Mitochondrial Genomic Landscape: A Portrait of the Mitochondrial Genome 40 Years after the First Complete Sequence
Abstract
:1. Introduction
- (i)
- Multipartite genomes. Multipartite mtDNAs are, in fact, widespread among eukaryotes [15,18]. The mtDNA of Trypanosoma brucei (Euglenozoa: Kinetoplastea) is organized as a kinetoplast, a compact network of maxicircles (~25 kb) and thousands of minicircles (~1 kb), where mitochondrial genes and regulatory small RNAs are located, respectively ([18,19,20,21] and references therein). The mtDNA in other euglenozoans, Diplonemea, is composed of dozens of circular chromosomes; they can be subdivided into two size classes, with chromosomes of the same class sharing approximately 95% of the sequence. The remainder constitutes the only coding region of the chromosome, where one or more exons are located, ranging from 40 to 540 bp in length and relying on a complex trans-splicing and post-transcriptional machinery [19,22]. Mitochondrial genomes from Alveolata, and specifically of dinoflagellates, are also highly fragmented and possibly constitute the most divergent mitochondrial genomes among eukaryotes along with diplonemeans [23,24].
- (ii)
- Chromosome architecture. Many examples are currently known of linear mtDNA [18,59]. Moreover, mtDNA is not always organized as a single chromosome; many species with multipartite mitochondrial genomes have been identified. Among Metazoans, linear chromosomes are known to be present in mammals with a wide array of concatenated forms ([60] and references therein); all medusozoans (cnidarians, excluding Anthozoa) analyzed so far show linear mtDNAs, which are further subdivided into multiple chromosomes in Hydrozoa and Cubozoa [40,42,43,61,62,63]. Linear, multipartite mtDNAs are also known to exist in calcareous sponges [13,40,41].
- (iii)
- Genome size. Genome size is highly variable among eukaryotes, ranging from 6 kb in apicomplexan [23,24,72,73] and <13 kb in some green algae [77], ctenophores [78,79], and some fungi [80]; through 43 kb in placozoans [78,81] and >70 kb in choanoflagellates and ciliates [57,76]; up to >200 kb in other green algae and fungi [82,83,84], and 11 Mb in flowering plants [38]. Moreover, phenomena of the punctuated expansion of mtDNA have been reported within clades with generally reduced genome size (e.g., frogs [85], ark shells [86,87]). In most cases, this variability is not related to gene content; rather, the expansion and reduction of the intergenic region appear to be the main drivers of genome size among eukaryotes (e.g., [38,56,58,82,83,88,89]).
- (iv)
- Gene content. Only three genes are located in the mtDNA of apicomplexans and their relatives [90,91,92], as well as in dinoflagellates [23,24]; only a dozen genes are encoded in euglenozoans’ mtDNAs [19], but up to ~100 have been identified in jakobids. The order Jakobida is included in the eukaryotic supergroup Discoba (see [93,94,95] and references therein); jakobids have been found to have up to ~100 mitochondrially-encoded genes [96], and, to the best of our knowledge, Andalucia godoyi has the most gene-rich mtDNA [97]. The choanoflagellate Monosiga brevicollis has an intermediate gene complement of 55 genes [57], while there are 47 for the ichthyosporean parasite Sphaerothecum destruens [98]. Conversely, a relatively constant gene content is known to be present in fungi and animals [56].
2. Materials and Methods
3. Results and Discussion
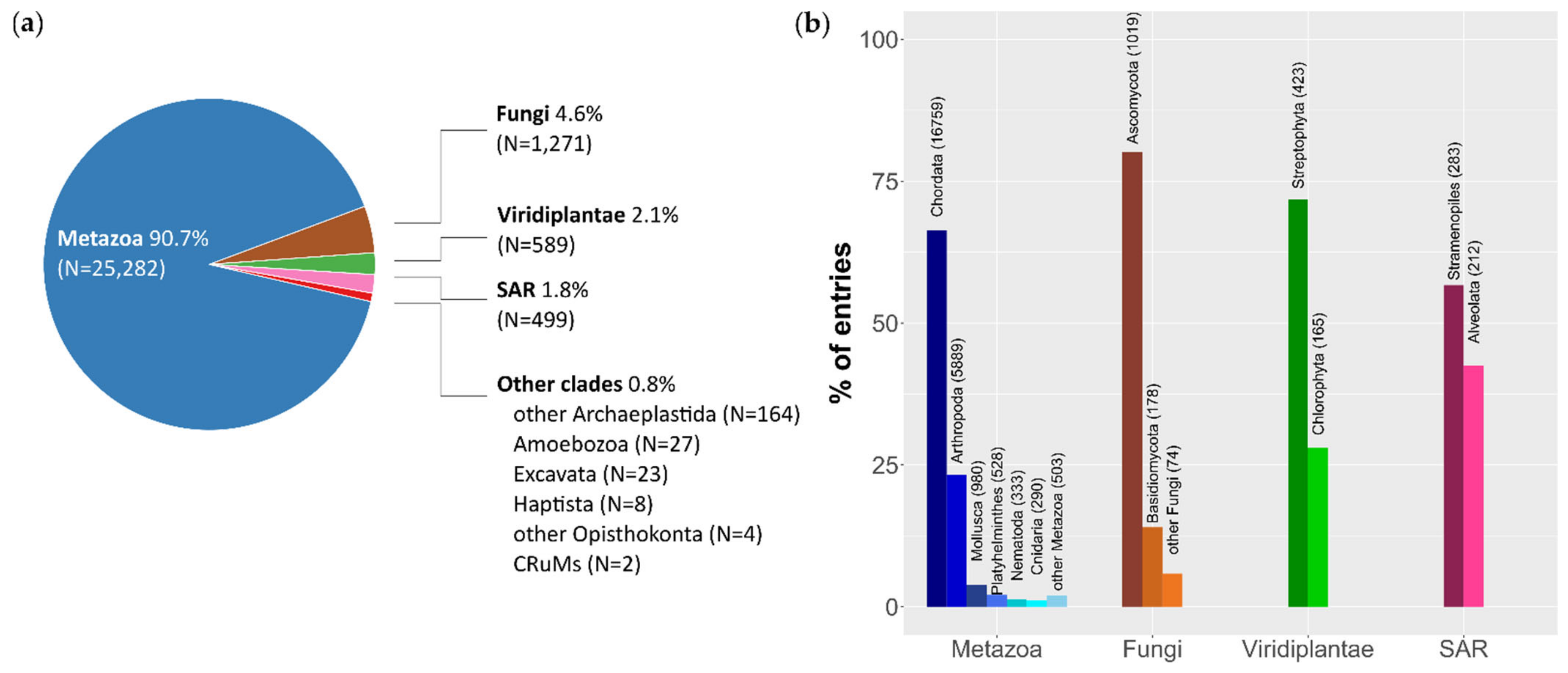
3.1. Dataset Composition
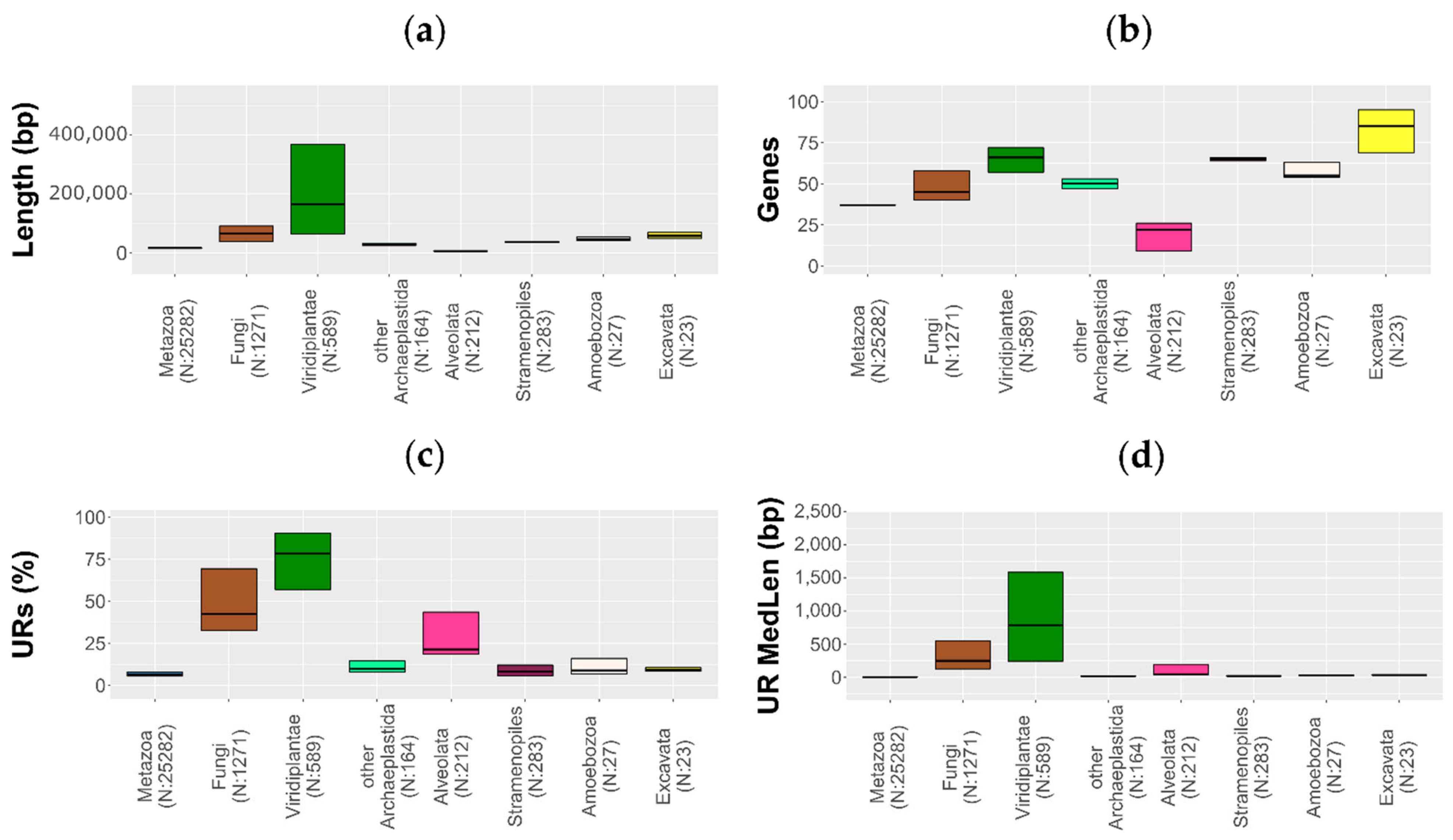
3.2. Mitogenome Reduction and Expansion
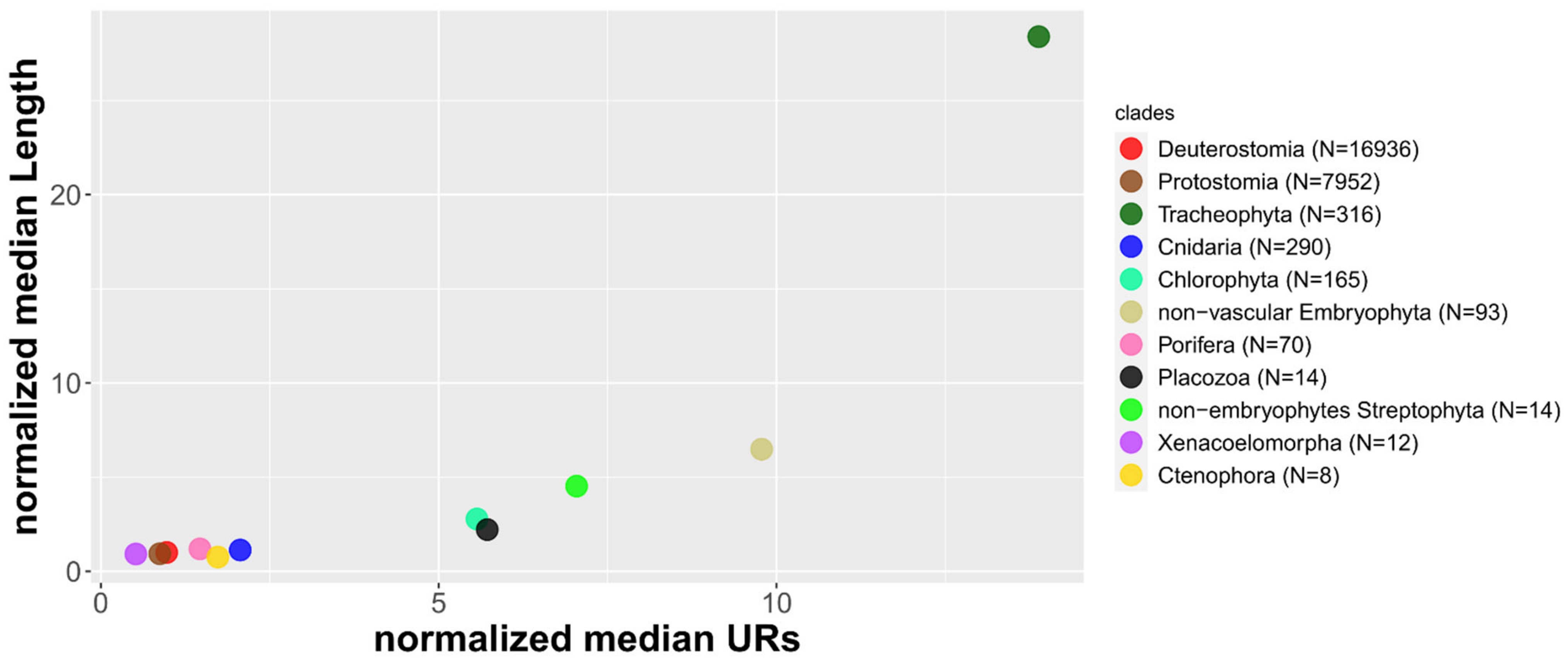
3.3. The Strand Asymmetry in Eukaryota
3.4. Codon Adaptation and A+T Content
4. Conclusions and Final Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sinclair, J.H.; Stevens, B.J. Circular DNA filaments from mouse mitochondria. Proc. Natl. Acad. Sci. USA 1966, 56, 508–514. [Google Scholar] [CrossRef] [Green Version]
- Van Bruggen, E.F.J.; Borst, P.; Ruttenberg, G.J.C.M.; Gruber, M.; Kroon, A.M. Circular mitochondrial DNA. Biochim. Biophys. Acta 1966, 119, 437–439. [Google Scholar] [CrossRef]
- Borst, P.; Kroon, A. Mitochondrial DNA: Physicochemical properties, replication, and genetic function. Int. Rev. Cytol. 1969, 26, 107–190. [Google Scholar]
- Williamson, D. The curious history of yeast mitochondrial DNA. Nat. Rev. Genet. 2002, 3, 1–7. [Google Scholar] [CrossRef]
- Altmann, R. Die Elementarorganismen und ihre Beziehungen zu den Zellen; Veit: Leipzig, Germany, 1890. [Google Scholar]
- Suyama, Y.; Miura, K. Size and structural variations of mitochondrial DNA. Proc. Natl. Acad. Sci. USA 1968, 60, 235–242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bendich, A.J. Reaching for the ring: The study of mitochondrial genome structure. Curr. Genet. 1993, 24, 279–290. [Google Scholar] [CrossRef]
- Hollenberg, C.P.; Borst, P.; Thuring, R.W.J.; Van Bruggen, E.F.J. Size, structure and genetic complexity of yeast mitochondrial DNA. Biochim. Biophys. Acta 1969, 186, 417–419. [Google Scholar] [CrossRef]
- Anderson, S.; Bankier, A.T.; Barrell, B.G.; de Bruijn, M.H.L.; Coulson, A.R.; Drouin, J.; Eperon, I.C.; Nierlich, D.P.; Roe, B.A.; Sanger, F.; et al. Sequence and organization of the human mitochondrial genome. Nature 1981, 290, 457–465. [Google Scholar] [CrossRef] [PubMed]
- Bibb, M.J.; Van Etten, R.A.; Wright, C.T.; Walberg, M.W.; Clayton, D.A. Sequence and gene organization of mouse mitochondrial DNA. Cell 1981, 26, 167–180. [Google Scholar] [CrossRef]
- Anderson, S.; de Bruijn, M.H.; Coulson, A.R.; Eperon, I.C.; Sanger, F.; Young, I.G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J. Mol. Biol. 1982, 156, 683–717. [Google Scholar] [CrossRef]
- Clary, D.O.; Wolstenholme, D.R. The Drosophila mitochondrial genome. Oxf. Surv. Eukaryot. Genes 1984, 1, 1–35. [Google Scholar]
- Lavrov, D.V.; Pett, W.; Voigt, O.; Wörheide, G.; Forget, L.; Lang, B.F.; Kayal, E. Mitochondrial DNA of Clathrina clathrus (Calcarea, Calcinea): Six linear chromosomes, fragmented rRNAs, tRNA editing, and a novel genetic code. Mol. Biol. Evol. 2013, 30, 865–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karnkowska, A.; Vacek, V.; Zubáčová, Z.; Treitli, S.C.; Petrželková, R.; Eme, L.; Novák, L.; Žárský, V.; Barlow, L.D.; Herman, E.K.; et al. A Eukaryote without a mitochondrial organelle. Curr. Biol. 2016, 26, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.; Keeling, P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.; Mentel, M.; van Hellemond, J.J.; Henze, K.; Woehle, C.; Gould, S.B.; Yu, R.Y.; van der Giezen, M.; Tielens, A.G.M.; Martin, W.F. Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 2012, 76, 444–495. [Google Scholar] [CrossRef] [Green Version]
- Hjort, K.; Goldberg, A.V.; Tsaousis, A.D.; Hirt, R.P.; Embley, T.M. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos. Trans. R Soc. Lond. B Biol. Sci. 2010, 365, 713–727. [Google Scholar] [CrossRef] [PubMed]
- Nosek, J.; Tomáska, L. Mitochondrial genome diversity: Evolution of the molecular architecture and replication strategy. Curr. Genet. 2003, 44, 73–84. [Google Scholar] [CrossRef] [PubMed]
- Burger, G.; Valach, M. Perfection of Eccentricity: Mitochondrial Genomes of Diplonemids. IUBMB Life 2018, 70, 1197–1206. [Google Scholar] [CrossRef] [Green Version]
- Lukeš, J.; Wheeler, R.; Jirsová, D.; David, V.; Archibald, J.M. Massive mitochondrial DNA content in diplonemid and kinetoplastid protists. IUBMB Life 2018, 70, 1267–1274. [Google Scholar] [CrossRef] [Green Version]
- Jensen, R.E.; Englund, P.T. Network news: The replication of kinetoplast DNA. Annu Rev. Microbiol. 2012, 66, 473–491. [Google Scholar] [CrossRef]
- Kaur, B.; Záhonová, K.; Valach, M.; Faktorová, D.; Prokopchuk, G.; Burger, G.; Lukeš, J. Gene fragmentation and RNA editing without borders: Eccentric mitochondrial genomes of diplonemids. Nucleic Acids Res. 2020, 48, 2694–2708. [Google Scholar] [CrossRef] [PubMed]
- Waller, R.F.; Jackson, C.J. Dinoflagellate mitochondrial genomes: Stretching the rules of molecular biology. BioEssays 2009, 31, 237–245. [Google Scholar] [CrossRef]
- Slamovits, C.H.; Saldarriaga, J.F.; Larocque, A.; Keeling, P.J. The highly reduced and fragmented mitochondrial genome of the earlybranching dinoflagellate Oxyrrhis marina shares characteristics with both apicomplexan and dinoflagellate mitochondrial genomes. J. Mol. Biol. 2007, 372, 356–368. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Waneka, G.; Sloan, D.B. The Tempo and Mode of Angiosperm Mitochondrial Genome Divergence Inferred from Intraspecific Variation in Arabidopsis thaliana. G3 2020, 10, 1077–1086. [Google Scholar] [CrossRef] [Green Version]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15, e1008373. [Google Scholar] [CrossRef] [Green Version]
- Gualberto, J.M.; Newton, K.J. Plant mitochondrial genomes: Dynamics and mechanisms of mutation. Annu Rev. Plant. Biol. 2017, 68, 225–252. [Google Scholar] [CrossRef]
- McCauley, D.E. Paternal leakage, heteroplasmy, and the evolution of plant mitochondrial genomes. New Phytol. 2013, 200, 966–977. [Google Scholar] [CrossRef] [PubMed]
- Bendich, A.J. The size and form of chromosomes are constant in the nucleus, but highly variable in bacteria, mitochondria and chloroplasts. BioEssays 2007, 29, 474–483. [Google Scholar] [CrossRef]
- Barr, C.M.; Neiman, M.; Taylor, D.R. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 2005, 168, 39–50. [Google Scholar] [CrossRef]
- Abdelnoor, R.V.; Yule, R.; Elo, A.; Christensen, A.C.; Meyer-Gauen, G.; Mackenzie, S.A. Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl. Acad. Sci. USA 2003, 100, 5968–5973. [Google Scholar] [CrossRef] [Green Version]
- Sloan, D.B. One ring to rule them all? Genome sequencing provides new insights into the ‘master circle’ model of plant mitochondrial DNA structure. New Phytol. 2013, 200, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P.; Case, A.L.; Floro, E.R.; Willis, J.H. Evidence against equimolarity of large repeat arrangements and a predominant master circle structure of the mitochondrial genome from a monkeyflower (Mimulus guttatus) lineage with cryptic CMS. Genome Biol. Evol. 2012, 4, 670–686. [Google Scholar] [CrossRef] [Green Version]
- Backert, S.; Borner, T. Phage T4-like intermediates of DNA replication and recombination in the mitochondria of the higher plant Chenopodium album (L.). Curr. Genet. 2000, 37, 304–314. [Google Scholar] [CrossRef]
- Bendich, A.J. Structural analysis of mitochondrial DNA molecules from fungi and plants using moving pictures and pulsedfield gel electrophoresis. J. Mol. Biol. 1996, 255, 564–588. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Shields, C.R. Tripartite structure of the Brassica campestris mitochondrial genome. Nature 1984, 307, 437–440. [Google Scholar] [CrossRef]
- Preuten, T.; Cincu, E.; Fuchs, J.; Zoschke, R.; Liere, K.; Börner, T. Fewer genes than organelles: Extremely low and variable gene copy numbers in mitochondria of somatic plant cells. Plant. J. 2010, 64, 948–959. [Google Scholar] [CrossRef]
- Sloan, D.B.; Alverson, A.J.; Chuckalovcak, J.P.; Wu, M.; McCauley, D.E.; Palmer, J.D.; Taylor, D.R. Rapid Evolution of Enormous, Multichromosomal Genomes in Flowering Plant Mitochondria with Exceptionally High Mutation Rates. PLoS Biol 2012, 10, e1001241. [Google Scholar] [CrossRef] [Green Version]
- Alverson, A.J.; Rice, D.W.; Dickinson, S.; Barry, K.; Palmer, J.D. Origins and recombination of the bacterial-sized multichromosomal mitochondrial genome of cucumber. Plant. Cell 2011, 23, 2499–2513. [Google Scholar] [CrossRef] [Green Version]
- Lavrov, D.V.; Pett, W. Animal Mitochondrial DNA as We Do Not Know It: Mt-Genome Organization and Evolution in Nonbilaterian Lineages. Genome Biol. Evol. 2016, 8, 2896–2913. [Google Scholar] [CrossRef]
- Lavrov, D.; Adamski, M.; Chevaldonné, P.; Adamska, M. Extensive mitochondrial mRNA editing and unusual mitochondrial genome organization in calcaronean sponges. Curr. Biol. 2016, 26, 86–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voigt, O.; Erpenbeck, D.; Wörheide, G. A fragmented metazoan organellar genome: The two mitochondrial chromosomes of Hydra magnipapillata. BMC Genom. 2008, 9, 350. [Google Scholar] [CrossRef] [Green Version]
- Kayal, E.; Bentlage, B.; Collins, A.G.; Kayal, M.; Pirro, S.; Lavrov, D.V. Evolution of linear mitochondrial genomes in medusozoan cnidarians. Genome Biol. Evol. 2012, 4, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, K.; Bessho, Y.; Kawasaki, M.; Hori, H. Mitochondrial genes are found on minicircle DNA molecules in the mesozoan animal Dicyema. J. Mol. Biol. 1999, 286, 645–650. [Google Scholar] [CrossRef]
- Witek, A.; Herlyn, H.; Ebersberger, I.; Mark Welch, D.B.; Hankeln, T. Support for the monophyletic origin of Gnathifera from phylogenomics. Mol. Phylogenet. Evol. 2009, 53, 1037–1041. [Google Scholar] [CrossRef] [PubMed]
- Suga, K.; Mark Welch, D.B.; Tanaka, Y.; Sakakura, Y.; Hagiwara, A. Two circular chromosomes of unequal copy number make up the mitochondrial genome of the rotifer Brachionus plicatilis. Mol. Biol. Evol. 2008, 25, 1129–1137. [Google Scholar] [CrossRef] [Green Version]
- Hwang, D.S.; Suga, K.; Sakakura, Y.; Park, H.G.; Hagiwara, A.; Rhee, J.S.; Lee, J.S. Complete mitochondrial genome of the monogonont rotifer, Brachionus koreanus (Rotifera, Brachionidae). Mitochondrial DNA 2014, 25, 29–30. [Google Scholar] [CrossRef]
- Gibson, T.; Blok, V.C.; Dowton, M. Sequence and characterization of six mitochondrial subgenomes from Globodera rostochiensis: Multipartite structure is conserved among close nematode relatives. J. Mol. Evol. 2007, 65, 308315. [Google Scholar] [CrossRef] [PubMed]
- Gibson, T.; Blok, V.C.; Phillips, M.S.; Hong, G.; Kumarasinghe, D.; Riley, I.T.; Dowton, M. The mitochondrial subgenomes of the nematode Globodera pallida are mosaics: Evidence of recombination in an animal mitochondrial genome. J. Mol. Evol. 2007, 64, 463471. [Google Scholar] [CrossRef]
- Sweet, A.D.; Johnson, K.P.; Cameron, S.L. Mitochondrial genomes of Columbicola feather lice are highly fragmented, indicating repeated evolution of minicircle-type genomes in parasitic lice. PeerJ 2020, 8, e8759. [Google Scholar] [CrossRef] [Green Version]
- Johnson, K.P.; Nguyen, N.; Sweet, A.D.; Boyd, B.M.; Warnow, T.; Allen, J.M. Simultaneous radiation of bird and mammal lice following the K-Pg boundary. Biol. Lett. 2018, 14, 20180141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Y.; Chu, Q.; Wei, D.D.; Qui, Y.J.; Shang, F.; Dou, W.; Wang, J.J. The mitochondrial genome of booklouse, Liposcelis sculptilis (Psocoptera: Liposcelididae) and the evolutionary timescale of Liposcelis. Sci. Rep. 2016, 6, 30660. [Google Scholar] [CrossRef] [Green Version]
- Dickey, A.M.; Kumar, V.; Morgan, J.K.; Jara-Cavieres, A.; Shatters, R.G., Jr.; McKenzie, C.L.; Osborne, L.S. A novel mitochondrial genome architecture in thrips (Insecta: Thysanoptera): Extreme size asymmetry among chromosomes and possible recent control region duplication. BMC Genomics 2015, 16, 439. [Google Scholar] [CrossRef] [Green Version]
- Wei, D.D.; Shao, R.; Yuan, M.L.; Dou, W.; Barker, S.C.; Wang, J.J. The multipartite mitochondrial genome of Liposcelis bostrychophila: Insights into the evolution of mitochondrial genomes in bilateral animals. PLoS ONE 2012, 7, e33973. [Google Scholar] [CrossRef] [Green Version]
- Song, F.; Li, H.; Lio, G.H.; Wang, W.; James, P.; Colwell, D.D.; Tran, A.; Gong, S.; Cai, W.; Shao, R. Mitochondrial genome fragmentation unites the parasitic lice of eutherian mammals. Syst. Biol. 2019, 68, 430440. [Google Scholar] [CrossRef] [Green Version]
- Bullerwell, C.E.; Gray, M.W. Evolution of the mitochondrial genome: Protist connections to animals, fungi and plants. Curr. Opin. Microbiol 2004, 7, 528–534. [Google Scholar] [CrossRef]
- Burger, G.; Forget, L.; Zhu, Y.; Gray, M.W.; Lang, B.F. Unique mitochondrial genome architecture in unicellular relatives of animals. Proc. Natl. Acad. Sci. USA 2003, 100, 892–897. [Google Scholar] [CrossRef] [Green Version]
- Freel, K.C.; Friedrich, A.; Schacherer, J. Mitochondrial genome evolution in yeasts: An all-encompassing view. FEMS Yeast Res. 2015, 15, fov023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pohjoismäki, J.L.O.; Goffart, S. Of circles, forks and humanity: Topological organisation and replication of mammalian mitochondrial DNA. BioEssays 2011, 33, 290–299. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.; Keeling, P.J. Gene conversion shapes linear mitochondrial genome architecture. Genome Biol. Evol. 2013, 5, 905–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ender, A.; Schierwater, B. Placozoa are not derived cnidarians: Evidence from molecular morphology. Mol. Biol. Evol. 2003, 20, 130–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bridge, D.; Cunningham, C.W.; Schierwater, B.; Desalle, R.; Buss, L.W. Class-level relationships in the phylum Cnidaria: Evidence from mitochondrial genome structure. Proc. Natl. Acad. Sci. USA 1992, 89, 8750–8753. [Google Scholar] [CrossRef] [Green Version]
- Warrior, R.; Gall, J. The mitochondrial DNA of Hydra attenuata and Hydra littoralis consists of two linear molecules. Arch. Sci. Geneve 1985, 38, 439–445. [Google Scholar]
- Dujon, B. Mitochondrial genetics revisited. Yeast 2020, 37, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Valach, M.; Farkas, Z.; Fricova, D.; Kovac, J.; Brejova, B.; Vinar, T.; Pfeiffer, I.; Kucsera, J.; Tomaska, L.; Lang, B.F.; et al. Evolution of linear chromosomes and multipartite genomes in yeast mitochondria. Nucleic Acids Res. 2011, 39, 4202–4219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerhold, J.M.; Aun, A.; Sedman, T.; Jõers, P.; Sedman, J. Strand invasion structures in the inverted repeat of Candida albicans mitochondrial DNA reveal a role for homologous recombination in replication. Mol. Cell 2010, 39, 851–861. [Google Scholar] [CrossRef]
- Ling, F.; Shibata, T. Recombination-dependent mtDNA partitioning. In vivo role of Mhr1p to promote pairing of homologous DNA. EMBO J 2002, 21, 4730–4740. [Google Scholar] [CrossRef] [PubMed]
- Kosa, P.; Valach, M.; Tomaska, L.; Wolfe, K.H.; Nosek, J. Complete DNA sequences of the mitochondrial genomes of the pathogenic yeasts Candida orthopsilosis and Candida metapsilosis: Insight into the evolution of linear DNA genomes from mitochondrial telomere mutants. Nucleic Acids Res. 2006, 34, 2472–2481. [Google Scholar] [CrossRef]
- Morin, G.B.; Cech, T.R. Mitochondrial telomeres: Surprising diversity of repeated telomeric DNA sequences among six species of Tetrahymena. Cell 1988, 52, 367–374. [Google Scholar] [CrossRef]
- Morin, G.B.; Cech, T.R. The telomeres of the linear mitochondrial DNA of Tetrahymena thermophila consist of 53 bp tandem repeats. Cell 1986, 46, 873–883. [Google Scholar] [CrossRef]
- Goddard, J.M.; Cummings, D.J. Mitochondrial DNA replication in Paramecium aurelia. Cross-linking of the initiation end. J. Mol. Biol. 1977, 109, 327–344. [Google Scholar] [CrossRef]
- Feagin, J.E.; Mericle, B.L.; Werner, E.; Morris, M. Identification of additional rRNA fragments encoded by the Plasmodium falciparum 6 kb element. Nucl Acids Res. 1997, 25, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Feagin, J.E. The 6-kb element of Plasmodium falciparum encodes mitochondrial cytochrome genes. Mol. Biochem. Parasitol. 1992, 52, 145–148. [Google Scholar] [CrossRef]
- Warren, J.M.; Simmons, M.P.; Wu, Z.; Sloan, D.B. Linear Plasmids and the Rate of Sequence Evolution in Plant Mitochondrial Genomes. Genome Biol. Evol. 2016, 8, 364–374. [Google Scholar] [CrossRef] [Green Version]
- Handa, H. Linear plasmids in plant mitochondria: Peaceful coexistences or malicious invasions? Mitochondrion 2008, 8, 15–25. [Google Scholar] [CrossRef]
- Swart, E.C.; Nowacki, M.; Shum, J.; Stiles, H.; Higgins, B.P.; Doak, T.G.; Schotanus, K.; Magrini, V.J.; Minx, P.; Mardis, E.R.; et al. The Oxytricha trifallax mitochondrial genome. Genome Biol. Evol. 2012, 4, 136–154. [Google Scholar] [CrossRef] [Green Version]
- Smith, D.R.; Hua, J.; Lee, R.W. Evolution of linear mitochondrial DNA in three known lineages of Polytomella. Curr. Genet. 2010, 56, 427–438. [Google Scholar] [CrossRef]
- Osigus, H.J.; Eitel, M.; Bernt, M.; Donath, A.; Schierwater, B. Mitogenomics at the base of Metazoa. Mol. Phylogenet. Evol. 2013, 69, 339–351. [Google Scholar] [CrossRef]
- Pett, W.; Ryan, J.F.; Pang, K.; Mullikin, J.C.; Martindale, M.Q.; Baxevanis, A.D.; Lavrov, D.V. Extreme mitochondrial evolution in the ctenophore Mnemiopsis leidyi: Insight from mtDNA and the nuclear genome. Mitochondrial DNA 2011, 22, 130–142. [Google Scholar] [CrossRef] [Green Version]
- Pramateftaki, P.V.; Kouvelis, V.N.; Lanaridis, P.; Typas, M.A. The mitochondrial genome of the wine yeast Hanseniaspora uvarum: A unique genome organization among yeast/fungal counterparts. FEMS Yeast Res. 2006, 6, 77–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernt, M.; Braband, A.; Schierwater, B.; Stadler, P.F. Genetic aspects of mitochondrial genome evolution. Mol. Phylogenet. Evol. 2013, 69, 328–338. [Google Scholar] [CrossRef] [Green Version]
- Araújo, D.S.; De-Paula, R.B.; Tomé, L.M.R.; Quintanilha-Peixoto, G.; Salvador-Montoya, C.A.; Del-Bem, L.-E.; Badotti, F.; Azevedo, V.A.C.; Brenig, B.; Aguiar, E.R.G.R.; et al. Comparative mitogenomics of Agaricomycetes: Diversity, abundance, impact and coding potential of putative open-reading frames. Mitochondrion 2021, 58, 1–13. [Google Scholar] [CrossRef]
- Repetti, S.I.; Jackson, C.J.; Judd, L.M.; Wick, R.R.; Holt, K.E.; Verbruggen, H. The inflated mitochondrial genomes of siphonous green algae reflect processes driving expansion of noncoding DNA and proliferation of introns. PeerJ 2020, 8, e8273. [Google Scholar] [CrossRef]
- Losada, L.; Pakala, S.B.; Fedorova, N.D.; Joardar, V.; Shabalina, S.A.; Hostetler, J.; Pakala, S.M.; Zafar, N.; Thomas, E.; Rodriguez-Carres, M.; et al. Mobile elements and mitochondrial genome expansion in the soil fungus and potato pathogen Rhizoctonia solani AG-3. FEMS Microbiol. Lett. 2020, 352, 165–173. [Google Scholar] [CrossRef] [Green Version]
- Hemmi, K.; Kakehashi, R.; Kambayashi, C.; Du Preez, L.; Minter, L.; Furuno, N.; Kurabayashi, A. Exceptional Enlargement of the Mitochondrial Genome Results from Distinct Causes in Different Rain Frogs (Anura: Brevicipitidae: Breviceps). Int J. Genomics 2020, 2020, 6540343. [Google Scholar] [CrossRef] [Green Version]
- Pu, L.; Liu, H.; Wang, G.; Li, B.; Xia, G.; Shen, M.; Yang, M. Complete mitochondrial genome of the cockle Anadara antiquata (Linnaeus, 1758). Mitochondrial DNA Part. B 2019, 4, 2293–2294. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kurokawa, T.; Sekino, M.; Tanabe, T.; Watanabe, K. Complete mitochondrial DNA sequence of the ark shell Scapharca broughtonii: An ultra-large metazoan mitochondrial genome. Comp. Biochem. Physiol. Part. D Genomics Proteomics 2013, 8, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Ghiselli, F.; Gomes-dos-Santos, A.; Adema, C.M.; Lopes-Lima, M.; Sharbrough, J.; Boore, J.L. Molluscan mitochondrial genomes break the rules. Phil. Trans. R Soc. B 2021, 376, 20200159. [Google Scholar] [CrossRef]
- Plazzi, F.; Puccio, G.; Passamonti, M. Comparative Large-Scale Mitogenomics Evidences Clade-Specific Evolutionary Trends in Mitochondrial DNAs of Bivalvia. Genome Biol. Evol. 2016, 8, 2544–2564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flegontov, P.; Michálek, J.; Janouškovec, J.; La, D.H.; Jirk, M.; Hajdušková, E.; Tomčala, A.; Otto, T.D.; Keeling, P.J.; Pain, A.; et al. Divergent mitochondrial ararespiratory chains in phototrophic relatives of apicomplexan parasites. Mol. Biol. Evol. 2015, 32, 1115–1131. [Google Scholar] [CrossRef]
- Rehkopf, D.H.; Gillespie, D.E.; Harrell, M.I.; Feagin, J.E. Transcriptional mapping and RNA processing of the Plasmodium falciparum mitochondrial mRNAs. Mol. Biochem. Parasitol. 2000, 105, 91–103. [Google Scholar] [CrossRef]
- Feagin, J.E. The extrachromosomal DNAs of apicomplexan parasites. Annu Rev. Microbiol. 1994, 48, 81–104. [Google Scholar] [CrossRef]
- Gray, M.W.; Burger, G.; Derelle, R.; Klimeš, V.; Leger, M.M.; Sarrasin, M.; Vlček, Č.; Roger, A.J.; Eliáš, M.; Lang, B.F. The draft nuclear genome sequence and predicted mitochondrial proteome of Andalucia godoyi, a protist with the most gene-rich and bacteria-like mitochondrial genome. BMC Biol. 2020, 18, 22. [Google Scholar] [CrossRef] [PubMed]
- Derelle, R.; Torruella, G.; Klimeš, V.; Brinkmann, H.; Kim, E.; Vlček, Č.; Lang, B.F.; Eliáš, M. Bacterial proteins pinpoint a single eukaryotic root. Proc. Natl Acad. Sci. USA 2015, 112, E693–E699. [Google Scholar] [CrossRef] [Green Version]
- Hampl, V.; Hug, L.; Leigh, J.W.; Dacks, J.B.; Lang, B.F.; Simpson, A.G.B.; Roger, A.J. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc. Natl. Acad. Sci. USA 2009, 106, 3859–3864. [Google Scholar] [CrossRef] [Green Version]
- Lang, B.F.; Burger, G.; O’Kelly, C.J.; Cedergren, R.; Golding, G.B.; Lemieux, C.; Sankoff, D.; Turmel, M.; Gray, M.W. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature 1997, 387, 493–497. [Google Scholar] [CrossRef]
- Burger, G.; Gray, M.W.; Forget, L.; Lang, B.F. Strikingly bacteria-like and gene-rich mitochondrial genomes throughout jakobid protists. Genome Biol Evol 2013, 5, 418–438. [Google Scholar] [CrossRef] [Green Version]
- Sana, S.; Hardouin, E.A.; Paley, R.; Zhang, T.; Andreou, D. The complete mitochondrial genome of a parasite at the animal-fungal boundary. Parasites Vectors 2020, 13, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solieri, L. Mitochondrial inheritance in budding yeasts: Towards an integrated understanding. Trends Microbiol 2010, 18, 521–530. [Google Scholar] [CrossRef]
- Foury, F.; Roganti, T.; Lecrenier, N.; Purnelle, B. The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett 1998, 32, 325–331. [Google Scholar] [CrossRef] [Green Version]
- Wolf, K.; Del Giudice, L. The variable mitochondrial genome of ascomycetes: Organization, mutations, alterations, and expression. Adv. Genet. 1988, 25, 185–308. [Google Scholar]
- Zamaroczy, M.; Bernardi, G. The primary structure of the mitochondrial genome of Saccharomyces cerevisiae—A review. Gene 1986, 47, 155–177. [Google Scholar] [CrossRef]
- Breton, S.; Milani, L.; Ghiselli, F.; Guerra, D.; Stewart, D.T.; Passamonti, M. A resourceful genome: Updating the functional repertoire and evolutionary role of animal mitochondrial DNAs. Trends Genet. 2014, 30, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [Green Version]
- Monnens, M.; Thijs, S.; Briscoe, A.G.; Clark, M.; Frost, E.J.; Littlewood, D.T.J.; Sewell, M.; Smeets, K.; Artois, T.; Vanhove, M.O.M. The first mitochondrial genomes of endosymbiotic rhabdocoels illustrate evolutionary relaxation of atp8 and genome plasticity in flatworms. Int J. Biol Macromol 2020, 162, 454–469. [Google Scholar] [CrossRef] [PubMed]
- Trindade Rosa, M.; Oliveira, D.S.; Loreto, E.L.S. Characterization of the first mitochondrial genome of a catenulid flatworm: Stenostomum leucops (Platyhelminthes). J. Zool Syst. Evol. Res. 2017, 55, 98–105. [Google Scholar] [CrossRef]
- Solà, E.; Álvarez-Presas, M.; Frías-López, C.; Littlewood, D.T.J.; Rozas, J.; Riutort, M. Evolutionary Analysis of Mitogenomes from Parasitic and Free-Living Flatworms. PLoS ONE 2015, 10, e0120081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sultana, T.; Kim, J.; Lee, S.H.; Han, H.; Kim, S.; Min, G.S.; Nadler, S.A.; Park, J.K. Comparative analysis of complete mitochondrial genome sequences confirms independent origins of plant-parasitic nematodes. BMC Evol. Biol. 2013, 13, 12. [Google Scholar] [CrossRef] [Green Version]
- Helfenbein, K.; Fourcade, H.; Vanjani, R.; Boore, J. The mitochondrial genome of Paraspadella gotoi is highly reduced and reveals that chaetognaths are a sister group to protostomes. Proc. Natl. Acad. Sci. USA 2004, 101, 10639–10643. [Google Scholar] [CrossRef] [Green Version]
- Papillon, D.; Perez, Y.; Caubit, X.; Le Parco, Y. Identification of chaetognaths as protostomes is supported by the analysis of their mitochondrial genome. Mol. Biol. Evol. 2004, 21, 2122–2129. [Google Scholar] [CrossRef] [Green Version]
- Von Nickisch-Rosenegk, M.; Brown, W.M.; Boore, J.L. Complete sequence of the mitochondrial genome of the tapeworm Hymenolepis diminuta: Gene arrangements indicate that Platyhelminths are Eutrochozoans. Mol. Biol. Evol. 2001, 18, 721–730. [Google Scholar] [CrossRef] [Green Version]
- Le, T.H.; Blair, D.; Agatsuma, T.; Humair, P.F.; Campbell, N.J.H.; Iwagami, M.; Littlewood, D.T.J.; Peacock, B.; Johnston, D.A.; Bartley, J.; et al. Phylogenies inferred from mitochondrial gene orders—a cautionary tale from the parasitic flatworms. Mol. Biol. Evol. 2000, 17, 1123–1125. [Google Scholar] [CrossRef]
- Okimoto, R.; Macfarlane, J.; Clary, D.; Wolstenholme, D. The mitochondrial genomes of two nematodes, Caenorhabditis elegans and Ascaris suum. Genetics 1992, 130, 471–498. [Google Scholar] [CrossRef] [PubMed]
- Barthelemy, R.; Seligmann, H. Cryptic tRNAs in chaetognath mitochondrial genomes. Comput. Biol. Chem. 2016, 62, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Lavrov, D.V.; Brown, W.M. Trichinella spiralis mtDNA: A nematode mitochondrial genome that encodes a putative ATP8, normally-structured tRNAs, and has a gene arrangement relatable to those of coelomate metazoans. Genetics 2001, 157, 621–637. [Google Scholar] [CrossRef] [PubMed]
- Schultz, D.T.; Eizenga, J.M.; Corbett-Detig, R.B.; Francis, W.R.; Christianson, L.M.; Haddock, S.H.D. Conserved novel ORFs in the mitochondrial genome of the ctenophore Beroe forskalii. PeerJ 2020, 8, e8356. [Google Scholar] [CrossRef] [Green Version]
- Arafat, H.; Alamaru, A.; Gissi, C.; Huchon, D. Extensive mitochondrial gene rearrangements in Ctenophora: Insights from benthic platyctenida. BMC Evol. Biol. 2018, 18, 65. [Google Scholar] [CrossRef]
- Cohen, P. New role for the mitochondrial peptide humanin: Protective agent against chemotherapy-induced side effects. J. Natl. Cancer Inst. 2014, 106, dju006. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Yen, K.; Cohen, P. Humanin: A harbinger of mitochondrial-derived peptides? Trends Endocrinol Metab 2013, 24, 222–228. [Google Scholar] [CrossRef] [Green Version]
- Plazzi, F.; Ribani, A.; Passamonti, M. The complete mitochondrial genome of Solemya velum (Mollusca: Bivalvia) and its relationships with Conchifera. BMC Genomics 2013, 14, 409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endo, K.; Noguchi, Y.; Ueshima, R.; Jacobs, H.T. Novel repetitive structures, deviant protein-encoding sequences and unidentified ORFs in the mitochondrial genome of the brachiopod Lingula anatina. J. Mol. Evol. 2005, 61, 36–53. [Google Scholar] [CrossRef]
- Signorovitch, A.; Buss, L.; Dellaporta, S. Comparative genomics of large mitochondria in Placozoans. PLoS Genet. 2007, 3, e13. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, Y.H.; Wang, X.; Zhang, H.X.; Lin, Q. The mitochondrial genome of a sea anemone Bolocera sp. exhibits novel genetic structures potentially involved in adaptation to the deep-sea environment. Ecol. Evol. 2017, 7, 4951–4962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFadden, C.S.; France, S.C.; Sánchez, J.A.; Alderslade, P. A molecular phylogenetic analysis of the Octocorallia (Cnidaria:Anthozoa) based on mitochondrial protein-coding sequences. Mol. Phylogenet Evol. 2006, 41, 513–527. [Google Scholar] [CrossRef]
- Kohn, A.B.; Citarella, M.R.; Kocot, K.M.; Bobkova, Y.V.; Halanych, K.M.; Moroz, L.L. Rapid evolution of the compact and unusual mitochondrial genome in the ctenophore, Pleurobrachia bachei. Mol. Phylogenet. Evol. 2012, 63, 203–207. [Google Scholar] [CrossRef] [Green Version]
- Osigus, H.J.; Eitel, M.; Schierwater, B. Deep RNA sequencing reveals the smallest known mitochondrial micro exon in animals: The placozoan cox1 single base pair exon. PLoS ONE 2017, 12, e0177959. [Google Scholar] [CrossRef] [PubMed]
- Žihala, D.; Eliáš, M. Evolution and Unprecedented Variants of the Mitochondrial Genetic Code in a Lineage of Green Algae. Genome Biol. Evol. 2019, 11, 2992–3007. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Kocot, K.M.; Tassia, M.G.; Cannon, J.T.; Bernt, M.; Halanych, K.M. Mitogenomics Reveals a Novel Genetic Code in Hemichordata. Genome Biol. Evol. 2018, 11, 29–40. [Google Scholar] [CrossRef] [Green Version]
- Kutyumov, V.A.; Predeus, A.V.; Starunov, V.V.; Maltseva, A.L.; Ostrovsky, A.N. Mitochondrial gene order of the freshwater bryozoan Cristatella mucedo retains ancestral lophotrochozoan features. Mitochondrion 2021, 59, 96–104. [Google Scholar] [CrossRef]
- Zhang, J.; Kan, X.; Miao, G.; Hu, S.; Sun, Q.; Tian, W. qMGR: A new approach for quantifying mitochondrial genome rearrangement. Mitochondrion 2020, 52, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Bernt, M.; Middendorf, M. A method for computing an inventory of metazoan mitochondrial gene order rearrangements. BMC Bioinform 2011, 12 (Suppl. 9), S6. [Google Scholar] [CrossRef] [Green Version]
- Zouros, E.; Rodakis, G.C. Doubly Uniparental Inheritance of mtDNA: An Unappreciated Defiance of a General Rule. Adv. Anat Embryol. Cell Biol. 2019, 231, 25–49. [Google Scholar] [PubMed]
- Gusman, A.; Lecomte, S.; Stewart, D.T.; Passamonti, M.; Breton, S. Pursuing the quest for better understanding the taxonomic distribution of the system of doubly uniparental inheritance of mtDNA. PeerJ 2016, 4, e2760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zouros, E. Biparental inheritance through uniparental transmission: The doubly uniparental inheritance (DUI) of mitochondrial DNA. Evol. Biol. 2013, 40, 1–31. [Google Scholar] [CrossRef]
- Passamonti, M.; Ghiselli, F. Doubly Uniparental Inheritance: Two mitochondrial genomes, one precious model for organelle DNA inheritance and evolution. DNA Cell Biol. 2009, 28, 79–89. [Google Scholar] [CrossRef]
- Breton, S.; Doucet-Beaupré, H.; Stewart, D.T.; Hoeh, W.R.; Blier, P.U. The unusual system of doubly uniparental inheritance of mtDNA: Isn’t one enough? Trends Genet. 2007, 23, 465–474. [Google Scholar] [CrossRef]
- Plazzi, F.; Puccio, G.; Passamonti, M. HERMES: An improved method to test mitochondrial genome molecular synapomorphies among clades. Mitochondrion 2021, 58, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, analysis and visualization of phylogenomic data. Mol. Biol Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [Green Version]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008. [Google Scholar]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Harrell, F.E., Jr. Hmisc: Harrell Miscellaneous. R Package Version 4.4-1. 2020. Available online: https://CRAN.R-project.org/package=Hmisc (accessed on 30 April 2021).
- Wei, T.; Simko, V. R Package “corrplot”: Visualization of a Correlation Matrix (Version 0.84). 2017. Available online: https://github.com/taiyun/corrplot (accessed on 30 April 2021).
- WoRMS Editorial Board. World Register of Marine Species. Available online: https://www.marinespecies.org (accessed on 30 April 2021).
- Chamberlain, S. Worrms: World Register of Marine Specie (WoRMS) Client. R package version 0.4.2. 2020. Available online: https://CRAN.R-project.org/package=worrms (accessed on 30 April 2021).
- Adl, S.M.; Bass, D.; Lane, C.E.; Lukeš, J.; Schoch, C.L.; Smirnov, A.; Agatha, S.; Berney, C.; Brown, M.W.; Burki, F.; et al. Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes. J. Eukaryot. Microbiol. 2019, 66, 4–119. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, J.; Liu, J.; Liu, A.; He, X.; Xiang, Q.; Li, Y.; Yin, H.; Luo, J.; Guan, G. Insights into the phylogenetic relationships and drug targets of Babesia isolates infective to small ruminants from the mitochondrial genomes. Parasites Vectors 2020, 13, 378. [Google Scholar] [CrossRef]
- Pedrola-Monfort, J.; Lázaro-Gimeno, D.; Boluda, C.G.; Pedrola, L.; Garmendia, A.; Soler, C.; Soriano, J.M. Evolutionary Trends in the Mitochondrial Genome of Archaeplastida: How Does the GC Bias Affect the Transition from Water to Land? Plants 2020, 9, 358. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Wang, B.; Li, L.; Qiu, Y.L.; Xue, J. Conservative and Dynamic Evolution of Mitochondrial Genomes in Early Land Plants. In Genomics of Chloroplasts and Mitochondria. Advances in Photosynthesis and Respiration (including Bioenergy and Related Processes); Bock, R., Knoop, V., Eds.; Springer: Dordrecht, The Netherlands, 2012; Volume 35, pp. 159–174. [Google Scholar]
- Liu, Y.; Xue, J.Y.; Wang, B.; Li, L.; Qiu, Y.L. The mitochondrial genomes of the early land plants Treubia lacunosa and Anomodon rugelii: Dynamic and conservative evolution. PLoS ONE 2011, 6, e25836. [Google Scholar] [CrossRef] [Green Version]
- Dellaporta, S.L.; Xu, A.; Sagasser, S.; Wolfgan, J.; Moreno, M.A.; Buss, L.W.; Schierwater, B. Mitochondrial genome of Trichoplax adhaerens supports Placozoa as the basal lower metazoan phylum. Proc. Natl. Acad. Sci. USA 2006, 103, 8751–8756. [Google Scholar] [CrossRef] [Green Version]
- Zardoya, R. Recent advances in understanding mitochondrial genome diversity. F1000Res. 2020, 9, F1000, Faculty Rev–270. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.G.; Colas Des Francs-Small, C.; Ostersetzer-Biran, O. Group II intron splicing factors in plant mitochondria. Front Plant. Sci. 2014, 5, 35. [Google Scholar] [CrossRef] [Green Version]
- Férandon, C.; Xu, J.; Barroso, G. The 135 kbp mitochondrial genome of Agaricus bisporus is the largest known eukaryotic reservoir of group I introns and plasmid-related sequences. Fungal. Genet. Biol. 2013, 55, 85–91. [Google Scholar] [CrossRef]
- Saccone, C.; De Giorgi, C.; Gissi, C.; Pesole, G.; Reyes, A. Evolutionary genomics in Metazoa: The mitochondrial DNA as a model system. Gene 1999, 238, 195–209. [Google Scholar] [CrossRef]
- Chevigny, N.; Schatz-Daas, D.; Lotfi, F.; Gualberto, J.M. DNA Repair and the Stability of the Plant Mitochondrial Genome. Int. J. Mol. Sci. 2020, 21, 328. [Google Scholar] [CrossRef] [Green Version]
- Xia, X. DNA replication and strand asymmetry in prokaryotic and mitochondrial genomes. Curr. Genom. 2012, 13, 16–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.J.; Clark-Walker, G.D. Unveiling the mystery of mitochondrial DNA replication in yeasts. Mitochondrion 2018, 38, 17–22. [Google Scholar] [CrossRef]
- Cupp, J.D.; Nielsen, B.L. Minireview: DNA replication in plant mitochondria. Mitochondrion 2014, 19 Pt. B, 231–237. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.Z.; Wang, J.; He, S.; Mayden, R.L. The complete mitochondrial genome of the Chinese hook snout carp Opsariichthys bidens (Actinopterygii: Cypriniformes) and an alternative pattern of mitogenomic evolution invertebrate. Gene 2007, 399, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Scouras, A.; Smith, M.J. The complete mitochondrial genomes of the sea lily Gymnocrinus richeri and the feather star Phanogenia gracilis: Signature nucleotide bias and unique nad4L gene rearrangement within crinoids. Mol. Phylogenet. Evol. 2006, 39, 323–334. [Google Scholar] [CrossRef]
- Hassanin, A. Phylogeny of Arthropoda inferred from mitochondrial sequences: Strategies for limiting the misleading effects of multiple changes in pattern and rates of substitution. Mol. Phylogenet. Evol. 2006, 38, 100–116. [Google Scholar] [CrossRef]
- Hassanin, A.; Léger, N.; Deutsch, J. Evidence for multiple reversals of asymmetric mutational constraints during the evolution of the mitochondrial genome of metazoa, and consequences for phylogenetic inferences. Syst. Biol. 2005, 54, 277–298. [Google Scholar] [CrossRef]
- Jemt, E.; Persson, Ö.; Shi, Y.; Mehmedovic, M.; Uhler, J.P.; Dávila López, M.; Freyer, C.; Gustafsson, C.M.; Samuelsson, T.; Falkenberg, M. Regulation of DNA replication at the end of the mitochondrial D-loop involves the helicase TWINKLE and a conserved sequence element. Nucleic Acids Res. 2015, 43, 9262–9275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Hewitt, M. Insect mitochondrial control region: A review of its structure, evolution and usefulness in evolutionary studies. Biochem. Syst. Ecol. 1997, 25, 99–120. [Google Scholar] [CrossRef]
- Wei, S.J.; Shi, M.; Sharkey, M.J.; van Achterberg, C.; Chen, X.X. Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to Holometabolous insects. BMC Genomics 2010, 11, 371. [Google Scholar] [CrossRef] [Green Version]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E.P. Codon usage bias from tRNA’s point of view: Redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 2004, 14, 2279–2286. [Google Scholar] [CrossRef] [Green Version]
- Ikemura, T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 1981, 146, 1–21. [Google Scholar] [CrossRef]
- Xia, X. Mutation and selection on the anticodon of tRNA genes in vertebrate mitochondrial genomes. Gene 2005, 345, 13–20. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Li, Q. Mutation and selection on the wobble nucleotide in tRNA anticodons in marine bivalve mitochondrial genomes. PLoS ONE 2011, 6, e16147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narakusumo, R.P.; Riedel, A.; Pons, J. Mitochondrial genomes of twelve species of hyperdiverse Trigonopterus weevils. PeerJ 2020, 8, e10017. [Google Scholar] [CrossRef]
- Mohandas, N.; Pozio, E.; La Rosa, G.; Korhonen, P.K.; Young, N.D.; Koehler, A.V.; Hall, R.S.; Sternberg, P.W.; Boag, P.R.; Jex, A.R.; et al. Mitochondrial genomes of Trichinella species and genotypes—A basis for diagnosis, and systematic and epidemiological explorations. Int. J. Parasitol. 2014, 44, 1073–1080. [Google Scholar] [CrossRef]
- Gibson, T.; Farrugia, D.; Barrett, J.; Chitwood, D.J.; Rowe, J.; Subbotin, S.; Dowton, M. The mitochondrial genome of the soybean cyst nematode, Heterodera glycines. Genome 2011, 54, 565–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, L.; Zhuo, K.; Lin, B.; Wang, H.; Liao, J. The complete mitochondrial genome of Meloidogyne graminicola (Tylenchina): A unique gene arrangement and its phylogenetic implications. PLoS ONE 2014, 9, e98558. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.C.; Wei, D.D.; Shao, R.; Dou, W.; Wang, J.J. The complete mitochondrial genome of the booklouse, Liposcelis decolor: Insights into gene arrangement and genome organization within the genus Liposcelis. PLoS ONE 2014, 9, e91902. [Google Scholar] [CrossRef] [Green Version]
- Froufe, E.; Bolotov, I.; Aldridge, D.C.; Bogan, A.E.; Breton, S.; Gan, H.M.; Kovitvadhi, U.; Kovitvadhi, S.; Riccardi, N.; Secci-Petretto, G.; et al. Mesozoic mitogenome rearrangements and freshwater mussel (Bivalvia: Unionoidea) macroevolution. Heredity 2020, 124, 182–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barroso-Lima, N.C.; Prosdocimi, F. The heavy strand dilemma of vertebrate mitochondria on genome sequencing age: Number of encoded genes or G+T content? Mitochondrial DNA A 2017, 27, 1–6. [Google Scholar] [CrossRef]
- Sun, S.; Li, Q.; Kong, L.; Yu, H. Multiple reversals of strand asymmetry in molluscs mitochondrial genomes, and consequences for phylogenetic inferences. Mol. Phylogenet Evol. 2018, 118, 222–231. [Google Scholar] [CrossRef]
- Min, X.J.; Hickey, D.A. DNA Asymmetric Strand Bias Affects the Amino Acid Composition of Mitochondrial Proteins. DNA Res. 2007, 14, 201–206. [Google Scholar] [CrossRef] [PubMed]


| A+T Content | Spearman’s Rho | p-Value | |
|---|---|---|---|
| Chlorophyta | 62.8% | ρ = 0.15 | p = 0.0506 |
| Non-embryophytes Streptophyta | 60.3% | ρ = −0.51 | p = 0.065 |
| Non-vascular Embryophyta | 58.9% | ρ = −0.64 | p = 0 |
| Tracheophyta | 55.0% | ρ = 0.07 | p = 0.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Formaggioni, A.; Luchetti, A.; Plazzi, F. Mitochondrial Genomic Landscape: A Portrait of the Mitochondrial Genome 40 Years after the First Complete Sequence. Life 2021, 11, 663. https://doi.org/10.3390/life11070663
Formaggioni A, Luchetti A, Plazzi F. Mitochondrial Genomic Landscape: A Portrait of the Mitochondrial Genome 40 Years after the First Complete Sequence. Life. 2021; 11(7):663. https://doi.org/10.3390/life11070663
Chicago/Turabian StyleFormaggioni, Alessandro, Andrea Luchetti, and Federico Plazzi. 2021. "Mitochondrial Genomic Landscape: A Portrait of the Mitochondrial Genome 40 Years after the First Complete Sequence" Life 11, no. 7: 663. https://doi.org/10.3390/life11070663
APA StyleFormaggioni, A., Luchetti, A., & Plazzi, F. (2021). Mitochondrial Genomic Landscape: A Portrait of the Mitochondrial Genome 40 Years after the First Complete Sequence. Life, 11(7), 663. https://doi.org/10.3390/life11070663








