Multi-Gene Phylogeny and Morphology Reveal Haplohelminthosporium gen. nov. and Helminthosporiella gen. nov. Associated with Palms in Thailand and A Checklist for Helminthosporium Reported Worldwide
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection, Isolation, and Identification
2.2. DNA Extraction and Amplification (PCR)
2.3. Phylogenetic Analyses
| Family | Species | Strain No. | GenBank Accession No. | References | |||
|---|---|---|---|---|---|---|---|
| ITS | LSU | SSU | tef1-α | ||||
| Corynesporaceae | Corynespora cassiicola | CBS 100,822 | - | GU301808 | GU296144 | GU349052 | [54] |
| Corynesporaceae | Corynespora cassiicola | CCP | KF810854 | - | GU296145 | - | [54,55] |
| Corynesporaceae | Corynespora smithii | CBS 139,925 | KY984299 | KY984299 | - | - | [21] |
| Corynesporaceae | Corynespora smithii | L120 | KY984297 | KY984297 | - | KY984435 | [21] |
| Corynesporaceae | Corynespora smithii | L130 | KY984298 | KY984298 | KY984419 | KY984436 | [21] |
| Corynesporaceae | Corynespora smithii | L139 | KY984300 | KY984300 | - | - | [21] |
| Cyclothyriellaceae | Cyclothyriella rubronotata | TR | KX650541 | KX650541 | - | KX650516 | [56] |
| Cyclothyriellaceae | Cyclothyriella rubronotata | TR9 * | KX650544 | KX650544 | KX650507 | KX650519 | [56] |
| Massariaceae | Byssothecium circinans | CBS 675.92 | - | GU205217 | GU205235 | GU349061 | [54] |
| Massarinaceae | Byssothecium circinans | CBS 675.92 | - | AY016357 | AY016339 | - | [57,58] |
| Massarinaceae | Haplohelminthosporium calami | MFLUCC 18-0074 * | MT928158 | MT928156 | MT928160 | - | This study |
| Massarinaceae | Helminthosporium aquaticum | MFLUCC 15-0357 | KU697302 | KU697306 | KU697310 | - | [20] |
| Massarinaceae | Helminthosporium aquaticum | DLUCC 0758 | MG098779 | MG098786 | MG098795 | MG98585 | [24] |
| Massarinaceae | Helminthosporium austriacum | L132 * | KY984301 | KY984301 | KY984420 | KY984437 | [21] |
| Massarinaceae | Helminthosporium austriacum | L169 | KY984303 | KY984303 | - | KY984439 | [21] |
| Massarinaceae | Helminthosporium austriacum | L137 | KY984302 | KY984302 | - | KY984438 | [21] |
| Massarinaceae | Helminthosporium caespitosum | L99 * | JQ044429 | JQ044448 | KY984421 | KY984440 | [21] |
| Massarinaceae | Helminthosporium caespitosum | L141 | KY984305 | KY984305 | - | - | [21] |
| Massarinaceae | Helminthosporium caespitosum | L151 | KY984306 | KY984306 | - | - | [21] |
| Massarinaceae | Helminthosporium dalbergiae | H 4628 | LC014555 | AB807521 | AB797231 | AB808497 | [19] |
| Massarinaceae | Helminthosporium endiandrae | CBS 138902 * | KP004450 | KP004478 | - | - | [59] |
| Massarinaceae | Helminthosporium endiandrae | CBS 138,902 | - | MH878637 | - | - | [60] |
| Massarinaceae | Helminthosporium endiandrae | SM64 | MT279335 | - | - | - | Unpublished |
| Massarinaceae | Helminthosporium endiandrae | SM61 | MT279339 | - | - | - | Unpublished |
| Massarinaceae | Helminthosporium endiandrae | SM64 | MT279340 | - | - | - | Unpublished |
| Massarinaceae | Helminthosporium endiandrae | SM61 | MT279336 | - | - | - | Unpublished |
| Massarinaceae | Helminthosporium endiandrae | AKRM1 | MN880136 | - | - | - | Unpublished |
| Massarinaceae | Helminthosporium erythrinicola | CBS 145,569 | MK876391 | MK876432 | - | - | [22] |
| Massarinaceae | Helminthosporium genistae | L128 | KY984308 | KY984308 | KY984422 | - | [21] |
| Massarinaceae | Helminthosporium genistae | L129 | KY984309 | KY984309 | KY984423 | - | [21] |
| Massarinaceae | Helminthosporium genistae | L142 * | KY984310 | KY984310 | - | - | [21] |
| Massarinaceae | Helminthosporium hispanicum | L109 * | KY984318 | KY984318 | KY984424 | KY984441 | [21] |
| Massarinaceae | Helminthosporium italicum | MFLUCC 17-0241 | KY797638 | KY815015 | - | KY815021 | [61] |
| Massarinaceae | Helminthosporium juglandinum | L97 | KY984322 | KY984322 | KY984425 | KY984445 | [21] |
| Massarinaceae | Helminthosporium juglandinum | L118 * | KY984321 | KY984321 | - | KY984444 | [21] |
| Massarinaceae | Helminthosporium leucadendri | CBS 135133 * | KF251150 | KF251654 | - | KF253110 | [62] |
| Massarinaceae | Helminthosporium magnisporum | H 4627 * | AB811452 | AB807522 | AB797232 | AB808498 | [19] |
| Massarinaceae | Helminthosporium massarinum | KT 1564 * | AB809629 | AB807524 | AB797234 | AB808500 | [19] |
| Massarinaceae | Helminthosporium massarinum | KT 838 | AB809628 | AB807523 | AB797233 | AB808499 | [19] |
| Massarinaceae | Helminthosporium microsorum | L94 | KY984327 | KY984327 | KY984426 | KY984446 | [21] |
| Massarinaceae | Helminthosporium microsorum | L95 | KY984328 | KY984328 | - | KY984447 | [21] |
| Massarinaceae | Helminthosporium microsorum | L96 * | KY984329 | KY984329 | KY984427 | KY984448 | [21] |
| Massarinaceae | Helminthosporium oligosporum | L92 | KY984332 | KY984332 | KY984428 | KY984450 | [21] |
| Massarinaceae | Helminthosporium oligosporum | L93 * | KY984333 | KY984333 | - | KY984451 | [21] |
| Massarinaceae | Helminthosporium oligosporum | L106 | KY984330 | KY984330 | - | KY984449 | [21] |
| Massarinaceae | Helminthosporium quercinum | L90 * | KY984339 | KY984339 | KY984429 | KY984453 | [21] |
| Massarinaceae | Helminthosporium quercinum | L91 | KY984340 | KY984340 | - | KY984454 | [21] |
| Massarinaceae | Helminthosporium solani | CBS 365.75 | KY984341 | KY984341 | KY984430 | KY984455 | [21] |
| Massarinaceae | Helminthosporium solani | CBS 640.85 | KY984342 | KY984342 | - | - | [21] |
| Massarinaceae | Helminthosporiella stilbacea | CPHmZC-01 | KX228298 | KX228355 | - | - | [63] |
| Massarinaceae | Helminthosporiella stilbacea | COAD 2126 | MG668862 | - | - | - | [64] |
| Massarinaceae | Helminthosporiella stilbacea | MFLUCC 15-0813 * | MT928159 | MT928157 | MT928161 | MT928151 | This study |
| Massarinaceae | Helminthosporium submersum | MFLUCC 16-1360 * | - | MG098787 | MG098796 | MG098586 | [24] |
| Massarinaceae | Helminthosporium submersum | MFLUCC 16-1290 | MG098780 | MG098788 | MG098797 | MG098587 | [24] |
| Massarinaceae | Helminthosporium submersum | DLUCC 0805 | MG098781 | MG098789 | MG098798 | - | [24] |
| Massarinaceae | Helminthosporium syzygii | CBS 145,570 * | MK876392 | MK876433 | - | - | [22] |
| Massarinaceae | Helminthosporium tiliae | L88 * | KY984345 | KY984345 | KY984431 | KY984457 | [21] |
| Massarinaceae | Helminthosporium tiliae | L89 | KY984346 | KY984346 | - | - | [21] |
| Massarinaceae | Helminthosporium tiliae | L171 | KY984343 | KY984343 | - | KY984456 | [21] |
| Massarinaceae | Helminthosporium velutinum | yone 38 | - | AB807527 | AB797237 | AB808502 | [19] |
| Massarinaceae | Helminthosporium velutinum | yone 63 | - | AB807528 | AB797238 | AB808503 | [19] |
| Massarinaceae | Helminthosporium velutinum | MFLUCC 15-0423 | KU697300 | KU697304 | KU697308 | - | [20] |
| Massarinaceae | Helminthosporium velutinum | MFLUCC 15-0428 | KU697299 | KU697303 | KU697307 | - | [20] |
| Massarinaceae | Helminthosporium velutinum | H 4626 | LC014556 | AB807530 | AB797240 | AB808505 | [19] |
| Massarinaceae | Helminthosporium velutinum | L117 | KY984349 | KY984349 | - | KY984460 | [21] |
| Massarinaceae | Helminthosporium velutinum | L126 | KY984350 | KY984350 | - | KY984461 | [21] |
| Massarinaceae | Helminthosporium velutinum | L131 * | KY984352 | KY984352 | KY984432 | KY984463 | [21] |
| Massarinaceae | Helminthosporium velutinum | CPC 26297= CBS 141,504 | KX306757 | KX306785 | - | - | [65] |
| Massarinaceae | Helminthosporium velutinum | yone 96 | LC014558 | AB807529 | AB797239 | AB808504 | [19] |
| Massarinaceae | Helminthosporium velutinum | H 4739 | LC014557 | AB807525 | AB797235 | AB808501 | [19] |
| Massarinaceae | Helminthosporium velutinum | L115 | KY984347 | KY984347 | - | KY984458 | [21] |
| Massarinaceae | Helminthosporium velutinum | L116 | KY984348 | KY984348 | - | KY984459 | [21] |
| Massarinaceae | Helminthosporium velutinum | L127 | KY984351 | KY984351 | - | KY984462 | [21] |
| Massarinaceae | Helminthosporium velutinum | L98 | KY984359 | KY984359 | KY984433 | KY984466 | [21] |
| Massarinaceae | Helminthosporium velutinum | H 4743 | - | AB807526 | AB797236 | - | [19] |
| Massarinaceae | Helminthosporium velutinum | MFLUCC 16-1096 | MG098783 | MG098791 | MG098799 | MG098588 | [24] |
| Massarinaceae | Helminthosporium velutinum | MFLUCC 16-1282 | MG098784 | MG098792 | MG098800 | MG098589 | [24] |
| Massarinaceae | Helminthosporium velutinum | MFLUCC 17-1707 | MG098785 | MG098793 | MG098801 | MG098590 | [24] |
| Massarinaceae | Helminthosporium velutinum | MFLUCC 17-1321 | - | MG098794 | MG098802 | MG098591 | [24] |
| Massarinaceae | Helminthosporium velutinum | S-076 | KU697301 | KU697305 | KU697309 | - | [20] |
| Massarinaceae | Helminthosporium velutinum | MFLUCC 15-0243 | KU697301 | KU697305 | KU697309 | - | [20] |
| Massarinaceae | Helminthosporium velutinum | MFLUCC 16-1300 | MG098782 | MG098790 | - | - | [24] |
| Massarinaceae | Massarina albocarnis | CBS119345 | LC194503 | LC194379 | LC194337 | LC194416 | [66] |
| Massarinaceae | Massarina cisti | CBS 266.62 * | LC014568 | AB807539 | AB797249 | AB808514 | [19] |
| Massarinaceae | Massarina cisti | CBS 266.62 | - | FJ795447 | FJ795490 | - | [67] |
| Massarinaceae | Massarina eburnea | CBS 473.64 | AF383959 | GU301840 | AF164367 | - | [60,68] |
| Massarinaceae | Massarina eburnea | JCM 14422 | LC014569 | AB521735 | AB521718 | AB808517 | [19] |
| Massarinaceae | Massarina igniaria | CBS 845.96 | - | FJ795452 | FJ795494 | - | [67] |
| Massarinaceae | Massarina pandanicola | MFLUCC 17-0596 | MG646958 | MG646947 | MG646979 | MG646986 | [4] |
| Massarinaceae | Massarina phragmiticola | CBS 110,446 | - | DQ813510 | DQ813512 | - | [69] |
| Massarinaceae | Neottiosporina paspali | CBS 331.37 | - | EU754172 | EU754073 | - | [70] |
| Massarinaceae | Pseudodidymosphaeria spartii | CBS 183.58 | - | GU205225 | GU205250 | - | [71] |
| Massarinaceae | Pseudodidymosphaeria spartii | MFLUCC 13-0273 | KP325434 | KP325436 | KP325438 | - | [72] |
| Massarinaceae | Pseudodidymosphaeria spartii | MFLUCC 14-1212 | KP325435 | KP325437 | KP325439 | - | [72] |
| Massarinaceae | Pseudosplanchnonema phorcioides | MFLUCC 14-0618 | KP683372 | KP683373 | KP683374 | - | [72] |
| Massarinaceae | Pseudosplanchnonema phorcioides | MFLUCC 13-0533 | - | KM875454 | KM875455 | - | [73] |
| Massarinaceae | Pseudosplanchnonema phorcioides | L16 | KY984360 | - | KY984434 | KY984467 | [21] |
| Massarinaceae | Pseudosplanchnonema phorcioides | MFLUCC 13-0611 | KP683375 | KP683376 | KP683377 | - | [21] |
| Massarinaceae | Semifissispora natalis | CPC 25383 | KT950846 | KT950858 | - | KT950878 | [21] |
| Massarinaceae | Semifissispora natalis | CBS 140659 | - | MH878157 | - | - | [21] |
| Massarinaceae | Semifissispora rotundata | CPC 549 | KT950847 | KT950859 | - | - | [21] |
| Massarinaceae | Semifissispora tooloomensis | CBS143431 | MG38607 | MG386124 | - | - | [21] |
| Massarinaceae | Stagonospora perfecta | KT 1726A | AB809642 | AB807579 | AB797289 | AB808555 | [19] |
| Massarinaceae | Stagonospora cf. paludosa | CBS 130,005 | KF251254 | KF251757 | - | - | [62] |
| Massarinaceae | Stagonospora duoseptata | CBS 135,093 | KF251255 | KF251758 | - | - | [62] |
| Massarinaceae | Stagonospora imperaticola | MFLUCC 15-0026 | KY706143 | KY706133 | KY706138 | KY706146 | [74] |
| Massarinaceae | Stagonospora multiseptata | MFLUCC 15-0449 | KX965735 | KX954404 | - | - | [74] |
| Massarinaceae | Stagonospora paludosa | CBS 135088 * | KF251257 | KF251760 | - | KF253207 | [62] |
| Massarinaceae | Stagonospora perfecta | CBS 135,099 | KF251258 | KF251761 | - | - | [62] |
| Massarinaceae | Stagonospora perfecta | KT 1726A | AB809642 | AB807579 | AB797289 | AB808555 | [19] |
| Massarinaceae | Stagonospora pseudocaricis | CBS 135,132 | KF251259 | KF251763 | - | - | [62] |
| Massarinaceae | Stagonospora pseudopaludosa | CPC 22,654 | KF777188 | KF777239 | - | - | [62] |
| Massarinaceae | Stagonospora pseudoperfecta | KT 889 * | AB809641 | AB807577 | AB797287 | AB808553 | [19] |
| Massarinaceae | Stagonospora sp. | CBS 135,096 | KF251263 | KF251766 | - | - | [62] |
| Massarinaceae | Stagonospora tainanensis | KT 1866 | AB809643 | AB807580 | AB797290 | AB808556 | [19] |
| Massarinaceae | Stagonospora trichophoricola | CBS 136,764 | KJ869110 | KJ869168 | - | - | [75] |
| Massarinaceae | Stagonospora uniseptata | CPC 22,150 | KF251266 | KF251769 | - | - | [62] |
| Massarinaceae | Stagonospora uniseptata | CBS 135,090 | KF251264 | KF251767 | - | - | [62] |
| Massarinaceae | Suttonomyces clematidis | MFLUCC 14-0240 | - | KP842917 | KP842920 | - | [76] |
| Massarinaceae | Suttonomyces rosae | MFLUCC 15-0051 | MG828973 | MG829085 | MG829185 | - | [77] |
| Periconiaceae | Periconia byssoides | H 4600 | LC014581 | AB807570 | AB797280 | AB808546 | [19] |
| Periconiaceae | Periconia digitata | CBS 510.77 | LC014584 | AB807561 | AB797271 | AB808537 | [19] |
| Periconiaceae | Periconia macrospinosa | CBS 135,663 | KP183999 | KP184038 | KP184080 | - | [78] |
| Periconiaceae | Periconia pseudodigitata | KT 1395 * | LC014591 | AB807564 | AB797274 | AB808540 | [19] |
3. Results and Discussion
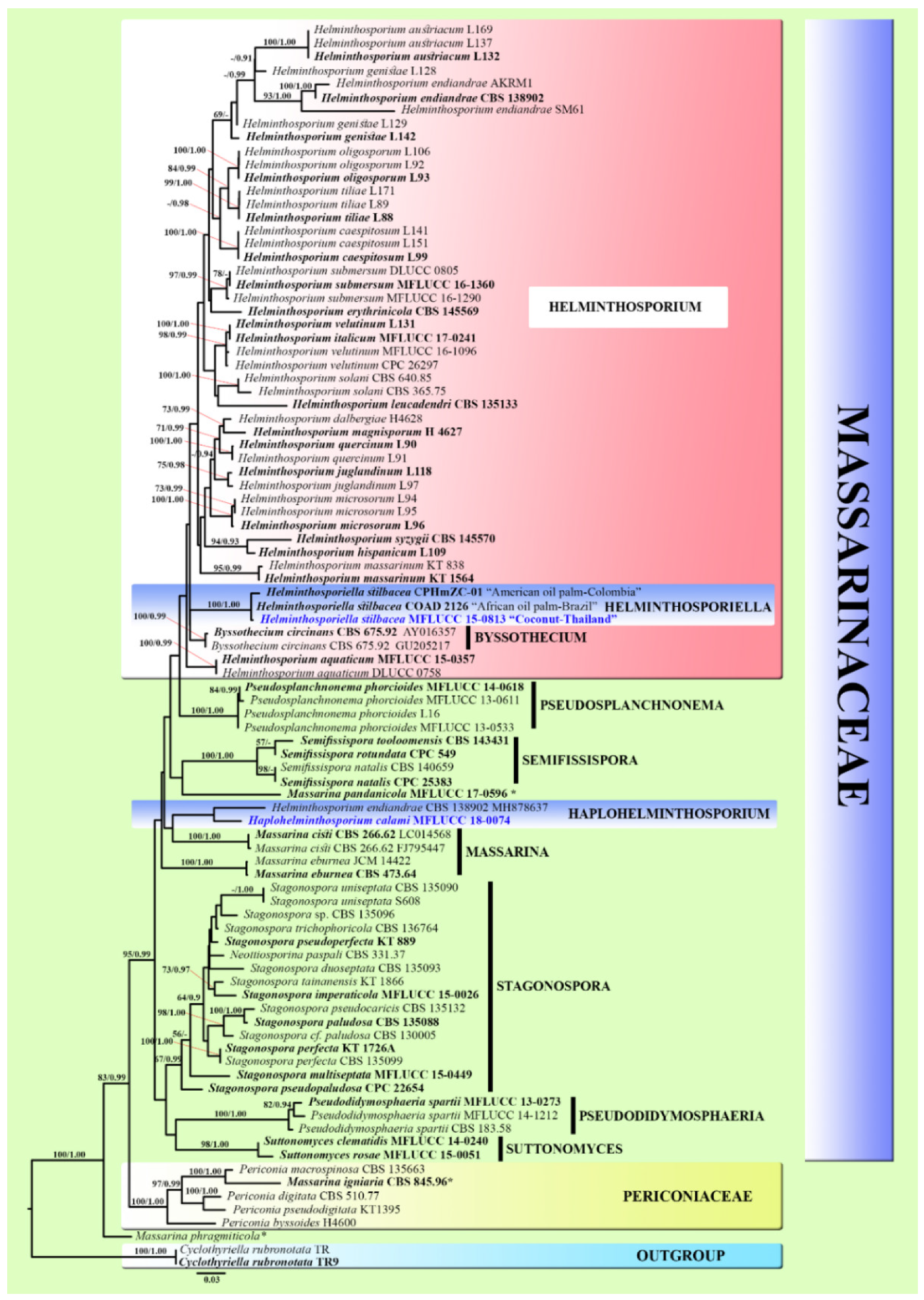
3.1. Phylogenetic Analyses
3.2. Taxonomy
3.2.1. Haplohelminthosporium Konta & K.D. Hyde, gen. nov
Haplohelminthosporium calami Konta & K.D. Hyde, sp. nov.
3.2.2. Helminthosporiella Konta & K.D. Hyde, gen. nov.
Helminthosporiella stilbacea Konta & K.D. Hyde, sp. nov.
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hyde, K.D.; Norphanphoun, C.; Chen, J.; Dissanayake, A.J.; Doilom, M.; Hongsanan, S.; Jayawardena, R.S.; Jeewon, R.; Perera, R.H.; Thongbai, B.; et al. Thailand’s amazing diversity: Up to 96% of fungi in northern Thailand may be novel. Fungal Divers. 2018, 93, 215–239. [Google Scholar] [CrossRef]
- Hyde, K.D.; de Silva, N.I.; Jeewon, R.; Bhat, D.J.; Phookamsak, R.; Doilom, M.; Boonmee, S.; Jayawardena, R.S.; Maharachchikumbura, S.S.N.; Senanayake, I.C.; et al. AJOM new records and collections of fungi: 1–100. AJOM 2020, 3, 22–294. [Google Scholar] [CrossRef]
- Dai, D.Q.; Phookamsak, R.; Wijayawardene, N.N.; Li, W.J.; Bhat, D.J.; Xu, J.C.; Taylor, J.E.; Hyde, K.D.; Chukeatirote, E. Bambusicolous fungi. Fungal Divers. 2017, 82, 1–105. [Google Scholar] [CrossRef]
- Tibpromma, S.; Hyde, K.D.; McKenzie, E.H.; Bhat, D.J.; Phillips, A.J.; Wanasinghe, D.N.; Samarakoon, M.C.; Jayawardena, R.S.; Dissanayake, A.J.; Tennakoon, D.S.; et al. Fungal diversity notes 840–928: Micro-fungi associated with Pandanaceae. Fungal Divers. 2018, 93, 1–160. [Google Scholar] [CrossRef]
- Pinruan, U.; Hyde, K.D.; Lumyong, S.; McKenzie, E.H.C.; Jones, E.G. Occurrence of fungi on tissues of the peat swamp palm Licuala longicalycata. Fungal Divers. 2007, 25, 157–173. [Google Scholar]
- Pinnoi, A.; Phongpaichit, S.; Hyde, K.D.; Jones, E.G. Biodiversity of fungi on Calamus (Palmae) in Thailand. Cryptogamie 2009, 30, 181–190. [Google Scholar]
- Konta, S.; Hongsanan, S.; Tibpromma, S.; Thongbai, B.; Maharachchikumbura, S.S.N.; Bahkali, A.H.; Hyde, K.D.; Boonmee, S. An advance in the endophyte story: Oxydothidaceae fam. nov. with six new species of Oxydothis. Mycosphere 2016, 7, 1425–1446. [Google Scholar] [CrossRef]
- Konta, S.; Hongsanan, S.; Eungwanichayapant, P.D.; Liu, J.K.; Jeewon, R.; Hyde, K.D.; Maharachchikumbura, S.S.N.; Boonmee, S. Leptosporella (Leptosporellaceae fam. nov.) and Linocarpon and Neolinocarpon (Linocarpaceae fam. nov.) are accommodated in Chaetosphaeriales. Mycosphere 2017, 8, 1943–1974. [Google Scholar] [CrossRef]
- Konta, S.; Hyde, K.D.; Eungwanichayapant, P.D.; Doilom, M.; Tennakoon, D.S.; Senwanna, C.; Boonmee, S. Fissuroma (Aigialaceae: Pleosporales) appears to be hyperdiverse on Arecaceae: Evidence from two new species from southern Thailand. Acta Bot. Bras. 2020, 34, 384–393. [Google Scholar] [CrossRef]
- Konta, S.; Maharachchikumbura, S.S.N.; Senanayake, I.C.; McKenzie, E.H.C.; Stadler, M.; Boonmee, S.; Phookamsak, R.; Jayawardena, R.S.; Senwanna, C.; Hyde, K.D.; et al. A new genus Allodiatrype, five new species and a new host record of diatrypaceous fungi from palms (Arecaceae). Mycosphere 2020, 11, 239–268. [Google Scholar] [CrossRef]
- Zhang, S.N.; Hyde, K.D.; Jones, E.B.G.; Cheewangkoon, R.; Liu, J.K. Additions to Fissuroma and Neoastrosphaeriella (Aigialaceae, Pleosporales) from palms. Mycosphere 2020, 11, 269–284. [Google Scholar] [CrossRef]
- Hongsanan, S.; Hyde, K.D.; Phookamsak, R.; Wanasinghe, D.N.; McKenzie, E.H.C.; Sarma, V.V.; Lücking, R.; Boonmee, S.; Bhat, J.D.; Liu, N.G.; et al. Refined families of Dothideomycetes: Orders and families incertae sedis in Dothideomycetes. Fungal Divers. 2020, 105, 17–318. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Al-Ani, L.K.T.; Tedersoo, L.; Haelewaters, D.; Rajeshkumar, K.C.; Zhao, R.L.; Aptroot, A.; Leontyev, D.V.; Saxena, R.K.; et al. Outline of Fungi and fungus-like taxa. Mycosphere 2020, 11, 1060–1456. [Google Scholar] [CrossRef]
- Munk, A. On Metasphaeria coccodes (Karst.) Sacc. (Massarinaceae n. fam.). Friesia 1956, 5, 303–308. [Google Scholar]
- Saccardo, P.A. Sylloge Fungorum; Johnson Reprint Corporation: New York, NY, USA, 1883; Volume 2. [Google Scholar]
- Link, H.F. Observationes in ordines plantarum naturales. Dissertatio I. Mag. der Ges. Nat. Freunde Berl. 1809, 3, 3–42. [Google Scholar]
- Gilman, J.C.; Abbott, E.V. A summary of the soil fungi. Iowa State Coll. J. Sci. 1927, 1, 225–343. [Google Scholar]
- Deshpande, K.S.; Deshpande, K.B. Contribution to the taxonomy of the genus Helminthosporium II. Sydowia 1969, 23, 69–76. [Google Scholar]
- Tanaka, K.; Hirayama, K.; Yonezawa, H.; Sato, G.; Toriyabe, A.; Kudo, H.; Hashimoto, A.; Matsumura, M.; Harada, Y.; Kurihara, Y.; et al. Revision of the Massarineae (Pleosporales, Dothideomycetes). Stud. Mycol. 2015, 82, 75–136. [Google Scholar] [CrossRef] [Green Version]
- Zhu, D.; Luo, Z.L.; Baht, D.J.; Mckenzie, E.H.; Bahkali, A.H.; Zhou, D.Q.; Su, H.Y.; Hyde, K.D. Helminthosporium velutinum and H. aquaticum sp. nov. from aquatic habitats in Yunnan Province, China. Phytotaxa 2016, 253, 179–190. [Google Scholar] [CrossRef] [Green Version]
- Voglmayr, H.; Jaklitsch, W.M. Corynespora, Exosporium and Helminthosporium revisited new species and generic reclassification. Stud. Mycol. 2017, 87, 43–76. [Google Scholar] [CrossRef]
- Crous, P.W.; Carnegie, A.J.; Wingfield, M.J.; Sharma, R.; Mughini, G.; Noordeloos, M.E.; Santini, A.; Shouche, Y.S.; Bezerra, J.D.P.; Dima, B.; et al. Fungal Planet description sheets: 868–950. Pers. Mol. Phylogeny Evol. Fungi 2019, 42, 291–473. [Google Scholar] [CrossRef] [PubMed]
- Rashmi, M.; Kushveer, J.S.; Sarma, V.V. A worldwide list of endophytic fungi with notes on ecology and diversity. Mycosphere 2019, 10, 798–1079. [Google Scholar] [CrossRef]
- Zhao, N.; Luo, Z.L.; Hyde, K.D.; Su, H.Y.; Bhat, D.J.; Liu, J.K.; Bao, D.F.; Hao, Y.E. Helminthosporium submersum sp. nov. (Massarinaceae) from submerged wood in north-western Yunnan Province, China. Phytotaxa 2018, 348, 269–278. [Google Scholar] [CrossRef]
- Drechsler, C. Some graminicolous species of Helminthosporium. J. Agric. Res. 1923, 24, 641–739. [Google Scholar]
- Drechsler, C. Phytopathological and taxonomical aspects of Ophilobolus, Pyrenophora, Helminthosporium and a new genus Cochliobolus. Phytopathology 1934, 24, 953–985. [Google Scholar]
- Shoemaker, R.A. Nomenclature of Drechslera and Bipolaris, grass parasites segregated from ‘Helminthosporium’. Can. J. Bot. 1959, 37, 879–887. [Google Scholar] [CrossRef]
- Misra, A.P.; Prakash, O. Helminthosporium species occurring on graminaceous hosts in India. Indian J. Mycol. Plant Pathol. 1972, 2, 95–97. [Google Scholar]
- Misra, A.P. Helminthosporium Species Occurring in Cereals and Other Gramineae; U.S.P.L. 480 Project No.A7-CR 133, Grant No. FG-IN-223Tirhut college of Agriculture, Dholi, Muzaffarpur, Bihar, India; Catholic Press: Ranchi, India, 1973; p. 289. [Google Scholar]
- Shenoy, B.D.; Jeewon, R.; Wu, W.P.; Bhat, D.J.; Hyde, K.D. Ribosomal and RPB2 DNA sequence analyses suggest that Sporidesmium and morphologically similar genera are polyphyletic. Mycol. Res. 2006, 110, 916–928. [Google Scholar] [CrossRef]
- Bärlocher, F. Molecular approaches promise a deeper and broader understanding of the evolutionary ecology of aquatic hyphomycetes. J. N. Am. Benthol. Soc. 2010, 29, 1027–1041. [Google Scholar] [CrossRef]
- Manamgoda, D.S.; Cai, L.; McKenzie, E.H.; Crous, P.W.; Madrid, H.; Chukeatirote, E.; Shivas, R.G.; Tan, Y.P.; Hyde, K.D. A phylogenetic and taxonomic re-evaluation of the Bipolaris-Cochliobolus-Curvularia complex. Fungal Divers. 2012, 56, 131–144. [Google Scholar] [CrossRef]
- Manamgoda, D.S.; Rossman, A.Y.; Castlebury, L.A.; Crous, P.W.; Madrid, H.; Chukeatirote, E.; Hyde, K.D. The genus Bipolaris. Stud. Mycol. 2014, 79, 221–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Crous, P.W.; Schoch, C.L.; Hyde, K.D. Pleosporales. Fungal Divers. 2012, 53, 1–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baschien, C.; Tsui, C.K.M.; Gulis, V.; Szewzyk, U.; Marvanová, L. The molecular phylogeny of aquatic hyphomycetes with affinity to the Leotiomycetes. Fungal Biol. 2013, 117, 660–672. [Google Scholar] [CrossRef] [PubMed]
- Woudenberg, J.H.; Groenewald, J.Z.; Binder, M.; Crous, P.W. Alternaria redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef] [Green Version]
- Ariyawansa, H.A.; Thambugala, K.M.; Manamgoda, D.S.; Jayawardena, R.; Camporesi, E.; Boonmee, S.; Wanasinghe, D.N.; Phookamsak, R.; Hongsanan, S.; Singtripop, C.; et al. Towards a natural classification and backbone tree for Pleosporaceae. Fungal Divers. 2015, 71, 85–139. [Google Scholar] [CrossRef]
- Senanayake, I.C.; Rathnayaka, A.R.; Marasinghe, D.S.; Calabon, M.S.; Gentekaki, E.; Lee, H.B.; Hurdeal, V.G.; Pem, D.; Dissanayake, L.S.; Wijesinghe, S.N.; et al. Morphological approaches in studying fungi: Collection, examination, isolation, sporulation and preservation. Mycosphere 2020, 11, 2678–2754. [Google Scholar] [CrossRef]
- Jayasiri, S.C.; Hyde, K.D.; Ariyawansa, H.A.; Bhat, J.; Buyck, B.; Cai, L.; Dai, Y.C.; Abd-Elsalam, K.A.; Ertz, D.; Hidayat, I.; et al. The Faces of Fungi database: Fungal names linked with morphology, phylogeny and human impacts. Fungal Divers. 2015, 74, 3–18. [Google Scholar] [CrossRef]
- Index Fungorum. 2021. Available online: http://www.indexfungorum.org/names/Names.asp (accessed on 30 April 2020).
- Dissanayake, A.J.; Bhunjun, C.S.; Maharachchikumbura, S.S.N.; Liu, J.K. Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 2020, 11, 2652–2676. [Google Scholar] [CrossRef]
- Vilgalys, R.; Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 1990, 172, 4238–4246. [Google Scholar] [CrossRef] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J.W. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; PCR Protocols: A Guide to Methods and Applications; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Rehner, S. Primers for Elongation Factor 1-α (EF1-α). 2001. Available online: http://ocid.NACSE.ORG/research/deephyphae/EF1primer.pdf (accessed on 1 November 2019).
- Rehner, S.A.; Buckley, E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: Evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 2005, 97, 84–98. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glez-Peña, D.; Gómez-Blanco, D.; Reboiro-Jato, M.; Fdez-Riverola, F.; Posada, D. ALTER: Program-oriented conversion of DNA and protein alignments. Nucleic Acids Res. 2010, 38, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. The CIPRES science gateway: A community resource for phylogenetic analyses. In Proceedings of the 2011 TeraGrid Conference: Extreme Digital Discovery, Association for Computing Machinery, New York, NY, USA, 18–21 July 2011; pp. 1–8. [Google Scholar] [CrossRef]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [Green Version]
- Nylander, J.A.A. MrModeltest 2.0.; Program distributed by the author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Rambaut, A. FigTree version 1.4.0. 2012. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 1 November 2019).
- Schoch, C.L.; Crous, P.W.; Groenewald, J.Z.; Boehm, E.W.A.; Burgess, T.I.; De Gruyter, J.; De Hoog, G.S.; Dixon, L.J.; Grube, M.; Gueidan, C.; et al. A class-wide phylogenetic assessment of Dothideomycetes. Stud. Mycol. 2009, 64, 1–15. [Google Scholar] [CrossRef]
- Déon, M.; Fumanal, B.; Gimenez, S.; Bieysse, D.; Oliveira, R.R.; Shuib, S.S.; Breton, F.; Elumalai, S.; Vida, J.B.; Seguin, M.; et al. Diversity of the cassiicolin gene in Corynespora cassiicola and relation with the pathogenicity in Hevea brasiliensis. Fungal Biol. 2014, 118, 32–47. [Google Scholar] [CrossRef]
- Jaklitsch, W.M.; Voglmayr, H. Hidden diversity in Thyridaria and a new circumscription of the Thyridariaceae. Stud. Mycol. 2016, 85, 35–64. [Google Scholar] [CrossRef] [Green Version]
- Lumbsch, H.T.; Lindemuth, R. Major lineages of Dothideomycetes (Ascomycota) inferred from SSU and LSU rDNA sequences. Mycol. Res. 2001, 105, 901–908. [Google Scholar] [CrossRef]
- Schoch, C.L.; Shoemaker, R.A.; Seifert, K.A.; Hambleton, S.; Spatafora, J.W.; Crous, P.W. A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 2006, 98, 1041–1052. [Google Scholar] [CrossRef]
- Crous, P.W.; Wingfield, M.J.; Schumacher, R.K.; Summerell, B.A.; Giraldo, A.; Gené, J.; Guarro, J.; Wanasinghe, D.N.; Hyde, K.D.; Camporesi, E.; et al. Fungal Planet description sheets: 281–319. Pers. Mol. Phylogeny Evol. Fungi. 2014, 33, 212–319. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.; Groenewald, M.; De Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef]
- Tian, Q.; Li, W.J.; Hyde, K.D.; Camporesi, E.; Bhat, D.J.; Chomnunti, P.; Xu, J.C. Molecular taxonomy of five species of microfungi on Alnus spp. from Italy. Mycol. Prog. 2018, 17, 255–274. [Google Scholar] [CrossRef]
- Quaedvlieg, W.; Verkley, G.J.M.; Shin, H.D.; Barreto, R.W.; Alfenas, A.C.; Swart, W.J.; Groenewald, J.Z.; Crous, P.W. Sizing up septoria. Stud. Mycol. 2013, 75, 307–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crous, P.W.; Wingfield, M.J.; Richardson, D.M.; Le Roux, J.J.; Strasberg, D.; Edwards, J.; Roets, F.; Hubka, V.; Taylor, P.W.J.; Heykoop, M.; et al. Fungal Planet description sheets: 400–468. Pers. Mol. Phylogeny Evol. Fungi. 2016, 36, 316–458. [Google Scholar] [CrossRef] [Green Version]
- Rosado, A.W.C.; de Jesus Boari, A.; Custódio, F.A.; Quadros, A.F.F.; Batista, I.C.A.; Pereira, O.L. Helminthosporiella stilbacea associated with African oil palm (Elaeis guineensis) in Brazil. For. Pathol. 2019, 49, e12529. [Google Scholar] [CrossRef]
- Hernández-Restrepo, M.; Schumacher, R.K.; Wingfield, M.J.; Ahmad, I.; Cai, L.; Duong, T.A.; Edwards, J.; Gené, J.; Groenewald, J.Z.; Jabeen, S.; et al. Fungal systematics and evolution: FUSE 2. Sydowia 2016, 68, 193–230. [Google Scholar] [CrossRef]
- Hashimoto, A.; Matsumura, M.; Hirayama, K.; Tanaka, K. Revision of Lophiotremataceae (Pleosporales, Dothideomycetes): Aquasubmersaceae, Cryptocoryneaceae, and Hermatomycetaceae fam. nov. Pers. Mol. Phylogeny Evol. Fungi 2017, 39, 51–73. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, H.K.; Fournier, J.; Crous, P.W.; Jeewon, R.; Pointing, S.B.; Hyde, K.D. Towards a phylogenetic clarification of Lophiostoma/Massarina and morphologically similar genera in the Pleosporales. Fungal Divers. 2009, 38, 225–251. [Google Scholar]
- Liew, E.C.; Aptroot, A.; Hyde, K.D. An evaluation of the monophyly of Massarina based on ribosomal DNA sequences. Mycologia 2002, 94, 803–813. [Google Scholar] [CrossRef]
- Kodsueb, R.; Lumyong, S.; Ho, W.H.; Hyde, K.D.; Mckenzie, E.H.; Jeewon, R. Morphological and molecular characterization of Aquaticheirospora and phylogenetics of Massarinaceae (Pleosporales). Bot. J. Linn. Soc. 2007, 155, 283–296. [Google Scholar] [CrossRef] [Green Version]
- De Gruyter, J.; Aveskamp, M.M.; Woudenberg, J.H.; Verkley, G.J.; Groenewald, J.Z.; Crous, P.W. Molecular phylogeny of Phoma and allied anamorph genera: Towards a reclassification of the Phoma complex. Mycol. Res. 2009, 113, 508–519. [Google Scholar] [CrossRef]
- Hu, F.J.; Jeewon, R.; Hyde, K.D. Relationships among Astrosphaeriella, Caryospora and Trematosphaeria. Ph.D. Thesis, The University of Hong Kong, Hong Kong, China, 2009. [Google Scholar]
- Thambugala, K.M.; Hyde, K.D.; Tanaka, K.; Tian, Q.; Wanasinghe, D.N.; Ariyawansa, H.A.; Jayasiri, S.C.; Boonmee, S.; Camporesi, E.; Hashimoto, A.; et al. Towards a natural classification and backbone tree for Lophiostomataceae, Floricolaceae, and Amorosiaceae fam. nov. Fungal Divers. 2015, 74, 199–266. [Google Scholar] [CrossRef]
- Chethana, T.; Liu, M.; Ariyawansa, H.A.; Konta, S.; Wanasinghe, D.N.; Zhou, Y.; Yan, J.; Camporesi, E.; Bulgakov, T.M.; Chukeatirote, E.; et al. Splanchnonema-like species in Pleosporales: Introducing Pseudosplanchnonema gen. nov. in Massarinaceae. Phytotaxa 2015, 231, 133–144. [Google Scholar] [CrossRef] [Green Version]
- Thambugala, K.M.; Wanasinghe, D.N.; Phillips, A.J.L.; Camporesi, E.; Bulgakov, T.S.; Phukhamsakda, C.; Dissanayake, A.; Tennakoon, D.S.; Tibpromma, S.; Chen, Y.Y.; et al. Mycosphere notes 1-50: Grass (Poaceae) inhabiting Dothideomycetes. Mycosphere 2017, 8, 697–796. [Google Scholar] [CrossRef]
- Crous, P.W.; Shivas, R.G.; Quaedvlieg, W.V.; van der Bank, M.; Zhang, Y.; Summerell, B.A.; Guarro, J.; Wingfield, M.J.; Wood, A.R.; Alfenas, A.C. Fungal Planet description sheets: 214–280. Pers. Mol. Phylogeny Evol. Fungi 2014, 32, 184–306. [Google Scholar] [CrossRef]
- Wijayawardene, N.N.; Hyde, K.D.; Bhat, D.J.; Goonasekara, I.D.; Nadeeshan, D.; Camporesi, E.; Schumacher, R.K.; Wang, Y. Additions to brown spored coelomycetous taxa in Massarinae, Pleosporales: Introducing Phragmocamarosporium gen. nov. and Suttonomyces gen. nov. Cryptogam. Mycol. 2015, 36, 213–224. [Google Scholar] [CrossRef]
- Wanasinghe, D.N.; Phukhamsakda, C.; Hyde, K.D.; Jeewon, R.; Lee, H.B.; Jones, E.G.; Tibpromma, S.; Tennakoon, D.S.; Dissanayake, A.J.; Jayasiri, S.C.; et al. Fungal diversity notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018, 89, 1–236. [Google Scholar] [CrossRef]
- Knapp, D.G.; Kovács, G.M.; Zajta, E.; Groenewald, J.Z.; Crous, P.W. Dark septate endophytic pleosporalean genera from semiarid areas. Pers. Mol. Phylogeny Evol. Fungi 2015, 35, 87–100. [Google Scholar] [CrossRef] [Green Version]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal identification using molecular tools: A primer for the natural products research community. J. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef]
- Species Fungorum. 2021. Available online: http://www.speciesfungorum.org/Names/Names.asp (accessed on 30 April 2021).
- Goh, T.K.; Hyde, K.D.; Lee, D.K. Generic distinction in the Helminthosporium-complex based on restriction analysis of the nuclear ribosomal RNA gene. Fungal Divers. 1998, 1, 85–107. [Google Scholar]
- Mukerji, K.G. Current Concepts in Botany; IK International Publishing House Pvt Ltd.: New Delhi, India, 2006; pp. 49–78. [Google Scholar]
- Hernández-Restrepo, M.; Madrid, H.; Tan, Y.P.; da Cunha, K.C.; Gene, J.; Guarro, J.; Crous, P.W. Multi-locus phylogeny and taxonomy of Exserohilum. Pers. Mol. Phylogeny Evol. Fungi 2018, 41, 71–108. [Google Scholar] [CrossRef] [PubMed]
- Cooke, W.B.; Shaw, C.G. Western fungi. III. Mycologia 1952, 44, 795–812. [Google Scholar] [CrossRef]
- Ellis, M.B. Dematiaceous hyphomycetes. III. Mycol. Pap. 1961, 82, 1–55. [Google Scholar]
- Ciferri, R. Observations on meliolicolous Hyphales from Santo Domingo. Sydowia 1955, 9, 296–335. [Google Scholar]
- Sydow, H.; Sydow, P. Novae fungorum species. Ann. Mycol. 1904, 2, 162–174. [Google Scholar]
- Jørstad, I. Parasitic fungi from the Canaries chiefly collected by J. Lid, with a note on Schizophyllum commune. Blyttia 1966, 24, 222–231. [Google Scholar]
- Ciferri, R. Mycoflora Domingensis Integrata. Quaderno del Laboratorio Crittogamico del Istituto Botanicodell’ Università di Pavia 1961, 19, 1–539. [Google Scholar]
- Petch, T. New Ceylon fungi. Ann. R. Bot. Gard. Perad. 1909, 4, 299–307. [Google Scholar]
- Thirumalachar, M.J. Some new or interesting fungi II. Sydowia 1950, 4, 66–73. [Google Scholar]
- Petrak, F.; Ciferri, R. Fungi Dominicani. II. Ann. Mycol. 1932, 30, 149–353. [Google Scholar]
- Petrak, F. Beiträgezur Pilzflora von Britisch Nord-Borneo. Sydowia 1954, 8, 12–26. [Google Scholar]
- Lavrov, N.P. Trud. tomsk. gos. Univ. Kuibysheva 1951, 110, 254. [Google Scholar]
- Jaczewski, A.L.A. Type species—Jaczewskiella altajensis. Mater. Mikol. Fitopat. Ross. 1915, 1, 41. [Google Scholar]
- Corda, A.C.J. Gliostroma. Icon. Fung. 1837, 1, 13. [Google Scholar]
- Unamuno, L.M. NotasMicológicas. II. Adiciones a los Hifales de la flora española. Boletín de la Real Soc. Española de Hist. Nat. 1932, 32, 161–169. [Google Scholar]
- Saccardo, P.A. Sylloge Fungorum. Mem. Reale Ist. Veneto Sci. 1902, 13, (reprint). [Google Scholar]
- Cooke, M.C. Ravenel’s American fungi. Grevillea 1878, 6, 129–146. [Google Scholar]
- Zaprometov, N.G. Fungal flora of the Kyrgyz SSR. Frunze Acad. Sci. Kyrg. SSR 1957, 1, 98. [Google Scholar]
- Deshpande, K.S.; Deshpande, K.B. Contribution to the taxonomy of genus Helminthosporium. I. Sydowia 1966, 20, 39–45. [Google Scholar]
- Diedicke, H. Aufzählungen der in der Umgebung Erfurts Beobachteten Micromyceten; Academische Loge Sincera Concordia: Erfurt, Germany, 1910; Volume 36, p. 221. [Google Scholar]
- Hennings, P. Fungi paraënses III. Hedwigia 1908, 48, 101–117. [Google Scholar]
- Sydow, H.; Sydow, P. Weitere Diagnosen neuer philippinischer Pilze. Ann. Mycol. 1916, 14, 353–375. [Google Scholar]
- Zhang, M.; Wu, H.Y.; Wang, Z.Y. Taxonomic studies of Helminthosporium from China 5. Two new species from Hunan and Sichuan Province. Mycotaxon 2010, 113, 95–99. [Google Scholar] [CrossRef]
- Dounine, M.S.; Yakimovitch, E.D. Sweet Potato Diseases and Their Control; Pan-Soviet Science Research Institute Cultural Soybean and Spice Crops: Moscow, Russia, 1934; p. 247. [Google Scholar]
- Subramanian, C.V.; Bhat, D.J. Hyphomycetes from South India I. Some new taxa. Kavaka 1987, 15, 41–74. [Google Scholar]
- Misra, A.P. Helminthosporium Species Occurring on Cereals and Other Gramineae; Tirhut College of Agriculture: Dholi, India, 1976; pp. 1–289. [Google Scholar]
- Matsushima, T. Matsushima Mycological Memoirs 7; Matsushima Fungus Collection: Kobe, Japan, 1993; Volume 7, pp. 1–141. [Google Scholar]
- Pidoplichko, N.M. New fungus species on coarse fodders. Mikrobiol. Zh. 1950, 12, 38. [Google Scholar]
- Viégas, A.P. Algunsfungos do Brasil: XIII—Hifomicetos. Bragantia 1946, 6, 353–442. [Google Scholar] [CrossRef] [Green Version]
- Ciferri, R. Mycoflora domingensis exsiccata (Cent. I, no. 1–100). Ann. Mycol. 1931, 29, 283–299. [Google Scholar]
- Bongini, V. Sur una malattia delle Cactacee. Difesa delle Piante. 1932, 9, 38. [Google Scholar]
- Steyaert, R.L. Contribution à l’étude des parasites des végétaux du Congo. Bull. Soc. R. Bot. Belg. 1948, 80, 11–58. [Google Scholar]
- Hennings, P.; Fungi, S. Paulensis IV a cl. Puttemanscollecti. Hedwigia 1909, 48, 1–20. [Google Scholar]
- Saccardo, P.A. Notae mycologicae. Ser. XXVII. Fungi sinenses aliquot a cl. Prof. Otto A. Reinking collecti et communicati. Philipp J. Sci. 1921, 18, 595–605. [Google Scholar]
- Ciferri, R. Mycoflora domingensis exsiccata. Ann. Mycol. 1938, 36, 198–245. [Google Scholar]
- McColloch; Pollack. Phytopathology 1946, 36, 991.
- Orillo, F.T. An undescribed species of Helminthosporium on kapok in the Philippines. Philipp. Agric. 1955, 38, 548–550. [Google Scholar]
- Olivier, C.; Berbee, M.L.; Shoemaker, R.A.; Loria, R. Molecular phylogenetic support from ribosomal DNA sequences for origin of Helminthosporium from Leptosphaeria-like loculoascomycete ancestors. Mycologia 2000, 92, 736–746. [Google Scholar] [CrossRef]
- Hennings, P. Fungi. Ann. Musée Congo Belge Bot. 1907, 2, 85–106. [Google Scholar]
- Petrak, F. Beiträgezur Pilzflora von Ekuador. Sydowia 1950, 4, 450–587. [Google Scholar]
- Stevens, F.L. Hawaiian fungi. Bull. Bernice Bishop Mus. 1925, 19, 1–189. [Google Scholar]
- Hughes, S.J. Revision es hyphomycetum aliquot cum appendice de nominibusrejiciendis. Can. J. Bot. 1958, 36, 727–836. [Google Scholar] [CrossRef]
- Sawada, K. Descriptive Catalogue of the Formosan Fungi V.; Report of the Department of Agriculture Government Research Institute of Formosa; Department of Agriculture Government Research Institute of Formosa: Formosa, Japan, 1931; Volume 51, pp. 1–131. [Google Scholar]
- Ciferri, R.; Fragoso, G. Bulletin of the Royal Spanish Society of Natural History; National Museum of Natural Sciences: Madrid, Spain, 1926; Volume 26, p. 340. [Google Scholar]
- Massee, G.E. Fungi exotici, III. Bull. Misc. Inf. R. Bot. Gard. Kew. 1901, 150–169. [Google Scholar]
- Zhang, M.; Zhang, T.Y. Taxonomic studies of Helminthosporium from China 4. Six new species and a key to Helminthosporium from China. Mycotaxon 2009, 109, 399–413. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, T.; Wu, W. Taxonomic studies of Helminthosporium from China III. Three new species in Guangdong Province. Mycotaxon 2007, 99, 137–142. [Google Scholar]
- Malençon, G.; Bertault, R. Champignons de la Péninsule Ibérique, IV. Les Iles Baleares. Acta Phytotax. Barcinon. 1972, 11, 1–64. [Google Scholar]
- Sawada, K. Descriptive catalogue of Taiwan (Formosan) fungi. Part XI. Spec. Publ. Coll. Agric. Natl. Taiwan Univ. 1959, 8, 1–268. [Google Scholar]
- Chowdhury, S. Notes on fungi from Assam. II. Lloydia 1957, 20, 133–138. [Google Scholar]
- Petrak, F. Petrak’s Lists 5; Commonwealth Agricultural Bureaux: Wallingford, UK, 1930; p. 214. [Google Scholar]
- Nisikado, Y. Ber. Ohara Inst. Landw. Forsch. Kurashiki. 1925, 2, 597–612.
- Matsushima, T. Matsushima Mycological Memoirs 5; Matsushima Fungus Collection: Kobe, Japan, 1987; Volume 5, pp. 1–100. [Google Scholar]
- Garbowski, L. Bull. Acad. Polon. Sci. Lett. Cracoviae Cl. Math. Nat. Ser. B. 1923 1924, 15.
- Saccardo, P.A. Notae mycologicae series XXIII. Fungi Philippinenses. Attidella Accad. Sci. Veneto-Trentino-Istriana 1917, 10, 57–94. [Google Scholar]
- Sturm, J.W. Deutschl. Fl. 3 Abt. (Pilze Deutschl.) 1831, 3, 21.
- Baccarini, P. Funghidell’Eritrea. Ann. Bot. 1906, 4, 269–277. [Google Scholar]
- Nishihara, N. Ann. Phytopath. Soc. Japan 1969, 35, 89.
- Sydow, H.; Sydow, P. Fungi novibrasilienses a cl. Ulelecti. Bull. del´Herb. Boissier 1901, 1, 77–85. [Google Scholar]
- Von Thümen, F. Die Pilze des Weinstockes; W. Braumüller: Wien, Austria, 1878; pp. 1–225. [Google Scholar]
- Berkeley, M.J. Notices of British fungi [208–256]. Ann. Mag. Nat. Hist. 1841, 6, 430–439. [Google Scholar] [CrossRef]
- Golovin, P.N. Novye vidy gribov Srednej Azii. Trudy Sredneaz. Univ. 1950, 14, 1–47. [Google Scholar]
- Berkeley, M.J.; Broome, C.E. Notices of British fungi (901–951). Ann. Mag. Nat. Hist. 1861, 7, 373–382. [Google Scholar]
- Roumeguère, C.; Saccardo, P.A. Fungi Algerienses Trabutiani—Sertulum II. Rev. Mycol. Toulouse 1881, 3, 26–32. [Google Scholar]
- Katsuki, S. Notes on parasitic fungi of Yaku island. J. Jap. Bot. 1953, 28, 279–288. [Google Scholar]
- Magnus, P. Ein neues Helminthosporium. Hedwigia 1903, 42, 222–225. [Google Scholar]
- Holubová-Jechová, V. Studies on hyphomycetes from Cuba VI. New and rare species with tretic and phialidic conidiogenous cells. Ceská Mykol. 1987, 41, 107–114. [Google Scholar]
- Zhang, M.; Zhang, T.Y. Flora Fungorum Sinicorum; Science Press: Beijing, China, 2009; Volume 30, pp. 1–272. [Google Scholar]
- Koorders, S.H. Botanische Untersuchungen. Verh. K. Ned. Akad. van Wet. Afd. Nat. 1907, 13, 1–264. [Google Scholar]
- Berkeley, M.J.; Broome, C.E. Notices of British fungi (1335–1401). Ann. Mag. Nat. Hist. 1873, 11, 339–349. [Google Scholar] [CrossRef]
- Ciferri, R. Notae mycologicae et phytopathologicae Serie II, Nr. 1-14. Riv. Patol. Veg. 1927, 17, 35–40. [Google Scholar]
- Rostrup, E. Flora of Koh Chang. Contributions to the knowledge of the vegetation of the gulf of Siam. Fungi. Bot. Tidsskr. 1902, 24, 355–363. [Google Scholar]
- Hennings, P. Fungi Amazonici IV. a cl. Ernesto Ulecollecti. Hedwigia 1905, 44, 57–71. [Google Scholar]
- Yates, H.S. Some recently collected Philippine fungi, II. Philipp. J. Sci. C. Botany 1918, 13, 361–384. [Google Scholar]
- Trotter, A. Supplementum Universale, Pars X. Myxomycetae, Myxobacteriaceae, Deuteromycetae, Mycelia sterilia. Sylloge Fungorum. 1931, 25, 1–1093. [Google Scholar]
- Curzi, M. Helminthosporium gibberosporum. C. R. Accad. Lincei. 1931, 6, 146. [Google Scholar]
- Stevens, F.L. Some meliolicolous parasites from Porto Rico. Bot. Gaz. Crawfordsville 1918, 65, 227–249. [Google Scholar] [CrossRef]
- Tucker, C.M. J. Agric. Res. 1926, 32, 391.
- Wildeman, E.de. Etude de Systématique et de Géographie Botanique sur la Flore du Bas- et du Moyen-Congo. Ann. Musée Congo Belge Bot.Sér. 5 1907, 2, 85–106. [Google Scholar]
- Stevens, F.L.; Dowell, R.I. A Meliola disease of cacao. Phytopathology 1923, 13, 247–250. [Google Scholar]
- Viégas, A.P. Algunsmicetos Brasileiros. Bragantia 1947, 7, 25–48. [Google Scholar] [CrossRef]
- Errampalli, D.; Saunders, J.M.; Holley, J.D. Emergence of silver scurf (Helminthosporium solani) as an economically important disease of potato. Plant Pathol. 2001, 50, 141–153. [Google Scholar] [CrossRef]
- Zhao, G.C.; Zhao, R.L. The Higher Microfungi from Forests of Yunnan Province; Yunnan Science and Technology Press: Kunming, China, 2012; p. 564. [Google Scholar]
- Siboe, G.M.; Kirk, P.M.; Cannon, P.F. New dematiaceous hyphomycetes from Kenyan rare plants. Mycotaxon 1999, 73, 283–302. [Google Scholar]
- Gornostai, V.I. Mikol. Fitopatol. 1972, 6, 154.
- Cheremisinov. Chaetomium subaffine Sergeeva. Notulae Syst. Sect. Crypt. Inst. Bot. Acad. Sci. U.S.S.R. 1951, 7, 158. [Google Scholar]
- Hansford, C.G. Contribution towards the fungus flora of Uganda. V. Fungi Imperfecti. Proc. Linn. Soc. Lond. 1943, 155, 34–67. [Google Scholar] [CrossRef]
- Chowdhury, S. Notes on fungi of Assam. Lloydia 1955, 18, 82–87. [Google Scholar]
- Crous, P.W.; Wingfield, M.J.; Burgess, T.I.; Hardy, G.S.J.; Gené, J.; Guarro, J.; Baseia, I.G.; García, D.; Gusmão, L.F.P.; Souza-Motta, C.M.; et al. Fungal Planet description sheets: 716–784. Pers. Mol. Phylogeny Evol. Fungi 2018, 40, 240–393. [Google Scholar] [CrossRef]
- Sydow, H. Novae fungorum species. XXV. Ann. Mycol. 1937, 35, 244–286. [Google Scholar]
- Von Szilvinyi, A. Mikrobiologische Boden untersuchungenim Lunzer Gebiet. Zent. Bakteriol. und Parasitenkd. Abt. 2 1941, 103, 133–189. [Google Scholar]
- Sousa da Câmara, M. Mycetes aliquot Lusitaniae IX. Agron. Lusit. 1949, 11, 39–73. [Google Scholar]
- Roldan, E.F. Philipp. J. Sci. 1936, 60, 121.
- Cooke, M.C. New British fungi [cont.]. Grevillea 1877, 6, 71–76. [Google Scholar]
- Shirouzu, T.; Harada, Y. Lignicolous dematiaceous hyphomycetes in Japan: Five new records for Japanese mycoflora, and proposals of a new name, Helminthosporium magnisporum, and a new combination, Solicorynespora foveolata. Mycoscience 2008, 49, 126–131. [Google Scholar] [CrossRef]
- Shirouzu, T.; Harada, Y. Notes on species of Helminthosporium and its allied genera in Japan. Mycoscience 2004, 45, 17–23. [Google Scholar] [CrossRef]
- Sydow, P.; Sydow, H. Weitere neue Micromyceten der Philippinen-Inseln. Ann. Mycol. 1920, 18, 98–104. [Google Scholar]
- Rangel, E.S. Arch. Jard. Bot. Rio de Janeiro. 1902, 2, 71.
- Castellani, E.; Ciferri, R. Scissioni di generi di licheni sulla base delle caratteristiche del fungo. Atti. Ist. Bot. Univ. Pavia Suppl. Agli. Ser. 5. 1950, 37. [Google Scholar]
- Miles, L.E. Trans. Ill. St. Acad. Sci. 1917, 10, 253.
- Ciferri, R.; González Fragoso, R. Hongosparásitos y saprofitos de la República Dominicana (4a serie). Boletín de la Real Soc. Española de Hist. Nat. 1926, 26, 192–202. [Google Scholar]
- Saccardo, D. Contribuzione alla micologia veneta e modenense. Malpighia 1898, 12, 201–228. [Google Scholar]
- Cooke, M.C. New British fungi. Grevillea 1888, 16, 77–81. [Google Scholar]
- Jaczewski, A.L.A. Alternative: Transactions of the Illinois Academy of Science. Microbiol. J. 1929, 9, 166. [Google Scholar]
- Wang, X.; Wu, H.; Zhang, M. A new species of Helminthosporium from Jiangsu, China. Mycotaxon 2014, 127, 1–4. [Google Scholar] [CrossRef]
- Sydow, H.; Sydow, P. Beitragzur Pilzflora Süd-Amerikas. Hedwigia 1903, 42, 105–106. [Google Scholar]
- Dearness, J.; House, H.D. New or noteworthy species of fungi. IV. Bull. N. Y. State Mus. 1925, 266, 57–98. [Google Scholar]
- Viennot-Bourgin, G. Champignons nouveaux de la Guinée. Bull. Soc. Mycol. Fr. 1959, 75, 33–37. [Google Scholar]
- Hughes, S.J. New Zealand Fungi 27. New species of Guedea, Hadrosporium, and Helminthosporium. N. Z. J. Bot. 1980, 18, 65–72. [Google Scholar] [CrossRef] [Green Version]
- Da Câmara, S.M. Contribution esadmyco floram Lusitaniae. Centuria XI. Bol. Agric. Lisboa 1936, 2, 1–80. [Google Scholar]
- Chevassut, G. Sur cinq espècesnouvelles et quelquesespècesrares de Micromycètes parasites du groupe des Adelomycetes (régions du Languedoc et de Franche-Comté) (Five new species and some rare species of parasitic Micromycetes from the Adelomycetes group (Languedoc and Franche-Comté) (en)). Bull. Trimest. Société Mycol. France 1992, 108, 101–106. [Google Scholar]
- Saccardo, P.A. Micromycetes Americani novi. J. Mycol. 1906, 12, 47–52. [Google Scholar]
- Hiroë, I.M. Trans. Tottori Soc. Agric. Sci. 1935, 5, 175.
- Hennings, P. Fungi Africae orientalis III. Bot. Jahrbücher für Syst. Pflanzengesch. und Pflanzengeogr. 1904, 34, 39–57. [Google Scholar]
- Savulescu, T.; Rayss, T. Contribution à l’étude de la mycoflore de Palestine. Ann. Crypt. Exot. 1935, 8, 49–87. [Google Scholar]
- Matsushima, T. Microfungi of the Solomon Islands and Papua-New Guinea; Matsushima Fungus Collection: Kobe, Japan, 1971; pp. 1–78. [Google Scholar]
- Castro, C.C.D.; Gutiérrez, A.H.; Sotão, H.M.P. Fungosconidiaisem Euterpe oleracea Mart.(açaizeiro) naIlha do Combu, Pará-Brasil. Acta. Bot. Bras. 2012, 26, 761–771. [Google Scholar] [CrossRef] [Green Version]
- Berg, A. Pamietn. Towarz. Nauk Sci. Paryzu. 1934, 260, 14.
- Liu, L.J. A New Species of Helminthosporium causing leaf spot disease of sugarcane in Puerto Rico. J. Agr. U. Puerto. Rico. 1971, 55, 12–22. [Google Scholar] [CrossRef]
- Cooke, W.B. Western fungi. II. Mycologia 1952, 44, 245–261. [Google Scholar] [CrossRef]
- Bourne, B.A. Mém. Mus. Hist. Nat. 1956, 1087.
- Lucas, M.T.; Sousa da Câmara, M. Fungi Lusitaniae. V. Agron. Lusit. 1953, 15, 153–182. [Google Scholar]
- Dearness, J. New or noteworthy North American fungi. Mycologia 1917, 9, 345–364. [Google Scholar] [CrossRef]
- Sydow, H.; Sydow, P. Aufzählungeiniger in den Provinzen Kwangtung und Kwangsi (Süd-China) gesammelter Pilze. Ann. Mycol. 1919, 17, 140–143. [Google Scholar]
- Fresenius, G. Beitr. Mykol. 1863, 3, 50. Available online: ia800500.us.archive.org/33/items/beitrgezurmyko00fres/beitrgezurmyko00fres.pdf (accessed on 12 May 2021).
- Berlese, A.N. Sur le développement de quelques champignons nouveaux ou critiques. Bull. Soc. Mycol. Fr. 1892, 8, 94–110. [Google Scholar]
- Woronichin, N.N. Contribution à la floremycologique du Caucase. Trav. du Musée Bot. de l’Académie des Sci. de Russ. 1927, 21, 87–243. [Google Scholar]
- Stevenson, J.A. Rep. P. Ricoinsul. Agric. Exp. Station. 1919, 1917-18, 137.
- Zhang, M.; Zhang, T.; Wu, W. Taxonomic studies of Helminthosporium from China II. Two new species in Sichuan Province. Mycosystema 2004, 23, 179–182. [Google Scholar] [CrossRef]
- Durieu de Maisonneuve, M.C. Expl. Sci. Alg. Fl. Algér. 1 (Livr. 9) 1848, 1, 321–360.
- Patterson, F.W. New species of fungi. Bull. Torrey Bot. Club. 1900, 27, 282–286. [Google Scholar] [CrossRef]
- Saccardo, P.A. Notae mycologicae. Series XXIV. I. Fungi Singaporenses Barkesiani. Bolletino dell’Orto Bot. Regia dell’Universita de Napoli. 1921, 6, 39–73. [Google Scholar]
- Peck, C.H. Report of the state botanist. Bull. N. Y. State Mus. 1911, 150, 5–100. [Google Scholar]
- Saccardo, P.A. Bulletino dell’orto Botanico della R.; Universitá di Napoli: Napoli, Italy, 1918; Volume 6, p. 23. [Google Scholar]
- Turconi, M. Sopra una nuovamalattia del cacao (Theobroma cacao L.). Atti dell’Istituto Bot. Univ. e Lab. Crittogam. di Pavia. 1920, 17, 1–8. [Google Scholar]
- Ciferri, R.; González Fragoso, R. Hongosparásitos y saprofitos de la RepúblicaDominicana (10a serie). Boletín de la Real Soc. Española de Hist. Nat. 1927, 27, 165–177. [Google Scholar]
- Hennings, P. Schädliche Pilze auf Kulturpflanzenaus Deutsch-Ostafrika. Notizbl. des Bot. Gart. und Mus. Berl. 1903, 3, 239–243. [Google Scholar]
- Berkeley, M.J.; Broome, C.E. Notices of British fungi (502–537). Ann. Mag. Nat. Hist. 1851, 7, 95–102. [Google Scholar]
- Hennings, P.C. Mission. E. Lauren. 1906, 3, 318.
- Alves-Barbosa, M.; Costa, P.M.; Malosso, E.; Castañeda-Ruiz, R.F. Two new species of Dictyosporium and Helminthosporium (Ascomycota) from the Brazilian Atlantic forest. Nova Hedwig 2017, 105, 65–73. [Google Scholar] [CrossRef]
- Sydow, H.; Sydow, P. Fungi Paraenses. Hedwigia 1910, 49, 78–84. [Google Scholar]
- Wakefield, E.M. New and rare British fungi. Bull. Misc. Inf. R. Bot. Gard. Kew. 1918, 1918, 229–233. [Google Scholar] [CrossRef] [Green Version]
- Ciferri, R.; González Fragoso, R. Parasitic and saprophytic fungi of the Dominican Republic, (11th Series.). Bol. de la Real Soc. Esp. de Hist. Nat. Madr. 1927, 27, 267–280. [Google Scholar]
- Batista, A.C.; Maia, H.S.; Lima, J.A.; Matta, E.A.F. Moniliales–descrição e revisão de algumasespécies. Atas do Inst. de Micol. Univ. de Pernamb. Recife 1960, 1, 247–274. [Google Scholar]



| Genes/loci | PCR Primer (Forward/Reverse) | PCR Conditions |
|---|---|---|
| LSU | LR0R/LR5 | a; 95 °C: 30 s, 55 °C: 50 s, 72 °C: 30 s (35 cycles); b |
| ITS | ITS5/ITS4 | |
| SSU | NS1/NS4 | |
| tef1-α | 983F/2218R |
| Species | Strain | LSU | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 34 | 74 | 270 | 400 | 412 | 419 | 427 | 480 | 484 | 490 | 491 | 524 | 644 | 843 | ||
| Haplohelminthosporium calami (this study) | MFLUCC 18-0074 | - | A | A | T | T | T | C | C | A | C | A | T | T | T | G |
| Helminthosporium endiandrae (Ex-type from the holotype) | CBS 138,902 (KP004478) | - | C | C | C | C | C | T | T | C | T | T | G | C | G | G |
| H. endiandrae (sister strain in Figure 1B and Figure 2) | CBS 138,902 (MH878637) | C | A | C | C | C | C | T | T | C | T | T | G | C | G | - |
| No. | Herbarium/ Culture No. | Host (Genus/Family) | Locality | Morphology | References | |||
|---|---|---|---|---|---|---|---|---|
| Mycelia (μmWide) | Conidiophores (μm) | Conidiogenous Cells (μm) | Conidia (μm) | |||||
| 1. | Herbarium: - Culture no.: CPHmZC-01 | On leaves of Elaeis oleifera/Arecaceae | Colombia | Hyaline to pale brown, smooth, branched, septate | Erect, brown to red-brown, synnematous, septate, compacted, 620–1400 × 19–54, individual hyphae 3–4 wide | Mono- or polytretic, integrated, determinate, terminal, cylindrical, 31–67 × 4.5–7 | Catenate, obclavate, subcylindrical, occasionally bifurcate, medium brown, 26–83 × 7–10, (1–)3–5(–6)-distoseptate | [63] |
| 2. | Herbarium: - Culture no.: COAD 2126 | On old leaves of Elaeis guineensis/Arecaceae | Brazil | Hyaline to pale brown, 2–4 | Erect, brown, septate, synnematous, 66–201(−770) × 2.5–6(−18) | Mono or polytretic, cylindrical, terminal, 18–59 × 4–7 | Catenate, subcylindrical, obclavate, brown, 32–83 × 4–11, 2–7-distoseptate | [64] |
| 3. | Herbarium: MFLU 20-0521 Culture no.: MFLUCC 15-0813 | On dead petiole of Cocos nucifera/Arecaceae | Thailand | Mostly immersed, dark brown | Solitarily, erect, unbranched, straight or flexuous, cylindrical, bulbous at base, dark brown, becoming pale brown at the apex, (60–)165–270(–310), (5–)7–9(–12) at the base, 5–8 μm wide at the apex, (4–)12–15 septate | Terminal and intercalary with well-defined pores, pale brown | Obpyriform to lageniform, straight or curved, light brown, (30–)45–60(–70) × 6–9, 5–8-distoseptate | This study |
| No. | Taxa | Host(Genus/Family) | Locality | Morphology | Sequence Data | References |
|---|---|---|---|---|---|---|
| 1 | H. abietis | Abies sp./Pinaceae | U.S.A./Washington | Conidiophores irregularly branched; Conidia 126–150 × 12–16 µm, fusiform, pointed at both ends, olive-green, 12–15-distoseptate | Absent | [84] |
| 2 | H. acaciae | On dead branches of Acacia farnesiana/ Fabaceae | Sierra Leone | Conidiophores 140–280 × 7–11 µm, dense, fasiculate, simple, straight or flexuous, sometimes swollen at at the tip, septate, smooth, thick-walled, brown, with well-difinded small pores at the apex; Conidia 31–(44–)49 × 10–(12–)14 µm in widest part, narrowing towards the apex to 3–5 µm, obclavate, straight or flexuous, smooth-walled subhyaline to pale brown, 3–6-distoseptate, with a small dark blackish-brown to black scar at the base | Absent | [85] |
| 3 | H. acalyphae | On leaves of Acalypha angustifolia/ Euphorbiaceae | Dominican Republic | Conidiophores 2.5–4 µm thick, erect, simple, superficial, brown-blackish, septate; Conidia 9–16 × 4–6 µm, one for each conidiophore, ovate-ellipsoid, olivaceous-brown or dull-brown, 2–3-distoseptate | Absent | [86] |
| 4 | H. accedens | On living leaves of Dolichos baumii/Fabaceae | Namibia | Conidiophores 250–300 × 5–9 µm, erect, olive-brown; Conidia 35–57 × 6.5–9 µm, solitary, oblong-fusoid, olive, 3–6-distoseptate | Absent | [87] |
| 5 | H. ahmadii | On dead branches of Quercus sp./Fagaceae | Pakistan | Conidiophores 220–650 × 12–15 µm, dense, fasiculate, simple, straight or flexuous, smooth, thick-walled, brown to dark brown, with small pores at the apex, septate; Conidia 95–(110–)150 × 25–30(–38) µm wide inthe broadest part, tapering towards the apex to 5–9 µm, obclavate, sometimes rostrate, straight or flexuous, smooth-walled, brown or dark brown, 5–15-distoseptate, with a dark blackish-brown to black scar at base | Absent | [85] |
| 6 | H. aichrysonis | On leaves of Aichryson dichotomum/ Crassulaceae | Spain | No information available | Absent | [88] |
| 7 | H. alatum | On dying leaves of Dioscorea alata/ Dioscoreaceae | Dominican Republic | No information available | Absent | [89] |
| 8 | H. albiziae | On leaves of Albizia lebbeck/Fabaceae | Sri Lanka | Conidiophores 70 ×7 µm; Conidia 42–56 × 12 µm, tapering to 4 µm diam. clavate, ends rounded, at the lower end, rough with minute warts, fuliginous, terminal cell paler, strgight or curved below, 3–4-distoseptate | Absent | [90] |
| 9 | H. albiziicola | Albizzia lebbek/Fabaceae | India | Conidiophores 28–44 × 4.5–6 µm, straight or slightly curved, one-septate at the base; Conidia 23.5–34 × 8–9 µm, pyriform, prolongate at the apex, rounded at the base, pale, cinnamon-brown, 3-distoseptate | Absent | [91] |
| 10 | H. allamandae | On living leaves of Allamanda cathartica/ Apocynaceae | Dominican Republic | Conidiophores 100–180 × 8–10 µm, solitary or aggregate, curved, simple, dark-brown; Conidia 66–110 × 17–20 µm, clavate, elongate-ellipsoid or subfusoid, erect or curved, gray-brown, 7–10-distoseptate | Absent | [92] |
| 11 | H. alphitoniae | On living leaves of Alphitonia sp./ Rhamnaceae | Malaysia/Mount Kinabalu | Conidiophores 250–500 ×5–8 µm, erect, dark-brown; Conidia 25–66 × 8–13 µm, obclavate, erect or curved, yellow-brown or pale olive, 1–6-distoseptate | Absent | [93] |
| 12 | H. aneurolepidii | On leaves of Aneurolepidium ramosum/Poaceae | Russia/West Siberia | No information available | Absent | [94] |
| 13 | H. anomalum | From soil | U.S.A./Iowa, Utah | No information available | Present | [17,63] |
| 14 | H. anonymicum | In culture: former Soviet Union | Russia | No information available | Absent | [95] |
| 15 | H. apiculatum | On dry tree of Betula sp. (Betulinum)/ Betulaceae | Czech Republic | Conidiophores fasiculate, flexuous, simple, hyaline; Conidia long, 37 μm, elliptical-fusiform, with color, multi-septate | Absent | [96] |
| 16 | H. appatternae | From leaves of Cynodon dactylon/Poaceae; from culture | India/Maharashtra | Conidiophores unbranched, of two types; determinate conidiophores uniform, 182 × 5.2 μm, single, olivaceous, 1–3 septate; indeterminante conidiophores narrower, 208–520 × 7.8 μm, paler and distantly septate at base, gradually broadened into a darker, close septate; Conidia 20.8–152.0 × 7.8 μm, 6–18-distoseptate | Absent | [18] |
| 17 | H. appendiculatum | On branches of the trees | Czechia | Conidiophores simple, fasciculate; Conidia 65 × 11 μm, clavate, curved, blunted, whitish, multi-septate | Absent | [96] |
| 18 | H. aquaticum | On submerged decaying wood | China/Yunnan | Conidiophores 410–580 × 13–17 μm, solitary or in groups of 2–4, erect, flexuous, unbranched, smooth, dark brown paler towards the apex, bulbous at base, 14–23 septate; Conidia 70–80 × 16–18 μm, single, obclavate, straight or curved, pale brown to brown, truncate and cicatrized at base, wider than apex, guttulate, 8–10-distoseptate | Present | [20] |
| 19 | H. arcautei | On living leaves Scorpiurus subvillosa/ Fabaceae | Spain | Conidiophores 35–50 × 7–8 μm, erect, simple, cylindrical, brownish-purple, 2–3 septate; Conidia 48–86 × 10.5–11 μm, cylindrical-fusoid, straight or slightly curved, light-brown chestnut, 3–8-distoseptate | Absent | [95,97] |
| 20 | H. asterinoides | On living leaves of Eugenia sp./Myrtaceae | Brazil | Conidiophores 5–7 μm thick, fasciculate, rhizoid; Conidia 22–24 × 5–6 μm, fusoid, curved, colorless at each bottom, 3-distoseptate | Absent | [98] |
| 21 | H. asterinum | On Liquidambar sp./ Altingiaceae | U.S.A./Florida | Conidiophores erect, simple, septate; Conidia 500–600 × 80 μm, clavate, 3–4-distoseptate | Present | [99] |
| 22 | H. astragali | On leaves of Astragalus siversianus/ Fabaceae | Kyrgyzstan | No information available | Absent | [100] |
| 23 | H. atypicum | On leaves of Triticum sp./Poaceae | India/Maharashtra | Conidiophores 3–7 septate, unbranched, and of two types; shorter conidiophore uniformly wide, 62.4–72.8 × 7.8 μm, brown; longer ones narrow at the base and paler, gradually broadening and darkening towards the apex, 440–680 × 5.2–10 μm; Conidia yellow to brown, darkening at maturity, of two kinds; normal ones 23–93.6 × 26 μm, elliptical with hemispherical edges, widest at the middle, 0–10-distoseptate; a typical conidia abundant, forked or geniculate, septation forked, brown to dark brown, 5–8-distoseptate | Absent | [101] |
| 24 | H. austriacum | On dead corticated twigs of Fagus sylvatica/Fagaceae | Austria/Döbling, Kahlenberg, Wien | Conidiophores 275–700(–920) μm long, 11.5–19 μm wide at the base, tapering to 7–11 μm near the apex, solitarily or fasciculate, erect, simple, sub-cylindrical, straight or flexuous, thick-walled, smooth, brown to dark brown, paler near the apex, with well-defined small pores at the apex, 1–12 septate; Conidia (30–)35–48(–97) × (10.0–)13.7–16.5(–19.8) μm, tapering to 4.5–6.0 μm at the distal end, obpyriform to lageniform, straight or curved, smooth, pale brown, (4–)5–7(–10)-distoseptate, with a blackish-brown 3–6 μm wide scar at the base | Present | [21] |
| 25 | H. avenae-pratensis | On sheaths of Avena pratensis/Poaceae | Germany | Conidiophores 300 × 8–11 μm, solitary or fasciculate, dark-chestnut, septate; Conidia 70–107 × 16–21 μm, cylindrical or obclavate, light brown, on both sides paler, 5–11-distoseptate | Absent | [102] |
| 26 | H. bactridis | On sheaths of Bactris sp./Arecaceae | Brazil/Pará | Conidiophores 200 × 3–4.5 μm, septate; Conidia 20–30 × 6–8 μm, fusoid, 6–7-distoseptate | Absent | [103] |
| 27 | H. bakeri | On dead stems of Premnavestita sp./ Lamiaceae | Philippines | Conidiophores 500–800 ×12 μm wide at base to below, 10 μm wide, erect, unbranched, dark; Conidia 80–150 × 17–22 μm, solitary, oblong, obclavate, 3–6-distoseptate | Absent | [104] |
| 28 | H. bambusicola | On dead culm of Bambusa sp./Poaceae | China/Sichuan | Conidiophores 55–247 × 4–6 μm, fasciculate or solitary, simple, cylindrical, straight or flexuous, thick walled, smooth, brown, paler towards the apex, with well-defined small pores, 1–2 septate; Conidia 36–66 × 6–11 μm narrowing towards the apex to 2–4.5 μm wide, obclavate, straight or slightly flexuous, thin-walled 1–1.5 μm thick, smooth, pale brown, paler towards the apex, 5–8-distoseptate, scar not distinct at the base | Absent | [105] |
| 29 | H. bataticola | On living leaves of Ipomoea batatas/ Convolvulaceae | Caucasus | No information available | Absent | [106] |
| 30 | H. bauhiniae | On dead twigs of Bauhinia tomentosa/ Fabaceae | Sierra Leone | Conidiophores 350–110 × 10–15 μm thick at the apex, 15–20 μm thick at the base, dense, fasciculate, simple, straight or flexuous, smooth-walled, dark brown, sometimes paler towards the apex, with well definded, small pores septate; Conidia 55-(86–)145 × 16–(17.2–)18 μm thick in broadest part, tapering to 3–4 μm the apex, obclavate, straight or flexuous, rostrate, smooth-walled, subhyaline to brown, 7–18-distoseptate, with a dark blackish brown to black scar ath the base | Absent | [85] |
| 31 | H. belgaumense | On litter, Calamus thwaitesii/Arecaceae | India/Karnataka | Conidiophores 140–250 × 6–9 μm, erect, straight to flexuous, unbranched, smooth, brown; Conidia 10–15 × 6–11 μm, solitary, dry, sub-spherical, dark brown, truncate at base, roundea at the apex, 1-distoseptate | Absent | [107] |
| 32 | H. bhawanii | On leaves of Eragrostis japonica/Poaceae | India/Bihar | No information available | Absent | [108] |
| 33 | H. bigenum | Palmae rotten petiole/Arecaceae | Peru | No information available | Absent | [109] |
| 34 | H. bondarzewii | From grains of Triticum sp. and Secale sp./ Poaceae | Russia, Ukraine | No information available | Present | [60,110] |
| 35 | H. cacaliae | Cacalia sonchifolia/Asteraceae | Brazil | No information available | Absent | [111] |
| 36 | H. cacaophilum | From unfermented Cacao beans, Theobroma cacao/Malvaceae | Dominican Republic/Santo Domingo | No information available | Absent | [112] |
| 37 | H. cactacearum | In young plants of Cereus species/Cactaceae | Italy | No information available | Absent | [113] |
| 38 | H. caespitiferum | Meliola spec. in leaf spots of living leafs of Omphalea pauciflora/ Euphorbiaceae | Dominican Republic/Santo Domingo | Conidiophores 150–300 × 6.5–8 μm, simple, dark-brown, septate; Conidia 18–42 × 8–11 μm, oblong to fusoid, dark-brown, constrict at septum, (3–)6–7-distoseptate | Absent | [92] |
| 39 | H. canephorae | Coffea canephora/Rubiaceae | Democratic Republic of the Congo/Zaire | No information available | Absent | [114] |
| 40 | H. cantareirense | On dead stems | Brazil/São Paulo | Conidiophores 7–12 μm thick, erect, fasciculate; Conidia 50–60 × 8–12 μm, clavate, brown, constrict at septum, 6–8-distoseptate | Absent | [115] |
| 41 | H. cantonense | On decaying culms of Bambusa vulgaris/Poaceae | China | Conidiophores 80–95 × 6 μm; Conidia 50–62 × 8 μm, obclavate, 7–9-distoseptate | Absent | [116] |
| 42 | H. caperoniae | On living leaves of Caperonia palustris/Euphorbiaceae | Dominican Republic | Conidiophores 100–300 × 3.5–5 μm, 2–5 fasciculate, simple, olive-brown; Conidia 22–55 × 4–6 μm, oblong-fusoid or subclavate, rarely cylindrical, yellow or gray-brown | Absent | [92] |
| 43 | H. carpocrinum | Parasite on perithecia of Meliola funebris on leaves of Omphalea sp./Euphorbiaceae (O. pauciflora) | Dominican Republic/Santo Domingo | Conidiophores 1–4 articulate, 200–350 μm long, very densely fasciculate, erect to sub-erect, straight or slightly irregularly curved, almost straight ot curved, dark-brown to blackish, tip light-colored; Conidia 22–25 ×8–10 μm, 1–4 to each conidiopore, easily falling, ellipsoid to ovoid, with narrowed ends, or basal end narrowed-truncate, apical end rounded to acute, not caudate, central cells from dark-brown to brownish, and cells light brown to yellowish, 2–5-distoseptate | Absent | [117] |
| 44 | H. carposaprum | On Lycopersicon esculentum/Solanaceae | British Guiana, Haiti, Mexico | No information available | Absent | [118] |
| 45 | H. ceibae | On leaves of Ceiba pentandra/Malvaceae | Philippines | No information available | Absent | [119] |
| 46 | H. chlorophorae | On dead twigs of Chlorophora regia/Moraceae | Sierra Leone | Conidiophores 120–270 × 7–10 μm thick at the base, often swollen towards the tip up to 12 μm, single or fasciculate, simple, straight or flexuous, smooth-walled, brown to dark brown, with 1–3 well-definded, small pores, septate; Conidia 52–(73–)102 × 8–(9.5–)11 μm, thick in the widest part narrowing gradually towards the apex to 3–5 μm, obclavate, straight or flexuous, smooth-walled, subhyaline to pale brown, 6–9-distoseptate, with a tather large dark blackish-brown to black scar at the base | Present | [85,120] |
| 47 | H. chrysobalani | On dry leaves of Chrysobalanus icaco/ Chrysobalanaceae | Dominican Republic/Bonao | Conidiophores up to 6 μm, fasciculate, erect, 2–3 septate; Conidia 25–50 × 3–4 μm, fusoid, 2–4-distoseptate | Absent | [121] |
| 48 | H. chusqueae | On living and dying leaves of Chusquea serrulata/Poaceae | Ecuador/Tungurahua | Conidiophores 200–350 × 4–6 μm, dense, erect, fasciculate, simple, straight or slightly curved, dark-brown or olive, septate; Conidia 32–50 × 9–11 μm, elongate-fusiform, blunt at both ends, curved, rarly straight, gray or olive-brown, 3–4-distoseptate | Absent | [122] |
| 49 | H. cibotii | On leaves of Cibotium sp./Cibotiaceae | U.S.A./Hawaii Islands | No information available | Absent | [123] |
| 50 | H. ciliare | - | - | No information available | Absent | [124] |
| 51 | H. citri | On leaves of Citrus poonensis, Citrus tankart, Citrus ponki, and of Citrus sinensis var. brasiliensis/Rutaceae | China/Taiwan | No information available | Absent | [125] |
| 52 | H. claviphorum | Rotten branch | Peru | No information available | Absent | [109] |
| 53 | H. cleosmatis | On living leaves of Clematis sp./ Ranunculaceae (in foliisvivis Cleosmati soctandri) | Dominican Republic | Conidiophores 140–250(–300) μm long, 4–5 μm wide, solitary, erect, simple, dark-brown, often becoming paler; Conidia 28–52 × 6.5–9 μm, clavate or fusoid, yellow or pale olive-brownish, (3–)4–5-distoseptate | Absent | [92] |
| 54 | H. clusiae | On leaves of Clusiarosa sp./Clusiaceae | Dominican Republic | Conidiophores 108–128 × 12–16.5 μm effuse, brown-black, irregular at based, or subbulbose, septate; Conidia 26–32 × 10–11.5 μm, fusoid, subfusoid or cylindrical, 4–8-distoseptate | Absent | [126] |
| 55 | H. coffeae | On leaves of Coffea liberica/Rubiaceae | Ghana | Conidiophores 300–400 × 7–8 μm, effuse, nigro-olivaceas, aggregate, erect, cylindrical, rect or flexuous, olives-brown, septate; Conidia 45–55 × 8–10 μm, obovate, 3–5-distoseptate | Absent | [127] |
| 56 | H. conidiophorellum | On dead branches of tree | China/Guangxi | Conidiophores 60–280 × 7.0–8.5 μm, fasciculate, simple, subcylindrical, straight or flexuous, thick-walled, smooth, dark brown, paler towards the apex, with 1–3 well-defined small pores at the apex, 1–2 septate; Conidia 100–147.5 μm long, 9.5–11 μm diam in the widest part, narrowing towards the apex to 3–4 μm diam, straight or slightly flexuous, smooth-walled, pale brown, sometimes verruculose at apex, 11–17-distoseptate, with a large dark blackish-brown scar at the base, 2–3 μm thick | Absent | [128] |
| 57 | H. constrictum | On dead branches of Trachycarpus fortunei/Arecaceae | China/Guangdong | Conidiophores single, simple, subcylindrical, straight or slightly flexuous, brown to dark brown, paler towards the apex, 1–3 septate; Conidia 57–120 × 9–12 µm, thick in the widest part, narrowing toward the apex to 2.5–5 µm, abruptly tapered to a truncate base, tretic, obclavate, straight or slightly flexuous, pale brown, paler toward to apex, 9–15-distoseptate, sometimes constricted at one or two septa | Absent | [129] |
| 58 | H. conviva | On Hyphoderma caliciferum, the genus of crust fungi in the family Meruliaceae. | Spain/Archipelago/Balearic/Baleares Islands | No information available | Absent | [130] |
| 59 | H. corchori | On leaves of Corchorus capsularis/Malvaceae | China/Taiwan | No information available | Absent | [131] |
| 60 | H. crassiseptum | Meliola abrupta | Dominican Republic | Conidiophores 30–50 × 2–3 µm, septate; Conidia 45–55(–65) × 12–14 µm, ovoid or elliptical, (2–)3-distoseptate | Absent | [86] |
| 61 | H. crotalariae | On leavesof Crotalaria juncea/Fabaceae | India/Assam | No information available | Absent | [132] |
| 62 | H. crus-galli | On living leaves of Echinochloa crus-galli (=Panicum crista-galli)/Poaceae | Japan | No information available | Absent | [133,134] |
| 63 | H. cubense | On rachis of Roystonea regia/Arecaceae | Cuba | No information available | Absent | [135] |
| 64 | H. cucumerinum | On living leaves of Cucumis sativus/ Zingiberaceae | Russia/Krym | No information available | Absent | [136] |
| 65 | H. curvulum | On decaying leaves of Zea mays/Poaceae | Philippines | Conidiophores 160–180 × 7–7.5 µm, fasciculate, filiform, septate; Conidia 25–35 × 8–9 µm, oblong-fusoid, narrow, 3(–4)-distoseptate | Absent | [137] |
| 66 | H. cuspidatum | On decaying branches of Afzelia rhomboidea/Fabaceae | Philippines | Conidiophores 800–900 × 8–9 µm, fasciculate, filiform, multiseptate; Conidia 100–130 × 11–12 µm, obclavate, 8–12-distoseptate | Absent | [137] |
| 67 | H. cylindricum | On rotten wood | Czech Republic/Bohemia | Conidiophores 100–130 × 4–5 µm, subfasciculate, filiform long, simple, fuliginous up paler, septate; Conidia 14–15 × 2.5 µm, cylindrical, apex rounded, base acuted, minute, pale fuliginous, 3-distoseptate | Absent | [138] |
| 68 | H. cymmartinii | On leaves of Cymbopogon martinii/Poaceae | India/Uttar Pradesh | No information available | Absent | [108] |
| 69 | H. cyperi | On Cyperus sp./ Cyperaceae | Greece | Conidiophores straight to subflexuous, greenish, paler at apex; Conidia 78 × 9 µm, fusoid, fuscidull, 5–8-distoseptate | Absent | [139] |
| 70 | H. dactylidis | On leaves of Dactylis glomerata/Poaceae | U.S.A./Pennsylvania | No information available | Absent | [140] |
| 71 | H. dalbergiae | On dead branches of Dalbergia sissoo/Fabaceae | Pakistan | Conidiophores 300–1300 × 10–12(–15) µm, dense, fasciculate, simple, flexuous, smooth-walled, brown to dark brown, sometimes paler towards the apex, with well-definded small pores, septate; Conidia 58–(93–)125 × 12–(13.2–)14 µm thick in broadest part, tapering to gradually towards the apex to 3–5 µm, obclavate, straight or flexuous, smooth-walled, straw-coloured to pale brownwith, 5–17-distoseptate, large dark blackish-brown to black scar at the base | Present | [85] |
| 72 | H. davillae | On leaves of Davilla rugosa/Dilleniaceae | U.S.A./San Francisco | Conidiophores 4–6 µm, thick filiform, flexuous, unbranched, elongate, brown, septate; Conidia 40–70 × 4–6 µm, elongate-obclavate, narrower and paler, (1–)2–4-distoseptate | Absent | [141] |
| 73 | H. decacuminatum | In the dry twigs on Vitis vinifera/Vitaceae | Italy | Conidiophores 4 µm thick, extremely short-articulated, irregular, dark reddish-brown; Conidia 40–45 × 10 µm, long clavate, decacumina to tip, or cut down in pedicellum narrowed, pale brown-gray, 4–5-distoseptate | Present | [60,142] |
| 74 | H. delicatulum | On stems of Umbelliferae or Apiaceae | UK/Great Britain | Conidiophores slender, subulate, multi-articultate, brown, paler at the tips; Conidia oblong, nearly colourless, with the apices very obtuse, consisting of about five swollen articulations, one or two of which have occasionally a vertical dissepiment | Absent | [143] |
| 75 | H. delphinii | On stems of Delphinium brunonianum/ Ranunculaceae | Russia | No information available | Absent | [144] |
| 76 | H. dendroideum | On Acer sp./ Sapindaceae | U.S.A./South Carolina | Conidiophores 1–2 short branchlets termintated, oblong, subfusiform, slightly curved, multiarticulate conidia; Conidia 60 µm long, each joint containing a globose nucleus | Absent | [145] |
| 77 | H. densum | - | - | No information available | Absent | [146] |
| 78 | H. desmodii | On Desmodium buergeri/ Fabaceae | Japan | No information available | Absent | [147] |
| 79 | H. diedickei | No information available | No information available | No information available | Absent | [148] |
| 80 | H. dimorphosporum | On decaying rotting stems of unknown liana | Cuba | Conidiophores 150–400 µm long, at the apex 9–12 µm, at the base 10–14 µm wide, single or fasciculate 2–10, simple, straight or flexuous, smooth, dark brown, paler towards the apex, septate; Conidia of two different types arising through pores a t the apex (1–4 pores) and late rally beneath the upper septa: (a) 19–24 × 8–10.5 µm, broadly ellipsoidal, ovoid or broadly fusiform, thick-walled, smooth, brown to dark brown, 1-distoseptate; (b) 24–65 µm long, 10–15 µm wide in the broadest part, tapering to 3.2–4.8 µm at the apex, obclavate, rostrate, straight or flexuous, pale brown, smooth, 6–9-distoseptate, with a dark brown scar at the base | Absent | [149] |
| 81 | H. dolichi | On living leaves of Dolichos sp./Fabaceae | Namibia | Conidiophores 250–350 × 4–6 µm, erect, olive-brown; Conidia 27–38 × 5.5–8 µm, solitary, oblong-subfusoid, olive, 2–3-distoseptate | Absent | [87] |
| 82 | H. dongxingense | Rhododendron sp. | China | No information available | Absent | [150] |
| 83 | H. elasticae | - | - | No information available | Absent | [151] |
| 84 | H. endiandrae | On leaves of Endiandra introrsa/Lauraceae | Australia/New South Wales, Nightcap National Park | Conidiophores 200–300 × 5–7 µm, solitary, erect, subcylindrical, straight to flexuous, unbranched, thick-walled, base bulbous, lacking rhizoids, brown, 8–16 septate; Conidia (35–)37–45(–57) × (7–)8(–9) µm, solitary or in short chains (2–3), obclavate, thick-walled, finely roughened, brown, 3(–4)-distoseptate | Present | [21,59] |
| 85 | H. eragrostiellae | On inflorescence and leaves of Eragrostis bifida/Poaceae | India/Uttar Pradesh | No information available | Absent | [108] |
| 86 | H. erythrinae | On leaves of Erythrina suberosa/Leguminosae | India/Karnataka | Conidiophores 32–42 × 4–5 μm, simple, brownish-yellow; Conidia 39–62 μm at base, straight or vermiform, rounded at the apex and flat at the base, pale cinnamon-brown, 4–8-distoseptate | Absent | [91] |
| 87 | H. erythrinicola | On leaves of Erythrina humeana/Fabaceae | South Africa/Eastern Cape | Conidiophores 500–1200 × 6–10 mm, fasciculate, subcylindrical, unbranched, brown, becoming pale brown at apex, multiseptate; Conidia (70–)80–90(–110) × (9–)10–11(–12) mm, obclavate, straight to curved, apex subobtuse, smooth, medium brown, (6–)7–8(–12)-distoseptate | Present | [22] |
| 88 | H. exasperatum | On Dianthus barbatus/ Caryophyllaceae | UK/Great Britain | Conidiophores flexuous, knotted above, each knot bearing oblong conidia; Conidia 30–45 × 10–12 μm | Absent | [152] |
| 89 | H. feijoae | On leaves of Acca sellowiana/Myrtaceae (syn: Feijoa sellowiana) | North America/Hispaniola island | No information available | Absent | [153] |
| 90 | H. ferrugineum | On leaves of Hiraea sp. and Heteropterys sp./Malpighiaceae | U.S.A./San Francisco | Conidiophores 8–9 μm thick, filiform, yellow, septate; Conidia 50–62 × 11–14 μm, obclavate, subhyaline, last 2 septate hyaline-yellow to yellow | Absent | [141] |
| 91 | H. fici | On leaves of Ficus retusa/Moraceae | Philippines, Thailand | Conidiophores fusciculate, long, nodulosis, septate; Conidia 18–20 × 5–6 μm, cylindrical, reddish-brown, 3-distoseptate | Absent | [137,154] |
| 92 | H. ficinum | On leaves of Ficus ulmifolia/Moraceae | Philippines | Conidiophores 250 × 6 μm, filiform, septate; Conidia 50–60 × 6–8 μm, obclavate, 4–5-distoseptate | Absent | [137] |
| 93 | H. filicicola | On leaves of Lygodium sp./Lygodiaceae and of Selaginella sp./Selaginellaceae | Peru | Conidiophores 400 × 3–5 μm thick, erect, simple, filiform, septate; Conidia 30–40 × 6–10 μm, cylindrical-fusoid or clavate, both side blunt, 3–5-distoseptate | Absent | [155] |
| 94 | H. flagellatum | On mycelium of Meliola, in leaves of Ardisia disticha/Myrsinaceae | Philippines | Conidiophores 2.5–4 μm thick, erect, sub-hylaline | Absent | [156] |
| 95 | H. flumeanum | On leaves of Bambusa sp./Bambuseae | Philippines | Conidiophores 90–100 × 6–7 μm, dense, fasticulate, filiform; Conidia 35–40 × 9–12 μm, obclavate, 3-distoseptate | Absent | [157] |
| 96 | H. fumagineum | On leaves of ficusulmifolia/ Moraceae | Philippines | Conidiophores 240–300 × 7 μm, filiform, septate; Conidia 35 × 9–10 μm, oblong-obclavate, 3-distoseptate | Absent | [137] |
| 97 | H. gibberosporum | Musa cavendishii/Musaceae | Somalia | No information available | Absent Present | [158] |
| 98 | H. glabroides | On Meliola glabroides, on Piper aduncum/ Piperaceae | Puerto Rico | Conidiophores 100–140 × 7 μm; Conidia 40–81 × 6–7 μm, 3–6-distoseptate | Absent | [159] |
| 99 | H. gleicheniae | On leaves of Dicranopteris linearis (=Gleichenia dichotoma)/ Gleicheniaceae | U.S.A./Hawaii Islands | No information available | Absent | [123] |
| 100 | H. gossypii | On living leaves and bracts of Gossypium sp./ Malvaceae | North America | Conidiophores 40–185 × 6.5–8.5 μm, singly or in groups of three to six, straight cylindrical to nodose or bent, brown, 5 septate; Conidia 35–118 × 11.7–18.4 μm, elliptical, curved, rarely straight, light to dark fuliginous, thick walled, rounded at the ends, 1–8-distoseptate | Absent | [160] |
| 101 | H. grewiae | On leaves of Grewia sp./Malvaceae | Democratic Republic of the Congo | Conidiophores 80–120 × 5–8 μm, fasciculate, septate; Conidia 35–45 × 8–10 μm, fusoid, 2–4-distoseptate | Absent | [161] |
| 102 | H. guangxiense | On dead branches of unidentified tree | China/Guangxi, Shanglin | Conidiophores 330–850 μm long, 15–20 μm wide just above the base and 8–13 μmwide toward the apex, fasciculate, simple, straight or flexuous, sub-cylindrical, thick-walled, smooth, brown, with 1–3 well-defined small pores at the apex, 1–4 septate; Conidia 76–110 μm long, 16–22 μm wide in the widest part, narrowing towards the apex to 3–6μm wide, straight or curved, obclavate, smooth, middle brown, paler towards the apex, 9–17-distoseptate, with a large dark blackish-brown scar at the base, 1.5–3.5 μm thick | Absent | [128] |
| 103 | H. guianense | Meliola guianensis parasitic on mycelium on living leaves of Theobroma cacao/ Malvaceae | Guyana | No information available | Absent | [162] |
| 104 | H. heringerianum | Tipuana speciosa/ Fabaceae | Brazil | No information available | Absent | [163] |
| 105 | H. hispanicum | On dead corticated twigs of Juglans regia/Juglandaceae | Asturias, Selviella, Spain | Conidiophores 130–540 μm long, 13–22.5 μm wide at the base, tapering to 8–15 μm near the apex, solitarily or in small groups, erect, simple, straight or flexuous, thick-walled, subcylindrical, smooth, dark to blackish brown, paler near the apex, with well-defined small pores at the apex, 1–2 septate; Conidia 69–99(–130) × (17–)18–21(–24) μm, obclavate, straight or flexuous, thin-walled, smooth, pale brown, (4–)6–11(–14)-distoseptate, with a blackish-brown 4–6 μm wide scar at the base | Present | [21] |
| 106 | H. hispaniolae | On living leaves of Manihot utilissima/ Euphorbiaceae | Dominican Republic/Haiti | Conidiophores sub-hyaline to light-grey, when old, with an almost hyaline tip; Conidia 14.8–(53.5–)81.4 × 7.4–(11–)14.8 μm, sub-hyaline to smoky, irregular, cylindric-elongate to ellipsoidal, straight or slightly curved, with the basal end applanate, 1–8-distoseptate | Absent | [112] |
| 107 | H. hunanense | On dead branches of unidentified tree | China/Zhangjiajie, Hunan | Conidiophores 70–226 × 5–7 above, 8.5–14 μm base, solitary or fasciculate, simple, cylindrical, straight or flexuous, thick-walled, smooth, brown, well-defined small pores at the apex, 1–3 septate; Conidia 56–127 × 10–14 base, apex 2–4 μm, obclavate, straight or curved, smooth, middle brown, paler towards the apex, 4–12-distoseptate, blackish-brown scar at the base, 1.5 μm thick | Absent | [67] |
| 108 | H. hygrophilae | On leaves of Hygrophila brasiliensis/Acanthaceae | Dominican Republic | No information available | Absent | [89] |
| 109 | H. insigne | On leaves of Mallotus philippensis/ Euphorbiaceae | Philippines | Conidiophores 600–800 × 50 μm, fasciculate, filiform, blackish, septate; Conidia 45–55 × 7–8 μm, obclavate, often curved, 4–5-distoseptate | Absent | [137] |
| 110 | H. insuetum | On living leaves of Philodendron sodiroi (=Piplocarpha sodiroi)/Araceae | Ecuador/Pichincha | Conidiophores 2.5–5 μm thick, olive brown or dark brown; Conidia 17–38 × 7–12 μm, oblong, ellipsoid or oblong-ellipsoid fusiform and often subclavate, rarely cylindrical, often straigtly, rarely curved, olive brown or dark-brown, (3–)5–7(–9)-distoseptate, scared or a little more often in the middle constricted | Absent | [122] |
| 111 | H. ipomoeae | On leaves of Ipomoea reptans/ Convolvulaceae | China/Taiwan | No information available | Absent | [130] |
| 112 | H. iranicum | On living leaves of Indigofera sp./Fabaceae | Iran/Bandar Abbas | Conidiophores 40–75 × 6-9 μm, dense, curved, rarely straight, dark-brown, septate; Conidia 36(–42) × 7–11 μm, oblong, narrowly ellipsoid or curved, obtuse at both ends, straight or curved, sometimes irregular, olive, 1–3-distoseptate | Absent | [164] |
| 113 | H. italicum | On dead branch of Alnus glutinosa/ Betulaceae | Italy | Conidiophores (190–)330–600 × (12–)16–18(−20) μm, aggregated, erect, straight or slightly flexuous, unbranched, cylindrical, dark brown, 13–25 septate; Conidia 58–78 × 15–19(−23) μm, obclavate, straight or curved, pale brown to brown, slightly truncate and black at base, rounded, narrowed, 6–11-distoseptate | Absent | [61] |
| 114 | H. juglandinum | On dead corticated twigs of Juglans regia/ Juglandaceae | Austria/Niederösterreich/Gießhübl, Italy | Conidiophores (175–)215–325(–455) μm long, 11–23 μm wide at the base, 8.5–14 μm wide near the slightly inflated apex, fasciculate, erect, simple, straight or flexuous, thick-walled, sub-cylindrical, smooth, brown to dark brown, darker to black at the apex, the latter with a well-defined apical pore; Conidia (69–)89–145(–205) × (15.0–)16.5–20.0(–25.0) μm, rostrate, straight or flexuous, thin-walled, smooth, pale brown, (5–)9–17(–20)-distoseptate, blackish-brown scar at the base | Present | [21] |
| 115 | H. juglandis | Juglans sp./ Juglandaceae | China, Yunnan | No information available | Absent | [165] |
| 116 | H. kakamegense | On dead attached twig of Uvariopsis congensis/ Annonaceae | Kenya | Conidiophores 250–550 × 8–12 μm, solitary, unbranched; Conidia 30–90 × 8–10 μm, in the broadest part, uniformly tapering to 2–4 μm wide at at the apex, solitary, simple straight or somewhat curved, obclavate, rostrate, subhyaline, smooth, 4–15-distoseptate | Absent | [166] |
| 117 | H. kalakadense | On dead unidentified twig | India/Tamil Nadu | Conidia 13–15 μm | Absent | [21] |
| 118 | H. kalopanacis | On dead wood of Kalopanax septemlobus/ Araliaceae | Russia/Primorye | No information available | Absent | [167] |
| 119 | H. kok-saghyz | In seeds of Taraxacum kok-saghyz/Asteraceae | Russia | No information available | Absent | [168] |
| 120 | H. kyllingae | Kyllinga sp./ Cyperaceae | Uganda | No information available | Absent | [169] |
| 121 | H. lablab | On leaves of Dolichos lablab/Fabaceae | China/Taiwan | No information available | Absent | [130] |
| 122 | H. leucadendri | On leaves of Leucadendron sp./ Proteaceae | South Africa/Western Cape Province, Helderberg Nature Reserve | On MEA and PDA Conidiophores 100–300 × 4–6(–7) μm, erect, subcylindrical, thick-walled, medium brown, multiseptate; Conidia (35–)70–110(–170) × (6–)7–8(–11) μm, obclavate to subcylindrical, straight to slightly curved, thick-walled, medium brown, (3–)4–6(–10)-distoseptate | Present | [21] |
| 123 | H. leucosykes | On Meliola, on leaves of Leucosyke capitellata/ Urticaceae | Philippines | Conidiophores 300 × 7–8 μm, erect, brown, septate; Conidia 30 × 8 μm, 3-distoseptate | Absent | [156] |
| 124 | H. ligustri | On dead branches of Ligustrum quihoui/ Oleaceae | China/Guangxi, Nanning | Conidiophores 127–700 μm long, 9.5–18 μm diam just above the base and 8.5–10 μm diam towards the apex, solitary, simple, straight or flexuous, smooth or verruculose, thickwalled, dark brown, with 1–3 well-defined small pores at the apex, 1–4 septate; Conidia 24–38.5 × 9.5–13 μm, obclavate, straight or slightly curved, rostrate or pseudorostrate, smoothwalled, pale brown, subhyaline towards the apex, 4–6-distoseptate, with a large dark blackish-brown scar at the base, 1–2 μm thick | Absent | [128] |
| 125 | H. litseae | Litsea polyantha/ Lauraceae | India/Assam | No information available | Absent | [170] |
| 126 | H. livistonae | On leaves of Livistona australis/Arecaceae | Australia/New South Wales, Murramarang National Park | Conidiophores 500 × 4–6 μm, erect, flexuous, cylindrical, smooth to rough-walled, medium brown, multiseptate; Conidia (25–)40–55(–65) × (7–)8–9 μm, subcylindrical, straight, smooth, medium brown, apex obutuse, base somewhat obconic, (3–)4–6(–7)-distoseptate | Present | [171] |
| 127 | H. longisinuatum | Palmae rotten trunk | Peru | Conidiophores 20–75 × 3.5–5 μm; Conidia 65–220(–1000) × 8–10.5 μm, solitary, long, narrowly obclavate, 9–22-distoseptate | Absent | [109] |
| 128 | H. lonicerae | On Lonicera sp./ Caprifoliaceae | Brazil | No information available | Absent | [111] |
| 129 | H. lophirae | On leaves of Lophiraalata sp./Ochnaceae | Sierra Leone | Conidiophores 110–200 × 3–4 μm thick, simple, bluntly rounded ends; Conidia solitary 15–29 × 3.5–4.5 μm, oblong or oblong-cylindrical, hook or curved, smooth, olive- brown, 1–2 guttulate, 1–3-distoseptate | Absent | [172] |
| 130 | H. lunzinense | No information available | No information available | No information available | Absent | [173] |
| 131 | H. lusitanicum | On Alnus glutinosa/ Betulaceae | Portugal | No information available | Absent | [174] |
| 132 | H. lycopersici | On Solanum lycopersicum/Solanaceae | Guinea | No information available | Absent | [175] |
| 133 | H. machaerii | On Machaerium sp./ Fabaceae | Brazil | No information available | Absent | [111] |
| 134 | H. macilentum | On rotten wood | UK/Great Britain | Conidiophores erect, simple, fusiform, 7–10 septate; Conidia 0.5-0.65 × 0.1 mm | Absent | [176] |
| 135 | H. magnisporum | On dead fallen branches of an unknown woody plant | Japan | Conidiophores 150–270 μm long, 9.5–13 μm thick at the apex, 8.5–13.5 μm thick at the base, single or fasciculate, straight or flexuous, smooth walled, brown to dark brown, sometimes paler toward the apex, septate; Conidia 100–203 × 12.5–22.5 μm tapering gradually to 2.5–5 μm thick near the apex, solitary, obclavate or rostrate, straight or flexuous, pale olive-brown to pale brown, paler toward the apex, 7–18-distoseptate, with a blackish-brown to black scar, 4–7 μm thick | Present | [177,178] |
| 136 | H. makilingense | On dead branches of Paramignya monophylla/ Rutaceae | Philippines | Conidiophores 400–600 × 7–9 μm, dense, erect, curved, brown, septate; Conidia 100–300 × 10–12 μm, obclavate, 12–18-distoseptate | Absent | [179] |
| 137 | H. manihotis | on living leaves of Manihot sp./ Euphorbiaceae | Brazil | Conidiophores 50–95 × 4–6 μm, 4–6 septate; Conidia 40–50 × 6–8 μm, vermiform, clavate to subfusoid, olives, 4–7-distoseptate | Absent | [180] |
| 138 | H. marantae | On leaves of Maranta arundinacea/ Marantaceae | China/Taiwan | No information available | Absent | [130] |
| 139 | H. massarinum | Berchemia racemose/ Rhamnaceae | Japan | Conidiophores 380–810 × 7–9 wide at the apex, 13.5–21 wide at the base μm, 15–25 septate; Conidia 17–56.5 × 5–9 μm, tretic, solitary or in short chains (5–6), obclavate, rostrate, pale brown, smooth, with or without guttules, 1–8-distoseptate | Present | [19] |
| 140 | H. mattiroloi | On branches of Sideroxylon oxyacantha/ Sapotaceae | Etiopia | No information available | Absent | [181] |
| 141 | H. mayaguezense | On culms and leaves of Paspalum conjugatum/ Poaceae | Puerto Rico | Conidiophores 300–500 × 18–22 μm; Conidia 135–155 × 35–45 μm, fusoid to clavate, 3–4-distoseptate | Absent | [182] |
| 142 | H. melastomacearum | On Meliolamelastomacearum, on Miconiaracemose/ Melastomataceae | Puerto Rico | Conidiophores 280 × 3 μm; Conidia 14–21 × 3.5–6 μm, ellipsoid, 3-distoseptate | Absent | [159] |
| 143 | H. meliae | On leaves of Melia azedarach/Meliaceae | Dominican Republic | Conidiophores 250–350 × 15–22 μm, simple, aggregated, branched, olive-brown to black, septate; Conidia 70–100 × 12–15 μm, elongate, fusoid, or clavate | Absent | [183] |
| 144 | H. melioloides | On leaves of Uvaria sp./Annonaceae | Philippines | Conidiophores 250–300 × 6–8 μm; Conidia 35–45 × 9–10 μm, obclavate, 3-distoseptate | Absent | [137] |
| 145 | H. microsorum | On twigs of Quercus ilex/ Fagaceae | England, Italy | Conidiophores 100–550 × 8–14 μm, fasciculate, simple, flexuous, cylindrical, smooth-walled, dark brown, with a pore at the apex and often 1–2, septate; Conidia 60–(114–)160 × 12–(17–)22 μm thick in broadest part, tapering to 4–10 μm near the apex, obclavate, smooth-walled, pale to mid golden-brown, 9–17-distoseptate, with 5–7 μm wide at the scar | Present | [184] |
| 146 | H. microsporum | From soil | India/Maharashtra | Conidiophores 234–468 × 10.8 μm, pale brown, 10–16 septate; Conidia 26–41 × 22 μm, fusoid, widest at the middle, brown, 2–7-distoseptate | Absent | [18] |
| 147 | H. minimum | On dead decorticatd branches | UK/Great Britain/England | Conidiophores erect, simple, septate; Conidia 12–14 × 3–4 μm, fusiform, obtuse at the ends, triseptate, scarcely constricted, hyaline | Absent | [185] |
| 148 | H. multiseptatum | On dead branches | China/Guangdong | Conidiophores 390–650 × 10–14 μm wide at the base, 7–9 μm at apex, simple, subcylindrical, straight or slightly flexuous, smooth-walled, brown to dark brown, paler towards the apex, with 1–3 well definded, small pores, 1–3 septate; Conidia 78–190 μm long, 11–16 μm thick in the widest part, narrowing toward the apex to 3–6 μm, tretic, straight or slightly flexuous, obclavate or whip-like, smooth-walled, pale brown paler toward the apex, 13–25-distoseptate, with a dark blackish-brown scar at the base | Absent | [129] |
| 149 | H. nadsonii | On fibers of Gossypium sp./ Malvaceae | Russia | No information available | Absent | [186] |
| 150 | H. nanjingense | On dead branches of an unidentified tree | China/Jiangsu, Nanjing | Conidiophores 250–470 × 6.9–7.7 μm, solitary or fasciculate, simple, straight or flexuous, thick-walled, sub-cylindrical, smooth, brown to dark brown, with well-defined small pores at the apex,1–4 septate; Conidia 64.5–170.5 μm long, 7.3–10.3 μm wide in the widest part, narrowing towards the apex to 5.0–6.8 μm wide, subulate or nearly whip-like, straight or curved, thin-walled, smooth, pale brown, 6–17-distoseptate, with a blackish-brown scar at the base, 1.4–2.7 μm thick | Present | [187] |
| 151 | H. naviculare | On leaves of Euphorbia sp./ Euphorbiaceae | Brazil/Tubarão | Conidiophores 6–8 μm thick, branched, often curved, yellow, septate; Conidia 50–84 × 11–16 μmnaviculiform, hyaline at length, very pale with brown | Absent | [188] |
| 152 | H. naviculatum | On dead herbaceous stems of Solidago sp./ Asteraceae | U.S.A./New York | No information available | Absent | [189] |
| 153 | H. newbouldiae | On leaves of Newbouldialaevis/ Bignoniaceae | Guinea | No information available | Absent | [190] |
| 154 | H. novae-zelandiae | On dead wood and bark of Vitex lucens/ Lamiaceae | New Zealand | Conidiophores 165 μm long, 4.8–7(–9) μm, erect, single or in groups, simple, or once-branched at the base, straight or flexuous, subcylindrical, brown to dark brown below, very pale brown to subhyaline above, 15 septate; Conidia 13.5–16.2 × 7.2–9.0 μm, solitary, obovoid, sometimes slightly, smooth, the 2 lower cells being brown and the distal cell paler with a dark band of wall overlying each septum, 2-distoseptate | Absent | [191] |
| 155 | H. obpyriforme | On dead branches of unidentified tree | China/Guangxi | Conidiophores 225–460 μm long, 9.5–13 μm diam just above the base and 6–8.5 μm diam towards the apex, arising singly from the upper cells of the stromata, simple, subcylindrical, straight or flexuous, dark brown, paler towards the apex, with well-defined small pores at the apex,1–3 septate; Conidia 47–74 μm long, 14–19 μm diam in the widest part, narrowing in diameter towards the apex to 2.5–5 μm, straight or slightly curved, obpyriform, smooth-walled, middle brown, paler towards the apex, 5–9-distoseptate, with a large dark blackish-brown scar at conidium base, 1–2 μm thick | Absent | [128] |
| 156 | H. ocoteae | On Meliola ocoteae, on Guareatrichilioides | Puerto Rico | Conidiphores 135–200 × 4 μm, septate; Conidia 20–28 × 4–6 μm, 3-distoseptate | Absent | [159] |
| 157 | H. oligosporum | Holotype of Sporidesmium olivaceum: on rotten branches of Tilia sp. Lectotype of Coryneumoligosporum, here designated: on rotten branches of Corylus sp. Epitype of Sporidesmiumolivaceum and of Coryneumoligosporum: on dead corticated twigs of Tiliacordata sp. | Austria, Czech Republic, Germany | From Epitype specimen [21] Conidiophores (17–)22–35(–46) × (8.0–)8.5–10.5(–11.5) μm, densely crowded, erect, simple, straight, cylindrical to slightly swollen at the apex, brown to dark brown, darker at the apex, smooth, 0–2 septate; Conidia (37–)59–80(–124) × (14.8–)15.8–18.0(–20.0) μm, tapering to 4–10.5 μm at the distal end, with 4–8 μm wide, dark brown to black scar at the base, obclavate, sometimes rostrate, straight or curved, smooth but occasionally wrinkled with age, pale brown to brown, paler toward the apex, 6–12(–16)-distoseptate | Present | [21,124] |
| 158 | H. olisipponense | Culture from the perithecia stage of Pyrenophora polytricha | - | No information available | Absent | [192] |
| 159 | H. oplismeni | On leaves of Oplismenus cotnpositus/Poaceae | China/Taiwan | No information available | Absent | [130] |
| 160 | H. orchidacearum | On leaves of Neottia ovata (=Listera ovata)/Orchidaceae | France | No information available | Absent | [193] |
| 161 | H. orthospermum | On rotten wood | U.S.A./New York | Conidiophores 50–60 × 5 μm, erect, simple, fasciculate, straight, dark, 3–4 septate; Conidia 60–80(–110) × 10–12 μm, cylindrical, straight, apex rounded, tuncated at base, 12–14-distoseptate | Absent | [194] |
| 162 | H. oryzae-microsporae | On Oryza sativa/Poaceae | Japan | No information available | Absent | [195] |
| 163 | H. ovoideum | On dead branches of tree | China/Jilin | Conidiophores 380–510 × 15–25 μm diam just above the base, 7.5–10 μm diam towards the apex, arising singly from the upper cells of the stromata, simple, subcylindrical, straight or flexuous, thick-walled, smooth, brown to dark brown, paler towards the apex, with 1–3 well-defined small pores at the apex, 1–6 septate; Conidia 27–61 × 13–21 μm diam in the widest part, narrowing towards the apex to 4.5–8.5 μm, straight, ovoid, to ellipsoidal, smooth-walled, moderately brown, paler towards the apex, 3–8-distoseptate, with a large dark blackish-brown scar at the base, 1.5–2.5 μm thick | Absent | [128] |
| 164 | H. pachystelae | On living leaves of Synsepalumm solo(=Pachystelam solo)/ Sapotaceae | Tanzania | Conidiophores 300–350 × 6–8 μm, erect, simple, septate; Conidia 35–50 × 10–13 μm, fusoid or oblong clavate or lanceolate, 3-5-distoseptate | Absent | [196] |
| 165 | H. palaestinum | On stems and flowers of Dianthus sp./ Caryophyllaceae | Israel | Conidiophores 30–160 × 6–8 μm, fasciculate, 8–16 aggregate, simple, bent, thick-walled, coffin terminal obtuse, thin, yellow or colorless, 5–7 septate; Conidia 60–120 × 9–12 μm, solitary, obclavate, rectiusculis or curved, pale-olive, minute-granule, thick-walled, towards colorless above, 5–7-distoseptate | Absent | [197] |
| 166 | H. palmigenum | On rotten fruit of Cocos nucifera/Arecaceae; On petriole and rachis from reference specimen | Brasil/Pará, Papua New Guinea | From reference specimen [190]; Conidiophores 132.5–195 × 5–6 μm, solitary, erect, simple, cylindrical, straight or flexuous, smooth, brown, light brown at the apex, 7–10 septate; Conidia 38–53 × 8–11 base, 3–4 μm apex, solitary, in small chains, obclavate or cylindrical, straight or slightly curved, simple, smooth, brown with light brown at apical cell, 6–10-distoseptate | Absent | [198,199] |
| 167 | H. panici | On leaves of Panicum maximum/Poaceae | Indonesia/Java | Conidiophores 115–180 × 8–10 μm; Conidia (35–)50–75 × (7–)10–13 μm, ellipsoidal-truncate, ellipsoidal-elongate, dull-brown, (1–)3(–4)-distoseptate | Absent | [159] |
| 168 | H. papulosum | On bark of Malus sylvestris or Pyrus communis/Rosaceae | West Virginia | No information available | Absent | [200] |
| 169 | H. parathesicola | On Meliola parathesicola, on Parathesis serrulate/Primulaceae | Puerto Rico | Conidiophores 120 × 4 μm, solitary; Conidia 17–20 × 4–6 μm, base truncate, apex beaked, beak often 7 μm long, 1–3-distoseptate | Absent | [159] |
| 170 | H. paulense | On leaves of Myrtaceae | Brazil/São Paulo | Conidiophores 3–4.5 μm thick, brown, septate; Conidia 15–24 × 4 μm, fusoid, brown, 3-distoseptate | Absent | [115] |
| 171 | H. penniseti | On leaves of Pennisetum glaucum (=Pennisetum typhoides)/Poaceae | India/Uttar Pradesh | No information available | Absent | [108] |
| 172 | H. philippinum | On decaying leaves of Arenga mindorensis/ Arecaceae | Philippines | Conidiophores 300–400 × 6–7 μm, fasciculate, filiform, curved, septate; Conidia 33–35 × 8–9 μm, obclavate, 4-distoseptate | Absent | [137] |
| 173 | H. philodendri | On Meliola philodendri, on Philodendron krebsii/Araceae | Puerto Rico | Conidiphores 400 × 3–4 μm; Conidia 24–35 × 5–9 μm, clavate, 3-distoseptate | Absent | [159] |
| 174 | H. phomatae | On bark of Acer pennsylvanicum/ Sapindaceae | U.S.A./New York | No information available | Absent | [189] |
| 175 | H. phyllantheum | On dead branches hanging down of Phyllanthus sp./Phyllanthaceae | Philippines | Conidiophores 180–200 × 4.7–6 μm, fillifrom, blackedned, septate; Conidia 80–90 × 9–10 μm, obclavate, long, 9–11-distoseptate | Absent | [137] |
| 176 | H. piperis | On leaves of Piper betle/Piperaceae | China/Taiwan | No information available | Absent | [130] |
| 177 | H. portoricense | On dead branches hanging down of Phyllanthus sp./Phyllanthaceae | Philippines | Conidiophores 25–250 × 2–5 μm; Conidia 30–60 × 6–10 μm, elongate-fusoid, olive-brown or brown, (2–)4-distoseptate | Absent | [86,201] |
| 178 | H. proliferatum | On grain of Triticum sp./Poaceae | India/Maharashtra | Colony on PDA; Conidiophores 292–510 × 7–13.8 μm, unbranched, pale, olivaceous, 5–20 septate; Conidia 23–126 × 11.5–13.8 μm; cylindrical, olivaceous, 3–13-distoseptate | Absent | [101] |
| 179 | H. pseudomicrosorum | On dead branches of unidentified tree | China/Changbaishan, Jilin | Conidiophores 155–288 × 11–15 μm, fasciculate, simple, cylindrical, straight or flexuous, smooth, dark brown, paler towards the apex, with 1–3 well-defined small, 1–4 septate; Conidia 82–142 × 17–27 μm in the widest part, narrowing towards the apex to 3–6 μm diam, tretic, straight or slightly flexuous, obclavate, smooth-walled, brown, paler towards the apex, 7–16-distoseptate, with a large dark blackish-brown scar at the base, 2–4 μm thick | Absent | [128] |
| 180 | H. pseudotsugae | On bark and resin exudations of Pseudotsuga taxifolia var. glauca/Pinaceae | U.S.A. | Conidiophores scattered on aerial hyphae with usually one at each cell; Conidia 65–105 × 14–15 µm, opaque, black or greenish black, smooth walls, with 8–14-distoseptate | Absent | [202] |
| 181 | H. purpurascens | On leaves of Panicum purpurascens/Poaceae | U.S.A./Florida | No information available | Absent | [203] |
| 182 | H. pyracanthae | Pyracantha sp./Rosaceae | Portugal | No information available | Absent | [204] |
| 183 | H. quercicola | On dead corticated branches of Quercus cf. reticulata/Fagaceae | U.S.A. | Conidiophores (115–)133–226(–300) μm long, 14–20 μm wide at the base, tapering to 10–15 μm near the apex, solitarily or fasciculate, simple, straight or flexuous, cylindrical, thick-walled, smooth, brown to dark brown, with well-defined small pores at the apex; Conidia 60–100 × 15–22 μm, straight or flexuous, obclavate, smooth-walled, brown, 8–10-distoseptate, with blackish-brown to black scar at the base | Absent | [21] |
| 184 | H. quercinum | On dead corticated twigs of Quercus petraea/Fagaceae | Austria/Niederösterreich, Spitzerberg | Conidiophores (40–)74–199(–332) μm long, 11–18 μm wide at the base, tapering to 8.5–13.5 μm near the apex, solitarily or fasciculate, simple, straight or flexuous, cylindrical, smooth, brown to dark brown, with well-defined small pores at the apex, 1–5 septa; Conidia (47–)78–130(–201) × (13.2–)15.3–18.0(–20.5) μm, straight or flexuous, rostrate, smooth-walled, brown, 8–13(–20)-distoseptate, with blackish-brown to black scar at the base | Present | [21] |
| 185 | H. repens | On bark of dead Acer grandidentatum/ Sapindaceae | U.S.A./Utah, Red Butte Canyon | Conidia 40–45(–60) × 8–9 μm, sub-oblong, 5–12-distosepate | Absent | [205] |
| 186 | H. reyesii | On dead branch of Guioa sp./ Sapindaceae | Philippines | Conidiophores 130 × 8–10 μm, erect, brown, septate; Conidia 34–112 × 8–13 μm, tereti-fusoid, brown, ends hyaline, 5–14-distosepate | Absent | [137] |
| 187 | H. rhodomyrti | On leaves of Rhodomyrtus tomentosa/Myrtaceae | China/Guangdong | Conidia 42–60 × 17–20 μm, fusoid, brown, 5–7-distosepate | Absent | [206] |
| 188 | H. rhopaloides | On decraying stem of Brassica oleracea/Brassicaceae | Britain, France, Germany, Italy, Portugal | Conidiophores short, dark brown-black, 12–14 septate; Conidia 0.04–0.1 mm long, 0.08 mm wide, straight or slightly curved | Absent | [207,208] |
| 189 | H. schelkownikowii | On branches | Armenia, Azerbaijan, Georgia, Russia | No information available | Absent | [209] |
| 190 | H. scolecoides | On dry woood | Germany/Reichenberg | Conidiophores simple, branched; Conidia 80 × 7.5 μm, torulus, fusciculate, septate, yellow | Absent | [96] |
| 191 | H. sechiicola | On Sechium edule/ Cucurbitaceae | Puerto Rico | No information available | Absent Present | [210] |
| 192 | H. sichuanense | On dead branches of tree | China/Sichuan | No information available | Absent Present | [211] |
| 193 | H. solani | On stem of Solanum nigrum/Solanaceae (type); Citrus linella; Leucaena glauac; Solanum dulcamara; S. nigrum; Solanum tuberosum | England, Guernsey, New Zealand, New Guinea, Sierra Leone, Wales | Conidiophores 120–600 × 9–15 μm thick near base, 6–9 μm thick near the apex, erect, simple, straight or flexuous, smooth or occasionally, brown to very dark brown, paler near apex, septate, with small pores at apex, 1–8 septate; Conidia (24–)39–85 × (7–)9–11 μm, straight or curved, obclavated, smooth-walled, subhyaline to brown, 2–8-distosepate, with a welll-defind dark brown to black scar at base | Present | [212] |
| 194 | H. solitarium | On leaves of Iris sp./Iridaceae | U.S.A./Minnesota | Conidiophores 60–150 × 6 μm, solitary, slightly fasciculate, erect, swollen at the base, lighter colored at the apex, dark brown, septate; Conidia 24–30 × 8–9 μm, oblong-elliptical, sometimes slightly curved, dark brown, at first 2–4 guttulate, 3–5-distosepate | Absent | [213] |
| 195 | H. spirotrichum | On withered leaves of Cyrtophyllum fragrans/Gentianaceae | Singapore | Conidiophores 190–220 × 6 μm, fasciculate, filiform, brown, septate; Conidia 23–25 × 9 μm, oblong-obclavate, gently curving, brown, 3-distosepate | Absent | [214] |
| 196 | H. spurirostrum | On dead branches of tree | China/Sichuan | No information available | Absent | [211] |
| 197 | H. subapiculatum | On dead wood of Sambucus callicarpa/Adoxaceae | U.S.A./Washington | Conidiophores 8–10 μm thick; Conidia 35–80 × 12–16 μm, oblong or subfusiform, 6–7-distosepate | Absent | [215] |
| 198 | H. subhyalinum | On living leaves of Phoenix hanceana/Arecaceae | China/Guangdong | Conidiophores 120–200 × 10–12 (basal), above 6–8.5 μm thick, simple or fasciculate, erect, subcylindrical, brown, pores 1–3 μm, septate; Conidia 72–125 × 9–11.5 μm, obclavate, straight or flexuous, subhyaline, apex 2.5–5 μm, black at tip, 6–9-distosepate, dark blackish-brown scar | Absent | [129] |
| 199 | H. submersum | On submerged decaying wood | China/Yunnan | Conidiophores 239–423 × 8.5–15.5 μm, solitary or in group of 2–4, unbranched, straight or curved, smooth, dark brown, paler towards to the apex, bulbous at base 9–14 septate; Conidia 41–55 × 14.5–18.5 μm, straight or curved, wider below than apex, truncate and dark at base, apically rostrate and pale, smooth, pale brown to mid-brown, guttulate, 6–10-distosepate | Present | [24] |
| 200 | H. subsimile | On withered and dead leaves of Bruguiera sexangula (=Bruguiera eriopetala)/Rhizophoraceae | Singapore | Conidiophores 200–250 × 8–9 μm, filiform-fasciculate, brown, septate; Conidia (38–)45–50 × 11–12(–14) μm, brown, 3-distoseptate | Absent | [216] |
| 201 | H. syzygii | On bark canker of Syzygium sp./Myrtaceae | South Africa/Eastern Cape Province | Conidiophores 150–400 × 10–15 mm, fasciculate, unbranched, clavate at apex, dark brown, multiseptate; Conidia (70–)80–100(–150) × (19–)22–23(–25) mm, obclavate, curved, apex subobtuse, warty, inner surface striate, medium brown, (7–)9–12-distoseptate | Present | [22] |
| 202 | H. theobromae | On leaves of Theobroma cacao/Malvaceae | Italy | Conidiophores 500–1000 μm, erect, 6–10 septate; Conidia 60–160 × 12–20 μm, obclavate to tereti-obclavate | Absent | [217] |
| 203 | H. theobromicola | On rotten leaves of Theobroma cacao/Malvaceae | Dominican Republic/Moca | Conidiophores 20–33 × 3.5–5 μm olives-brown, septate; Conidia 46–58 × 10–13.5 μm, elliptic-oblong or subfusoid, irregular, 3–5-distoseptate | Absent | [218] |
| 204 | H. tritici | On seedhead of Triticum vulgare/Poaceae | Tanzania | Conidiophores 3.5–5 μm, thick fasciculate, erect, sepate; Conidia 12–25 × 4–7 μm, subcylindrical-oblong, clavate or fusoid, 2–4-distoseptate, constrict at septum | Absent | [219] |
| 205 | H. tritikernelis | On kernels of Triticum aestivum/Poaceae | India/Bihar | No information available | Absent | [108] |
| 206 | H. turbinatum | On unidentified wood | Great Britain | Conidiophores simple, slender; Conidia elongated, turbinatis, tuncatus, apiculate, brown, 4–7-distoseptate | Absent | [220] |
| 207 | H. ubangiense | On leaves of Coffea sp./Rubiaceae | Democratic Republic of the Congo/Ubangi River | Conidiophores (2–)3–6 μm, fasciculate, erect, branched, septate; Conidia 30–60 × 5–8 μm, fusoid, 3–4-distoseptate | Absent | [221] |
| 208 | H. ustilaginoideum | On flowers of Panicum spicatum/Poaceae | Democratic Republic of the Congo | Conidiophores 3–3.5 μm thick, fasciculate; Conidia 10–50 × 3.5–4.5 μm, cylindrical or subfusoid, blunted, 1–5-distoseptate | Absent | [121] |
| 209 | H. varium | On decaying leaves of unidentified plants | Brazil/Pernambuco | Conidiophores 150–200 × 10–14 μm, erect, unbranched, straight or flexuous, cylindrical, slightly inflated at the apex, smooth, brown, 5–7 septate; Conidia 29–58 × 4–7 μm, cylindrical-obclavate, subcylindrical, oblong or navicular, dry, trick-walled, with wall verrucose or verruculose, gray-brown, lumina pale yellow, (0–)1–4-distoseptate | Absent | [222] |
| 210 | H. varroniae | On leaves of Varronia sp./Boraginaceae | Puerto Rico | Conidiophores 160–200 × 4 μm; Conidia 27–44 × 6–7 μm, 3-distoseptate | Absent | [223] |
| 211 | H. velutinum | Fagus sylvatica dead corticated twigs/saprobic on decaying wood submerged in stream | Austria, Wien, Döbling, Kahlenberg/China | From reference specimen [20]; Conidiophores 530–655 × 16–18 μm, erect or flexuousk, unbranched, dark brown, 17–23 septate; Conidia 67–79 × 15–19 μm, single, obclavate, straight or curved, smooth, pale brown to brown, 7–9-distoseptate, rounded at apex, guttulate when young, non-guttulate at maturity | Present | [16,20] |
| 212 | H. viticis | On leaves of Vitis sp./Vitaceae | Brazil/Pará | Conidiophores 80 × 2–3 µm, fasciculate, septate; Conidia 12–20 × 2.5–3.5 µm cylindrical, 1–3-distoseptate | Absent | [223] |
| 213 | H. wagateae | On leaves of Moullava spicata (=Wagatea spicata)/Fabaceae | India/Karnataka | Conidiophores 81–125 × 1.5 –2.5 µm, yellowish-brown, multiseptate; Conidia 15.5–28 × 3–4 µm, clavate-cylindric, cinnamon-yellow, rounded at both ends, 2–4-distoseptate | Absent | [91] |
| 214 | H. warpuriae | On stem of Warpuria clandestina/Acanthaceae | Great Britain/England | Conidiophores 300–500 × 6–8 µm; Conidia 115–190 × 12–14 µm, obclavate, 8–11-distoseptate | Absent | [224] |
| 215 | H. xanthosomatis | On leaves of Xanthosoma violaceum/Araceae | Dominican Republic/Moca | Conidiophores 35–90 µm long, septate; Conidia 185 × 24 µm, fusoid, subfusoid to subclavate, 3–7(-10)-distoseptate | Absent | [225] |
| 216 | H. xylopiifolii | On Asterina, on Xylopia sericea/Annonaceae | Brazil/Pernambuco | Conidiophores 85–305 × 5.5–8 µm, erect, 3–5 septate; Conidia 38–62 × 8–13.5 µm, cylindrical or clavate, 3–6-distoseptate | Absent | [226] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Konta, S.; Hyde, K.D.; Karunarathna, S.C.; Mapook, A.; Senwanna, C.; Dauner, L.A.P.; Nanayakkara, C.M.; Xu, J.; Tibpromma, S.; Lumyong, S. Multi-Gene Phylogeny and Morphology Reveal Haplohelminthosporium gen. nov. and Helminthosporiella gen. nov. Associated with Palms in Thailand and A Checklist for Helminthosporium Reported Worldwide. Life 2021, 11, 454. https://doi.org/10.3390/life11050454
Konta S, Hyde KD, Karunarathna SC, Mapook A, Senwanna C, Dauner LAP, Nanayakkara CM, Xu J, Tibpromma S, Lumyong S. Multi-Gene Phylogeny and Morphology Reveal Haplohelminthosporium gen. nov. and Helminthosporiella gen. nov. Associated with Palms in Thailand and A Checklist for Helminthosporium Reported Worldwide. Life. 2021; 11(5):454. https://doi.org/10.3390/life11050454
Chicago/Turabian StyleKonta, Sirinapa, Kevin D. Hyde, Samantha C. Karunarathna, Ausana Mapook, Chanokned Senwanna, Lucas A. P. Dauner, Chandrika M. Nanayakkara, Jianchu Xu, Saowaluck Tibpromma, and Saisamorn Lumyong. 2021. "Multi-Gene Phylogeny and Morphology Reveal Haplohelminthosporium gen. nov. and Helminthosporiella gen. nov. Associated with Palms in Thailand and A Checklist for Helminthosporium Reported Worldwide" Life 11, no. 5: 454. https://doi.org/10.3390/life11050454
APA StyleKonta, S., Hyde, K. D., Karunarathna, S. C., Mapook, A., Senwanna, C., Dauner, L. A. P., Nanayakkara, C. M., Xu, J., Tibpromma, S., & Lumyong, S. (2021). Multi-Gene Phylogeny and Morphology Reveal Haplohelminthosporium gen. nov. and Helminthosporiella gen. nov. Associated with Palms in Thailand and A Checklist for Helminthosporium Reported Worldwide. Life, 11(5), 454. https://doi.org/10.3390/life11050454








