossRef]
- Tamura, H.; Schild, L.; Enomoto, N.; Matsui, N.; Marumo, F.; Rossier, B.C. Liddle disease caused by a missense mutation of beta subunit of the epithelial sodium channel gene. J. Clin. Investig. 1996, 97, 1780–1784. [Google Scholar] [CrossRef]
- Ikeda, M.; Ikeda, A.; Longan, L.C.; Longnecker, R. The Epstein-Barr virus latent membrane protein 2A PY motif recruits WW domain-containing ubiquitin-protein ligases. Virology 2000, 268, 178–191. [Google Scholar] [CrossRef]
- Winberg, G.; Matskova, L.; Chen, F.; Plant, P.; Rotin, D.; Gish, G.; Ernberg, R.; Ingham, I.; Pawson, T. Latent membrane protein 2A of Epstein-Barr virus binds WW domain E3 protein-ubiquitin ligases that ubiquitinate B-cell tyrosine kinases. Mol. Cell. Biol. 2000, 20, 8526–8535. [Google Scholar] [CrossRef]
- Pang, M.F.; Lin, K.W.; Peh, S.C. The signaling pathways of Epstein-Barr virus-encoded latent membrane protein 2A (LMP2A) in latency and cancer. Cell. Mol. Biol. Lett. 2009, 14, 222–247. [Google Scholar] [CrossRef] [PubMed]
- Matskova, L.V.; Helmstetter, C.; Ingham, R.J.; Gish, G.; Lindholm, C.K.; Ernberg, I.; Pawson, T.; Winberg, G. The Shb signalling scaffold binds to and regulates constitutive signals from the Epstein-Barr virus LMP2A membrane protein. Oncogene 2007, 26, 4908–4917. [Google Scholar] [CrossRef] [PubMed]
- Fish, K.; Sora, R.P.; Schaller, S.J.; Longnecker, R.; Ikeda, M. EBV latent membrane protein 2A orchestrates p27kip1 degradation via Cks1 to accelerate MYC-driven lymphoma in mice. Blood 2017, 130, 2516–2526. [Google Scholar] [CrossRef]
- Seo, M.D.; Park, S.J.; Kim, H.J.; Lee, B.J. Identification of the WW domain-interaction sites in the unstructured N-terminal domain of EBV LMP 2A. FEBS Lett. 2007, 581, 65–70. [Google Scholar] [CrossRef]
- Morales, B.; Ramirez-Espain, X.; Shaw, A.Z.; Martin-Malpartida, P.; Yraola, F.; Sánchez-Tilló, E.; Farrera, C.; Celada, A.; Royo, M.; Macias, M.J. NMR structural studies of the ItchWW3 domain reveal that phosphorylation at T30 inhibits the interaction with PPxY-containing ligands. Structure 2007, 15, 473–483. [Google Scholar] [CrossRef]
- Park, S.J.; Seo, M.D.; Lee, S.K.; Ikeda, M.; Longnecker, R.; Lee, B.J. Expression and characterization of N-terminal domain of Epstein-Barr virus latent membrane protein 2A in Escherichia coli. Protein Expr. Purif. 2005, 41, 9–17. [Google Scholar] [CrossRef]
- Wishart, D.S.; Sykes, B.D. The 13C chemical-shift index: A simple method for the identification of protein secondary structure using 13C chemical shift data. J. Biomol. NMR 1994, 4, 171–180. [Google Scholar] [CrossRef]
- Güntert, P. Automated NMR protein structure calculation with CYANA. Methods Mol. Biol. 2004, 278, 353–378. [Google Scholar] [CrossRef]
- Gabriel, C.; Frank, D.; Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 1999, 13, 289–302. [Google Scholar] [CrossRef]
- Brunger, A.T.; Adams, P.D.; Clore, G.M.; Delano, W.L.; Gros, P.; Jiang, R.W.; Grosse-kunstleve, J.S.; Kuszewski, J.; Nilges, N.; Pannu, N.S.; et al. Crystallography and NMR system(CNS): A new software system for macromolecular structure determination. ACTA Cryst. 1998, D54, 905–921. [Google Scholar] [CrossRef]
- Koradi, R.; Billeter, M.; Wüthrich, K. A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996, 14, 51–55. [Google Scholar] [CrossRef]
- Mooers, B.H.M. Shortcuts for faster image creation in PyMOL. Protein Sci. 2020, 29, 268–276. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Rullmannn, J.A.; MacArthur, M.W.; Kaptein, R.; Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 1996, 8, 477–486. [Google Scholar] [CrossRef]
- Delaglio, F.; Grzesiek, S.; Vuister, G.W.; Zhu, G.; Pfeifer, J.; Bax, A. NMR Pipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 1995, 6, 277–293. [Google Scholar] [CrossRef]
- Johnson, B.A.; Blevins, R.A. NMR View: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR 1994, 4, 603–614. [Google Scholar] [CrossRef]
- Grzesiek, S.; Stahl, S.J.; Wingfield, P.T.; Bax, A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry 1996, 35, 10256–10261. [Google Scholar] [CrossRef]
- Tovchigrechko, A.; Vakser, I.A. GRAMM-X public web server for protein-protein docking. Nucl. Acids Res. 2006, 34, W310–W314. [Google Scholar] [CrossRef]
- Chong, P.A.; Lin, H.; Wrana, J.L.; Forman-Kay, J.D. An expanded WW domain recognition motif revealed by the interaction between Smad7 and the E3 ubiquitin ligase Smurf2. J. Biol. Chem. 2006, 281, 17069–17075. [Google Scholar] [CrossRef]
- Kowalski, J.A.; Liu, K.; Kelly, J.W. NMR solution structure of the isolated Apo Pin1 WW domain: Comparison to the x-ray crystal structures of Pin1. Biopolymers 2002, 63, 111–121. [Google Scholar] [CrossRef]
- Wahl, L.C.; Watt, J.E.; Yim, H.T.T.; De Bourcier, D.; Tolchard, J.; Soond, S.M.; Blumenschein, T.M.A.; Chantry, A. Smad7 Binds Differently to Individual and Tandem WW3 and WW4 Domains of WWP2 Ubiquitin Ligase Isoforms. Int. J. Mol. Sci. 2019, 20, 4682. [Google Scholar] [CrossRef] [PubMed]
- Iglesias-Bexiga, M.; Palencia, A.; Corbi-Verge, C.; Martin-Malpartida, P.; Blanco, F.J.; Macias, M.J.; Cobos, E.S.; Luque, I. Binding site plasticity in viral PPxY Late domain recognition by the third WW domain of human NEDD4. Sci. Rep. 2019, 9, 15076. [Google Scholar] [CrossRef] [PubMed]
- Spagnol, G.; Kieken, F.; Kopanic, J.L.; Li, H.; Zach, S.; Stauch, K.L.; Grosely, R.; Sorgen, P.L. Structural Studies of the Nedd4 WW Domains and Their Selectivity for the Connexin43 (Cx43) Carboxyl Terminus. J. Biol. Chem. 2016, 291, 7637–7650. [Google Scholar] [CrossRef] [PubMed]
- Kanelis, V.; Bruce, M.C.; Skrynnikov, N.R.; Rotin, D.; Forman-Kay, J.D. Structural determinants for high-affinity binding in a Nedd4 WW3 domain-Comm PY motif complex. Structure 2006, 14, 543–553. [Google Scholar] [CrossRef]
- Kato, Y.; Miyakawa, T.; Kurita, J.; Tanokura, M. Structure of FBP11 WW1-PL ligand complex reveals the mechanism of proline-rich ligand recognition by group II/III WW domains. J. Biol. Chem. 2006, 281, 40321–40329. [Google Scholar] [CrossRef]
- Verdecia, M.A.; Bowman, M.E.; Lu, K.P.; Hunter, T.; Noel, J.P. Structural basis for phosphoserine-proline recognition by group IV WW domains. Nat. Struct. Biol. 2000, 7, 639–643. [Google Scholar] [CrossRef]
- Merchant, M.; Caldwell, R.G.; Longnecker, R. The LMP2A ITAM is essential for providing B cells with development and survival signals in vivo. J. Virol. 2000, 74, 9115–9124. [Google Scholar] [CrossRef]
- Ikeda, M.; Longnecker, R. The c-Cbl proto-oncoprotein downregulates EBV LMP2A signaling. Virology 2009, 385, 183–191. [Google Scholar] [CrossRef]
- Fruehling, S.; Swart, R.; Dolwick, K.M.; Kremmer, E.; Longnecker, R. Tyrosine 112 of latent membrane protein 2A is essential for protein tyrosine kinase loading and regulation of Epstein-Barr virus latency. J. Virol. 1998, 72, 7796–7806. [Google Scholar] [CrossRef]
- Khan, S.; He, Y.; Zhang, X.; Yuan, Y.; Pu, S.; Kong, Q.; Zheng, G.; Zhou, D. PROteolysis TArgeting Chimeras (PROTACs) as emerging anticancer therapeutics. Oncogene 2020, 39, 4909–4924. [Google Scholar] [CrossRef]
- Jin, J.; Wu, Y.; Chen, J.; Shen, Y.; Zhang, L.; Zhang, H.; Chen, L.; Yuan, H.; Chen, H.; Zhang, W.; et al. The peptide PROTAC modality: A novel strategy for targeted protein ubiquitination. Theranostics 2020, 10, 10141–10153. [Google Scholar] [CrossRef]



| Experimental Constraints | |
|---|---|
| NOE constraints total | 305 |
| Intra-residue (i = j) | 114 |
| Sequential (|i-j| = 1) | 100 |
| Medium range (1 < |i-j| < 5) | 30 |
| Long range (|i-j| ≥ 5) | 61 |
| Dihedral constraints | |
| Φ | 15 |
| ψ | 16 |
| RMSD from idealized geometry | |
| Bonds (Å) | 0.002 ± 0.00007 |
| Angles (°) | 0.3455 ± 0.0062 |
| RMSD to the mean structure (residues 7–16, 20–34) | |
| #Backbone atoms (N, Cα, CO) | 0.41 ± 0.09 |
| #All heavy | 0.99 ± 0.11 |
| CNS energy (kcal/mol) a | |
| Eoverall | 55.72 ± 0.97 |
| Ebond | 2.71 ± 0.19 |
| Eangle | 21.44 ± 0.77 |
| Eimproper | 2.66 ± 0.31 |
| Evdw | 20.77 ± 0.96 |
| Enoe | 8.11 ± 0.59 |
| Ecdih | 0.02 ± 0.03 |
| Violations per conformer | |
| Distance constraints (>0.1 Å) | 0 |
| Dihedral angle constraints (>5 Å) | 0 |
| van der Waals (<1.6 Å) | 0 |
| Ramanchandran plot (%) b | |
| Most favored region | 70.7 |
| Additionally allowed region | 20.2 |
| Generously allowed region | 8.6 |
| Disallowed region | 0.5 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, M.-D.; Seok, S.-H.; Kim, J.-H.; Choi, J.W.; Park, S.J.; Lee, B.-J. Molecular Interactions between Two LMP2A PY Motifs of EBV and WW Domains of E3 Ubiquitin Ligase AIP4. Life 2021, 11, 379. https://doi.org/10.3390/life11050379
Seo M-D, Seok S-H, Kim J-H, Choi JW, Park SJ, Lee B-J. Molecular Interactions between Two LMP2A PY Motifs of EBV and WW Domains of E3 Ubiquitin Ligase AIP4. Life. 2021; 11(5):379. https://doi.org/10.3390/life11050379
Chicago/Turabian StyleSeo, Min-Duk, Seung-Hyeon Seok, Ji-Hun Kim, Ji Woong Choi, Sung Jean Park, and Bong-Jin Lee. 2021. "Molecular Interactions between Two LMP2A PY Motifs of EBV and WW Domains of E3 Ubiquitin Ligase AIP4" Life 11, no. 5: 379. https://doi.org/10.3390/life11050379






