Enhanced Expression of Thaumatin-like Protein Gene (LeTLP1) Endows Resistance to Trichoderma atroviride in Lentinula edodes
Abstract
1. Introduction
2. Materials and Methods
2.1. L. edodes and T. atroviride Strains
2.2. Evaluation of the Resistance of L. edodes to T. atroviride
2.3. Responses of the Highly Resistant and Susceptible L. edodes Strains to Infection with T. atroviride
2.4. Comparative Transcriptomics Analysis
2.4.1. RNA Extraction, Library Preparation, and Sequencing
2.4.2. Transcriptome Data Analysis
2.5. qRT-PCR
2.6. Construction of LeTLP1-Overexpression and LeTLP1-Silencing Vectors and Fungal Transformation
2.7. Evaluation of the Level of Resistance against T. atroviride in LeTLP1-Overexpression and LeTLP1-Silenced Transformants
2.8. Prokaryotic Expression of LeTLP1 and LeTLP (Y55)
2.9. Assays of the Antifungal Activity of LeTLP1
2.10. Statistical Analysis
3. Results
3.1. Evaluation of Lentinula edodes Strains That Were Resistant to Trichoderma atrovide
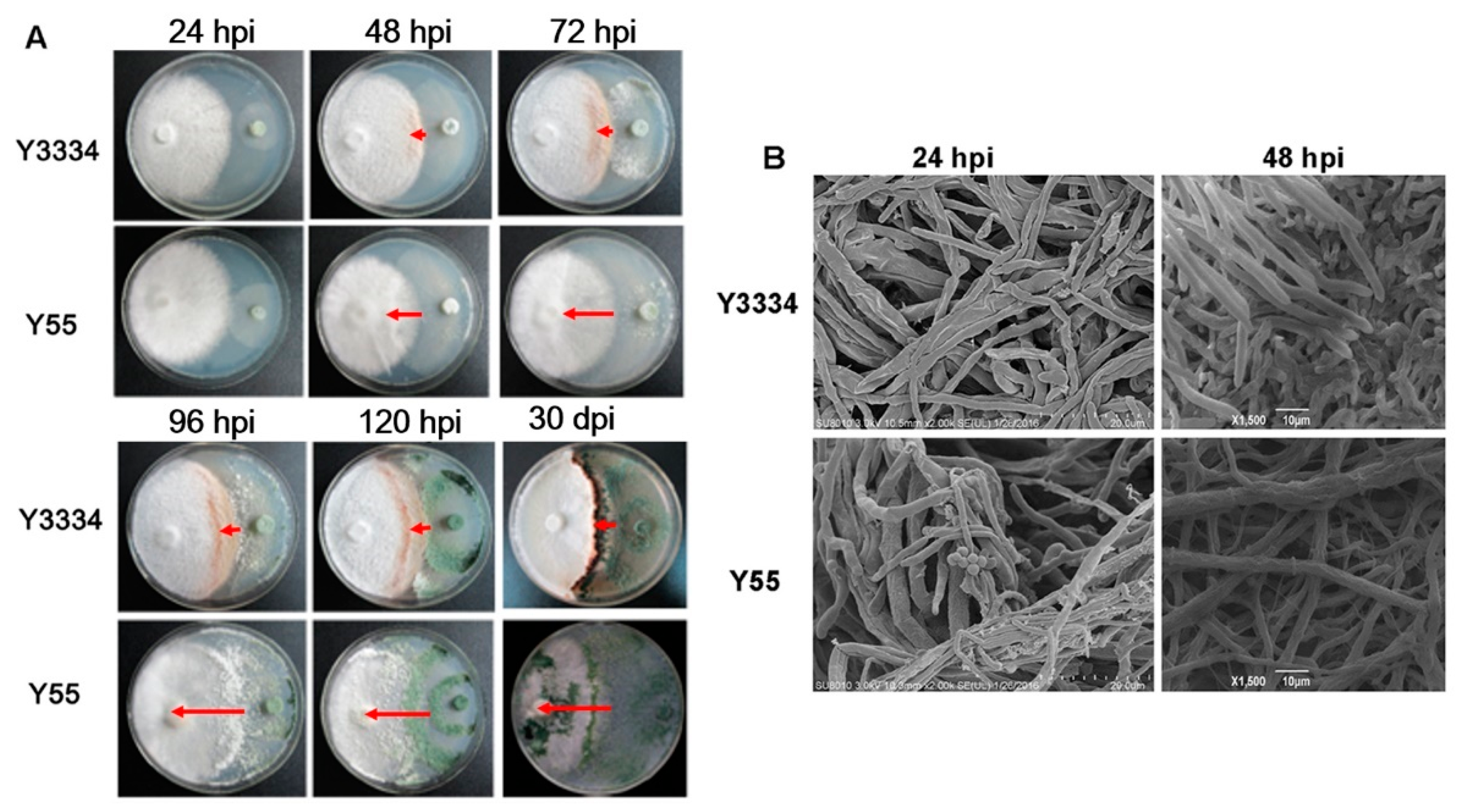
3.2. Significant Differences in the Responses of the Highly Resistant and Susceptible L. edodes strains to T. atroviride Infection
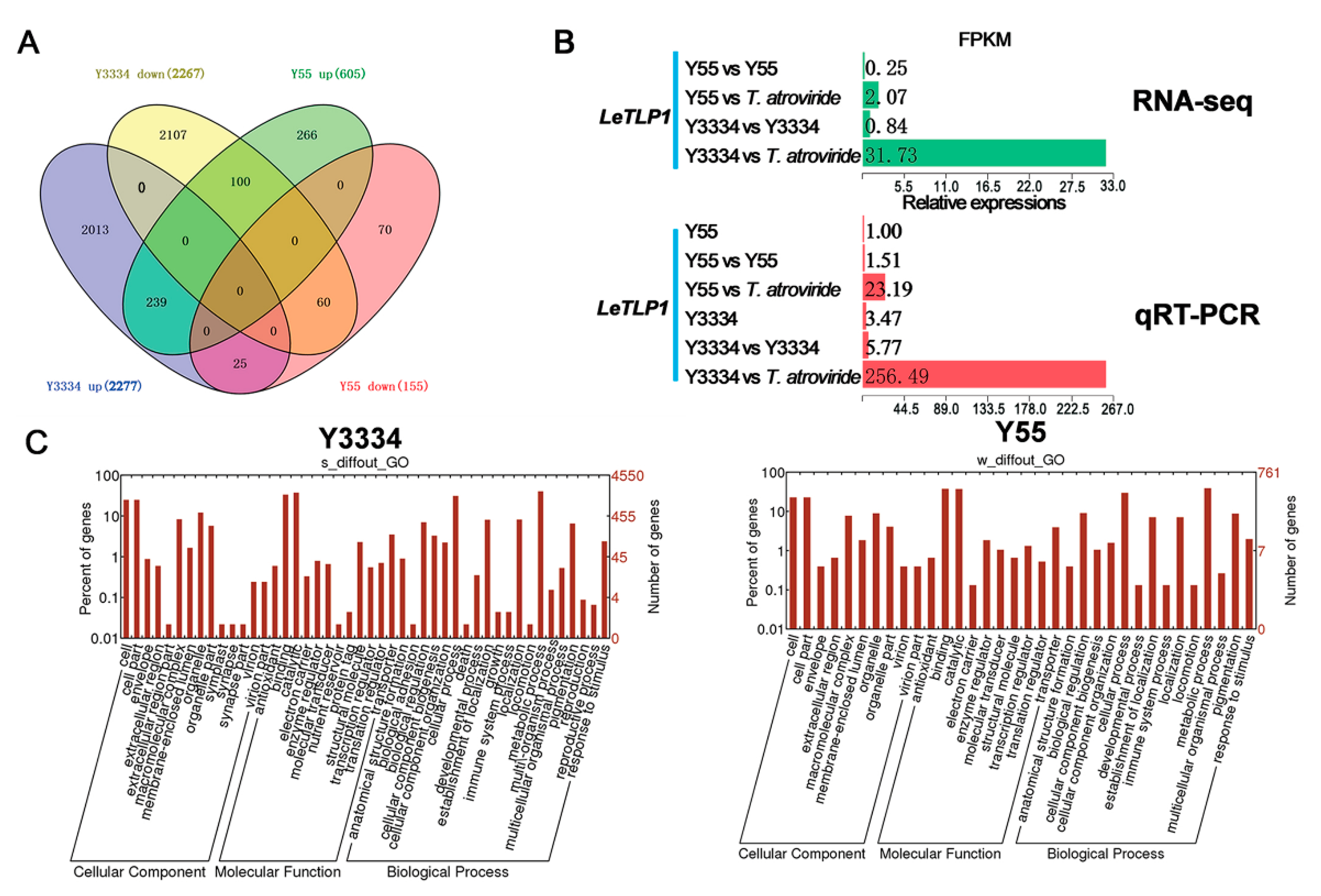
3.3. Transcriptional Responses of the Highly Resistant and Susceptible L. edodes strains to T. atroviride
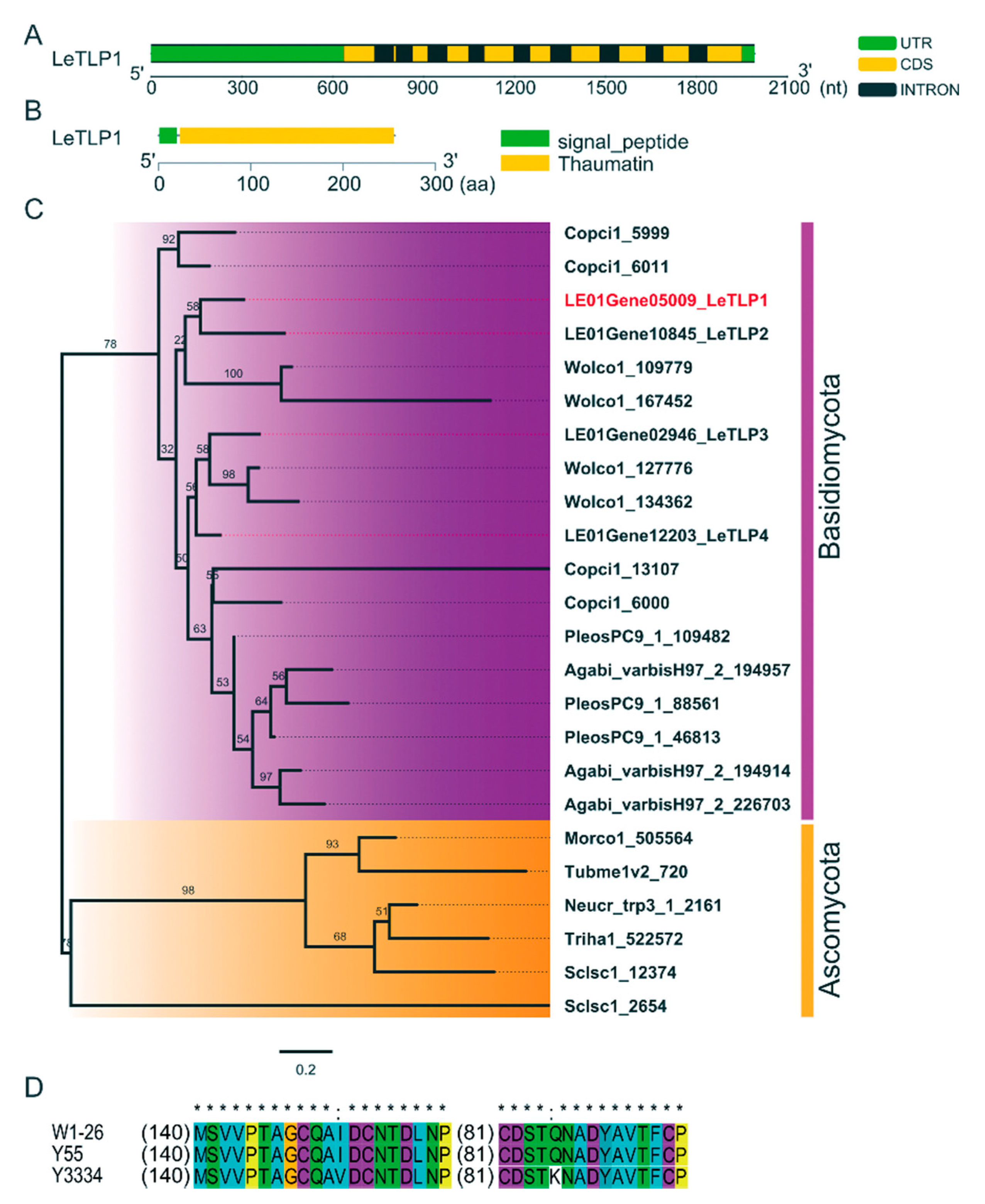
3.4. Molecular Characterization of LeTLP1
3.5. Effects of LeTLP1 Overexpression on the Resistance of L. edodes against T. atroviride
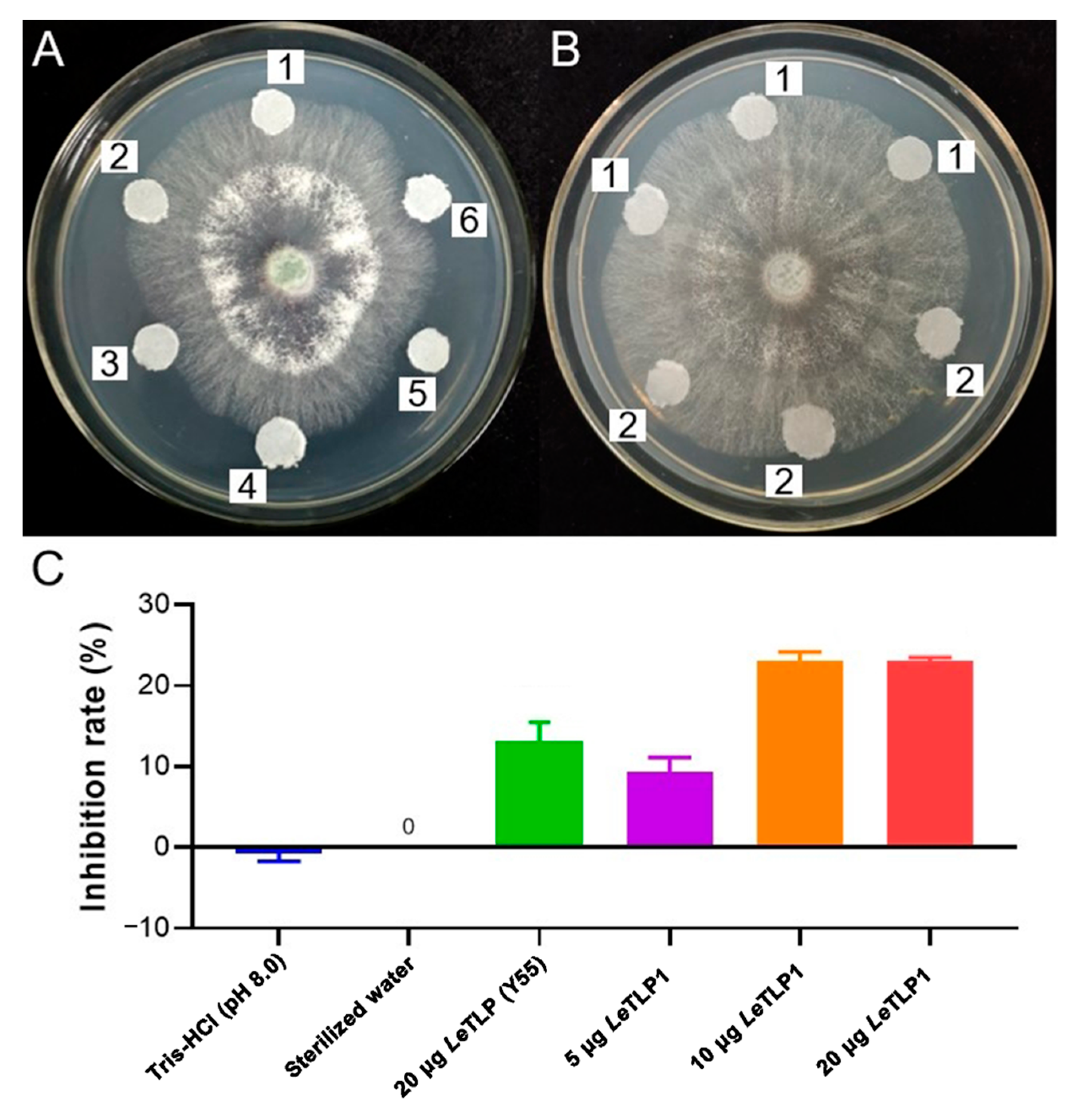
3.6. Antifungal Activity of LeTLP1 against T. atroviride
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kwan, H.S.; Au, C.H.; Wong, M.C.; Qin, J.; Kwok, I.S.W.; Nong, W.Y.; Chum, W.W.Y. Genome sequence and genetic linkage analysis of Shiitake mushroom Lentinula edodes. Nat. Preced. 2012, 1, 1. [Google Scholar] [CrossRef]
- Quaicoe, E.H.; Amoah, C.; Obodai, M.; Odamtten, G.T. Nutrient requirements and environmental conditions for the cultivation of the medicinal mushroom (Lentinula edodes) (Berk.) in Ghana. Int. J. Sci. Technol. Res. 2014, 3, 45–50. [Google Scholar]
- Finimundy, T.; Dillon, A.; Henriques, J.; Ely, M. A review on general nutritional compounds and pharmacological properties of the Lentinula edodes mushroom. Int. J. Food. Sci. Nutr. 2014, 5, 1095–1105. [Google Scholar]
- Li, C.; Gong, W.B.; Zhang, L.; Yang, Z.Q.; Nong, W.Y.; Bian, Y.B.; Hoi-Shan, K.; Man-Kit, C.; Xiao, Y. Association mapping reveals genetic loci Associated with Important Agronomic Traits in Lentinula edodes, Shiitake Mushroom. Front. Microbiol. 2017, 8, 237. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Diego, M.; Eva, T.C.; Noelia, J.C.; Gonzalo, P.; María, T.; Alejandro, R.R.; Carlota, L.; Cristina, S.R. In vitro and in vivo testing of the hypocholesterolemic activity of ergosterol- and β-glucan-enriched extracts obtained from shiitake mushrooms (Lentinula edodes). Food Funct. 2019, 10, 7325–7332. [Google Scholar]
- Diego, M.; María, T.; Carlota, L.; Gonzalo, P.; Adriana, J.P.; Cristina, S.R. Effect of traditional and modern culinary processing, bioaccessibility, biosafety and bioavailability of eritadenine, a hypocholesterolemic compound from edible mushrooms. Food Funct. 2018, 9, 6360–6368. [Google Scholar]
- Diego, M.; Renata, R.; Marisol, A.V.; Hellen, A.; Cristina, S.R.; Susana, S.; Marcello, I.; Fhernanda, S. Isolation and comparison of α- and β-D-glucans from shiitake mushrooms (Lentinula edodes) with different biological activities. Carbohydr. Polym. 2019, 229, 115521. [Google Scholar]
- Pan, W.; Jiang, P.; Zhao, J.; Shi, H.; Yu, Y. β-glucan from Lentinula edodes prevents cognitive impairments in high-fat diet-induced obese mice: Involvement of colon-brain axis. J. Transl. Med. 2021, 19, 54. [Google Scholar] [CrossRef] [PubMed]
- Ohmasa, M.; Cheong, M.L. Effects of culture conditions of Lentinula edodes, Shiitake mushroom, on the disease resistance of Lentinula edodes against Trichoderma harzianum in the sawdust cultures. In Proceedings of the 3rd International Conference on Mushroom Biology and Mushroom Products, Sydney, Australia, 11–15 October 1999. [Google Scholar]
- Julian, A.V.; Reyes, R.G.; Eguchi, F. Reference Module in Earth Systems and Environmental Sciences, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 13–20. [Google Scholar]
- Ohga, S.; Royse, D.J. Transcriptional regulation of laccase and cellulase genes during growth and fruiting of Lentinula edodes on supplemented sawdust. FEMS Microbiol. Lett. 2001, 201, 111–115. [Google Scholar] [CrossRef]
- Wang, G.; Cao, X.; Ma, X.; Guo, M.; Liu, C.; Yan, L.; Bian, Y. Diversity and effect of Trichoderma spp. associated with green mold disease on Lentinula edodes in China. MicrobiologyOpen 2016, 5, 709–718. [Google Scholar] [CrossRef]
- Luković, J.; Milijašević-Marčić, S.; Hatvani, L.; Kredics, L.; Szűcs, A.; Vágvölgyi, C.; Nataša, D.; Ivana, V.; Potočnik, I. Sensitivity of Trichoderma strains from edible mushrooms to the fungicides prochloraz and metrafenone. J. Environ. Sci. Health B 2020, 56, 54–63. [Google Scholar] [CrossRef]
- Jiang, H.F.; Wang, F.L.; Tan, Q. A preliminary study on Trichoderma spp. and dominant T. species in Lentinula edodes growing. Shanghai ACTA Agric. 1995, 11, 85–90. [Google Scholar]
- Wu, X.P.; Wu, X.J.; Hu, F.P.; He, H.Z.; Xie, B.G. Identification of Trichoderma species associated with cultivated edible fungi. J. Agric. Biotechnol. 2008, 16, 1048–1055. [Google Scholar]
- Tokimoto, K. Physiological studies on antagonism between Lentinula edodes and Trichoderma spp. in bedlogs of the former. Rep. Tottori. Mycol. Inst. 1985, 23, 1–54. (In Japanese) [Google Scholar]
- Seaby, D. Trichoderma as a weed mould or pathogen in mushroom cultivation. Trichoderma Gliocladium 1998, 2, 267–272. [Google Scholar]
- Lee, H.M.; Bak, W.C.; Lee, B.H.; Park, H.; Ka, K.H. Breeding and Screening of Lentinula edodes species resistance to Trichoderma spp. Mycobiology 2008, 4, 270–272. [Google Scholar] [CrossRef]
- Park, M.S.; Bae, K.S.; Yu, S.H. Two new species of Trichoderma associated with green mold of oyster mushroom cultivation in Korea. Mycobiology 2006, 34, 111–113. [Google Scholar] [CrossRef]
- Cao, X.T.; Bian, Y.B.; Xu, Z.Y. First Report of Trichoderma oblongisporum Causing Green Mold Disease on Lentinula edodes (shiitake) in China. Plant Dis. 2014, 98, 1440. [Google Scholar] [CrossRef]
- Jhune, C.S.; You, C.H.; Cha, D.Y. Effects of thiabendazole on green mold, Trichoderma spp. during cultivation of oyster mushroom, Pleurotus spp. Korean J. Mycol. 1990, 18, 89–96. [Google Scholar]
- Rezaei, D.Y.; Mohammadi, G.E. Studies of the effects of benomyl and carbendazim on Trichoderma green mould control in button mushroom farms. J. Agric. Sci. 2007, 16, 157–165. [Google Scholar]
- Abdi, S.; Sobhan, A.S.; Sobhan, A.S. Monitoring of Benomyl Residue in Mushroom Marketed in Hamadan City. Sci. J. Hamadan Univ. Med. Sci. 2015, 22, 137–143. [Google Scholar]
- Shuping, D.S.S.; Eloff, J.N. The use of plants to protect plants and food against fungal pathogens: A review. Afr. J. Tradit. Complement. Altern. Med. 2017, 14, 120–127. [Google Scholar] [CrossRef]
- Guo, M.P.; Ye, Z.M.; Shen, G.Y.; Bian, Y.B.; Xu, Z.Y. Effects of mycovirus LeV-HKB on resistance of heat stress challenged Lentinula edodes mycelia against Trichoderma atroviride. Acta Edulis Fungi 2020, 27, 143. [Google Scholar]
- Mata, G.; Savoie, J.M. Screening of Lentinula Edodes strains by laccase induction and resistence to Trichoderma spp. Rev. Mex. Micol. 1998, 14, 29–32. [Google Scholar]
- Savoie, J.M.; Mata, G. The antagonistic action of Trichoderma spp. hyphae to Lentinula edodes hyphae changes ignocellulotytic activities during cultivation in wheat straw. World J. Microbiol. Biotechnol. 1999, 15, 369–373. [Google Scholar] [CrossRef]
- Ragucci, S.; Landi, N.; Russo, R.; Valletta, M.; Pedone, P.V.; Chambery, A.; Maro, A.D. Ageritin from pioppino mushroom: The prototype of ribotoxin-like proteins, a novel family of specific ribonucleases in edible mushrooms. Toxins 2021, 13, 263. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Z.R.; Li, C.; Bian, Y.B.; Xiao, Y. Evaluating genetic diversity and constructing core collections of Chinese Lentinula edodes cultivars using ISSR and SRAP markers. J. Basic Microbiol. 2015, 55, 749–760. [Google Scholar] [CrossRef]
- Jiang, Q. Preliminary Research on Resistance Differentiation of Lentinula edodes Resources to Trichoderma spp. Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2017. [Google Scholar]
- Liu, J.; Ma, C.J.; Xiang, X.J.; Li, C.; Bian, Y.B.; Zhang, J.; Xiao, Y. Constructing Core Collections of Chinese Wild Lentinula edodes Strains Based on SRAP Markers. Acta Edulis Fungi 2017, 24, 7–17. [Google Scholar]
- Wang, G.Z.; Zhou, S.S.; Luo, Y.; Ma, C.J.; Gong, Y.H.; Zhou, Y.; Gao, S.S.; Huang, Z.C.; Yan, L.L.; Hu, Y.; et al. The heat shock protein 40 LeDnaJ regulates stress resistance and indole-3-acetic acid biosynthesis in Lentinula edodes. Fungal Genet. Biol. 2018, 118, 37–44. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J. The Sequence Alignment-Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Lalitha, S. Primer premier 5. Biotech Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.L.; Xu, R.P.; Dai, S.H.; Wang, G.Z.; Bian, Y.B.; Gong, Y.H.; Zhou, Y. Overexpression of a laccase gene Lelcc1 and phenotypic characterizations in Lentinula edodes. Mycosystema 2019, 38, 831–840. [Google Scholar]
- Imtiaj, A.; Lee, T.S. Screening of antibacterial and antifungal activities from Korean wild mushrooms. World J. Agrc. Sci. 2007, 3, 316–321. [Google Scholar]
- Ulhoa, C.J.; Peberdy, J.F. Purification and some properties of the extracellular chitinase produced by Trichoderma harzianum. Enzym. Microb. Technol. 1992, 14, 236–240. [Google Scholar] [CrossRef]
- Kobayashi, T.; Oguro, M.; Akiba, M.; Taki, H.; Kitajima, H.; Ishihara, H. Mushroom yield of cultivated shiitake (Lentinula edodes) and fungal communities in logs. J. For. Res. 2020, 25, 269–275. [Google Scholar] [CrossRef]
- Velazhahan, R.; Chen-Cole, K.; Anuratha, C.S.; Muthukrishnan, S. Induction of thaumatin-like proteins (TLPs) in Rhizoctonia solani-infected rice and characterization of two new cDNA clones. Physiol. Plant. 1998, 102, 21–28. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Watanabe, H.; Nagai, M.; Nakade, K.; Takahashi, M.; Sato, T. Lentinula edodes tlg1 encodes a thaumatin-like protein that is involved in lentinan degradation and fruiting body senescence. Plant Physiol. 2006, 141, 793–801. [Google Scholar] [CrossRef]
- Koiwa, H.; Kato, H.; Nakatsu, T.; Oda, J.; Yamada, Y.; Sato, F. Crystal structure of tobacco PR-5d protein at 1.8 Å resolution reveals a conserved acidic cleft structure in antifungal thaumatin-like proteins. J. Mol. Biol. 1999, 286, 1137–1145. [Google Scholar] [CrossRef]
- Trudel, J.; Grenier, J.; Potvin, C.; Asselin, A. Several thaumatin like proteins bind to β-1,3-glucans. Plant Physiol. 1998, 118, 1431–1438. [Google Scholar] [CrossRef]
- Kumar, R.; Mukherjee, P.K. Trichoderma virens Alt a 1 protein may target maize PR5/thaumatin-like protein to suppress plant defence: An in silico analysis. Physiol. Mol. Plant Pathol. 2020, 112, 101551. [Google Scholar] [CrossRef]
- Grenier, J.; Potvin, C.; Asselin, A. Some fungi express β-1,3-glucanases similar to thaumatin-like proteins. Mycologia 2000, 92, 841–848. [Google Scholar]
- Philipp, H.F.; Alga, Z. β-glucan: Crucial component of the fungal cell wall and elusive MAMP in plants. Fungal Genet. Biol. 2016, 90, 53–60. [Google Scholar]
- Brown, S.M.; Free, S.J. The structure and synthesis of the fungal cell wall. BioEssays 2006, 28, 799–808. [Google Scholar]
- Ojola, P.O.; Nyaboga, E.N.; Njiru, P.N.; Orinda, G.O. Overexpression of rice thaumatin-like protein (Ostlp) gene in transgenic cassava results in enhanced tolerance to Colletotrichum gloeosporioides f. sp. manihotis. J. Genet. Eng. Biotechnol. 2018, 16, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Zhou, Y.; Movahedi, A.; Wei, H.; Zhuge, Q. Thaumatin-like protein (Pe-TLP) acts as a positive factor in transgenic poplars enhanced resistance to spots disease. Physiol. Mol. Plant Pathol. 2020, 112, 101512. [Google Scholar] [CrossRef]


| Primer Name | Primer Sequence (5′ to 3′) |
|---|---|
| O-Legpd Promotor-F | catATTCAAGCAGTCAATGGATTGGA |
| O-Legpd Promotor-R | tctagaggatccccgggtaccCGAAGTTTGAGGTGGTTGCG |
| O-LeTLP1-F | ccacctcaaacttcggaattcTCAGGGGCAAAATGTAACAGCATA |
| O-LeTLP1-R | ccattgactgcttgaatATGATGAAGAATTCTATCATTATCTCTGC |
| R-LeTLP1-F | agctcttcacgGATCCACTGCAGCAGTTGGG |
| R-LeTLP1-R | tgcttgaatTGGCCGGCAATCTTCACG |
| R-Leactin-F | ccacctcaaacttcggaattcGCAGTATTTATACCTACGGAGC |
| R-Leactin-R | cagtggatcCGTGAAGAGCTGCGAGTGTTG |
| Strains | AC about 7 Days | AC about 30 Days | Strains | AC about 7 Days | AC about 30 Days |
|---|---|---|---|---|---|
| c2 | + | ++ | Y30 | + | ++ |
| c6 | + | ++ | Y37 | + | ++ |
| c9 | + | ++ | Y39 | + | ++ |
| c10 | + | ++ | Y51 | + | ++ |
| c20 | + | ++ | Y55 | − | − |
| c23 | + | ++ | Y70 | + | ++ |
| c27 | + | ++ | Y76 | + | ++ |
| c28 | + | ++ | Y78 | + | ++ |
| c31 | + | ++ | Y79 | + | ++ |
| c42 | + | ++ | Y84 | + | ++ |
| c47 | + | ++ | Y88 | + | ++ |
| c48 | + | ++ | Y89 | + | ++ |
| c49 | + | ++ | Y91 | + | ++ |
| c50 | + | ++ | Y94 | + | ++ |
| c51 | + | ++ | Y100 | + | ++ |
| c64 | + | ++ | Y104 | + | ++ |
| c67 | + | +++ | Y110 | + | ++ |
| c82 | + | ++ | Y111 | + | ++ |
| c85 | + | ++ | Y113 | + | ++ |
| c87 | + | ++ | Y118 | + | ++ |
| c88 | + | ++ | Y121 | + | +++ |
| Y1 | + | ++ | Y234 | + | ++ |
| Y5 | + | ++ | Y358 | + | ++ |
| Y7 | + | ++ | Y366 | + | ++ |
| Y8 | + | ++ | Y1515 | + | +++ |
| Y11 | + | ++ | Y1518 | + | ++ |
| Y14 | + | ++ | Y3334 | + | +++ |
| Y29 | + | +++ | Y3353 | + | ++ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, X.; Fan, X.; Wang, G.; Xu, R.; Yan, L.; Zhou, Y.; Gong, Y.; Xiao, Y.; Bian, Y. Enhanced Expression of Thaumatin-like Protein Gene (LeTLP1) Endows Resistance to Trichoderma atroviride in Lentinula edodes. Life 2021, 11, 863. https://doi.org/10.3390/life11080863
Ma X, Fan X, Wang G, Xu R, Yan L, Zhou Y, Gong Y, Xiao Y, Bian Y. Enhanced Expression of Thaumatin-like Protein Gene (LeTLP1) Endows Resistance to Trichoderma atroviride in Lentinula edodes. Life. 2021; 11(8):863. https://doi.org/10.3390/life11080863
Chicago/Turabian StyleMa, Xiaolong, Xiaolin Fan, Gangzheng Wang, Ruiping Xu, Lianlian Yan, Yan Zhou, Yuhua Gong, Yang Xiao, and Yinbing Bian. 2021. "Enhanced Expression of Thaumatin-like Protein Gene (LeTLP1) Endows Resistance to Trichoderma atroviride in Lentinula edodes" Life 11, no. 8: 863. https://doi.org/10.3390/life11080863
APA StyleMa, X., Fan, X., Wang, G., Xu, R., Yan, L., Zhou, Y., Gong, Y., Xiao, Y., & Bian, Y. (2021). Enhanced Expression of Thaumatin-like Protein Gene (LeTLP1) Endows Resistance to Trichoderma atroviride in Lentinula edodes. Life, 11(8), 863. https://doi.org/10.3390/life11080863






