Overexpression of BQ323636.1 Modulated AR/IL-8/CXCR1 Axis to Confer Tamoxifen Resistance in ER-Positive Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures, Transfection and Stable Cell Lines Establishment
2.2. Plasmids and siRNA
2.3. Chemicals
2.4. Cell Viability Assay
2.5. RNA Extraction, Reverse Transcription and qPCR
2.6. Western Blot
2.7. IL-8 Measurement and AKT Activity Assay
2.8. Luciferase Reporter Assay
2.9. Xenograft
2.10. Immunohistochemistry
2.11. In Silico Analysis
2.12. Statistical Analysis
3. Results
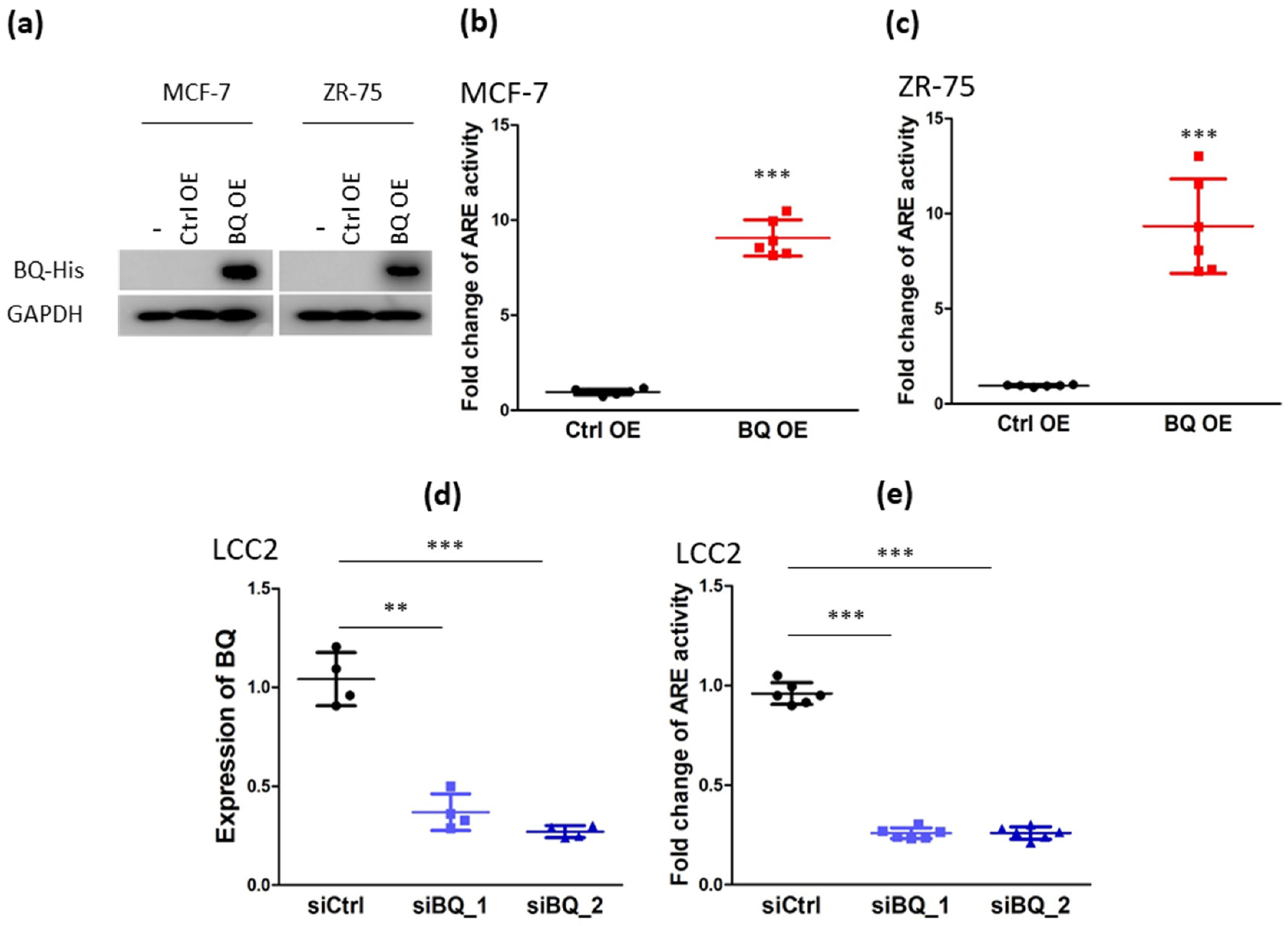
3.1. Overexpression of BQ Could Activate AR Signalling and Thus Modulate the Response to Tamoxifen in Breast Cancer
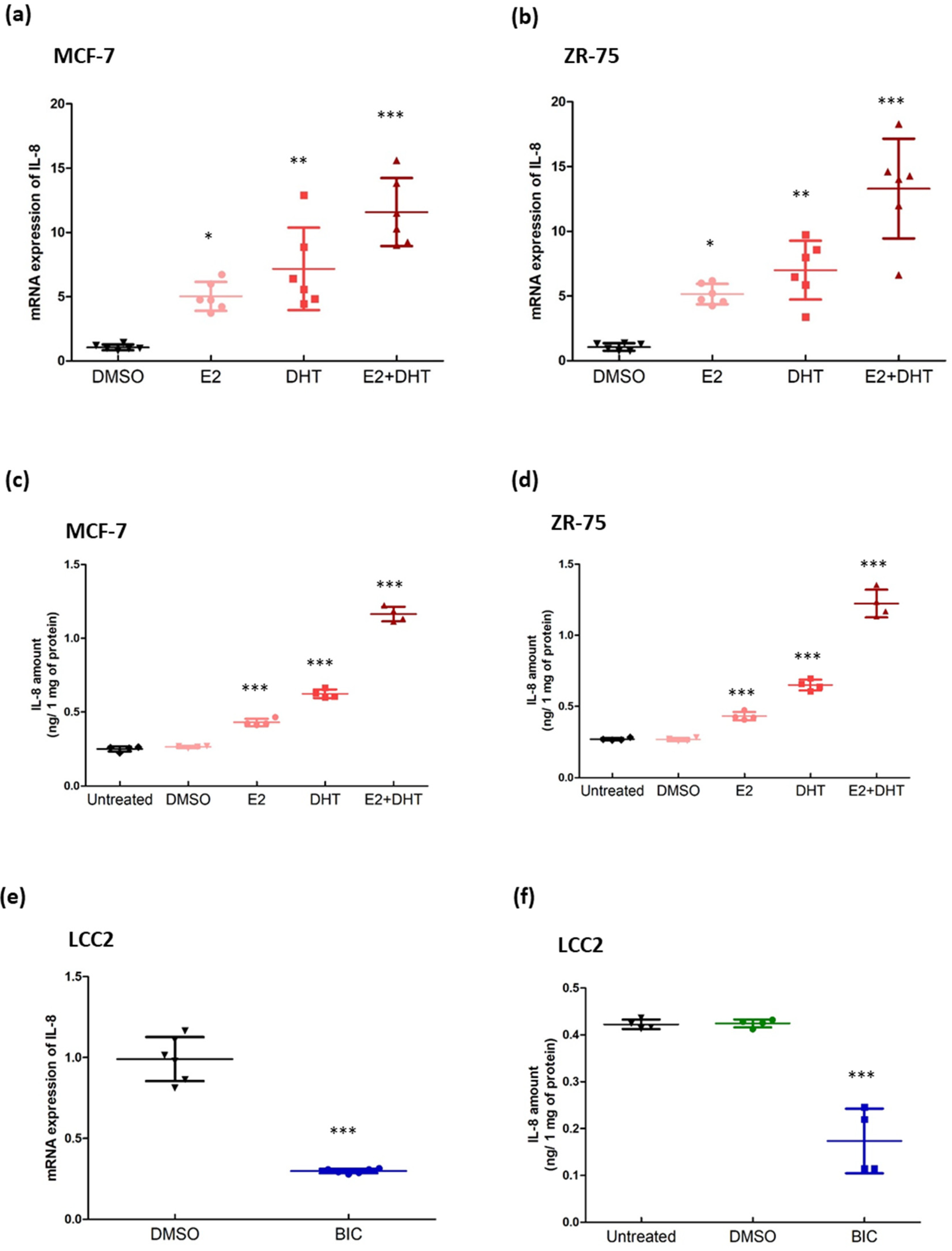
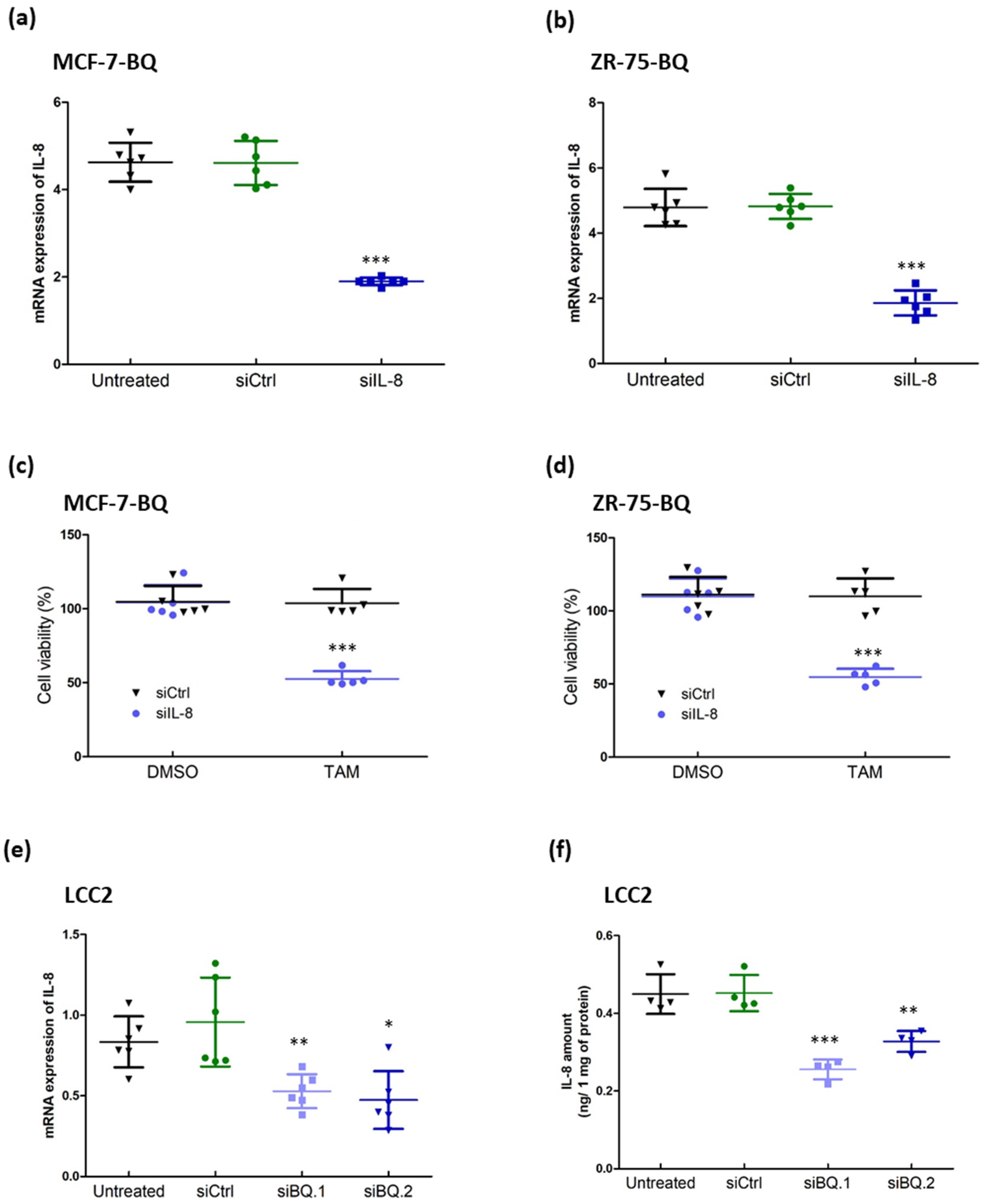
3.2. Identification of IL-8 as a Candidate to Modulate Tamoxifen Resistance in Breast Cancer
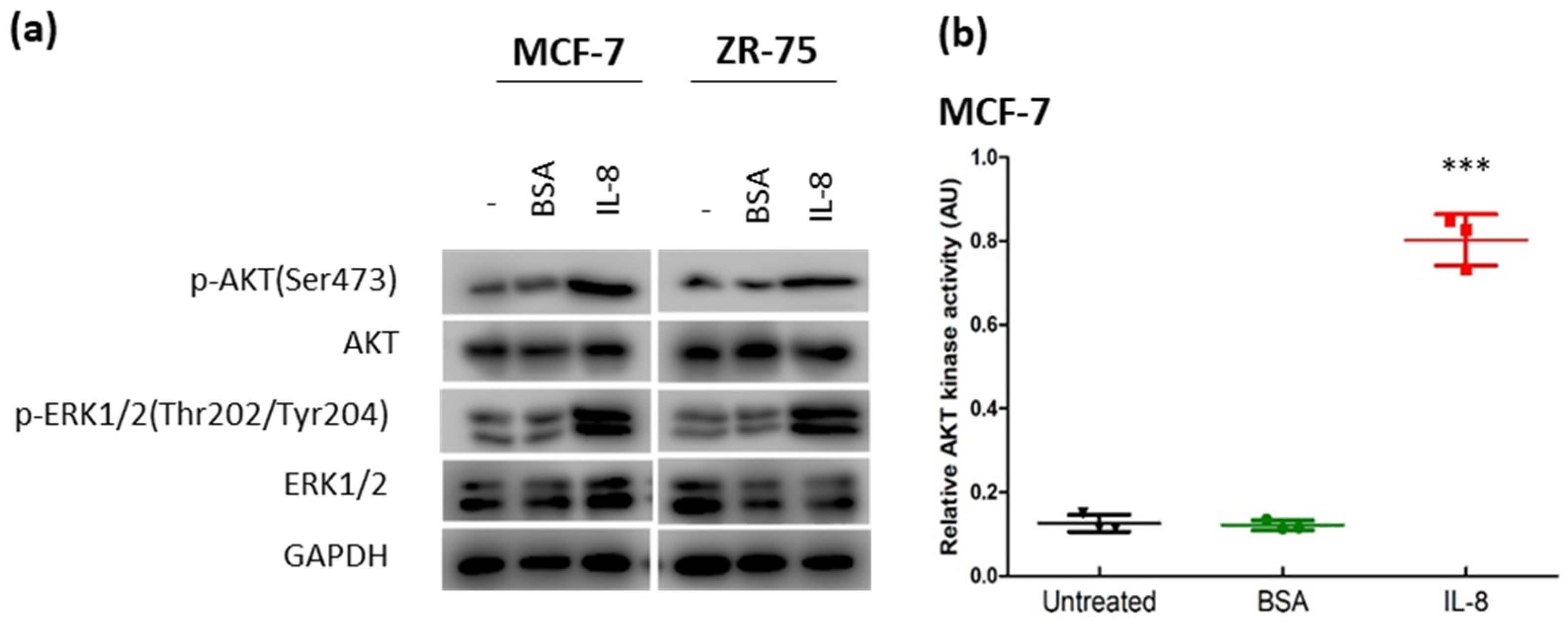
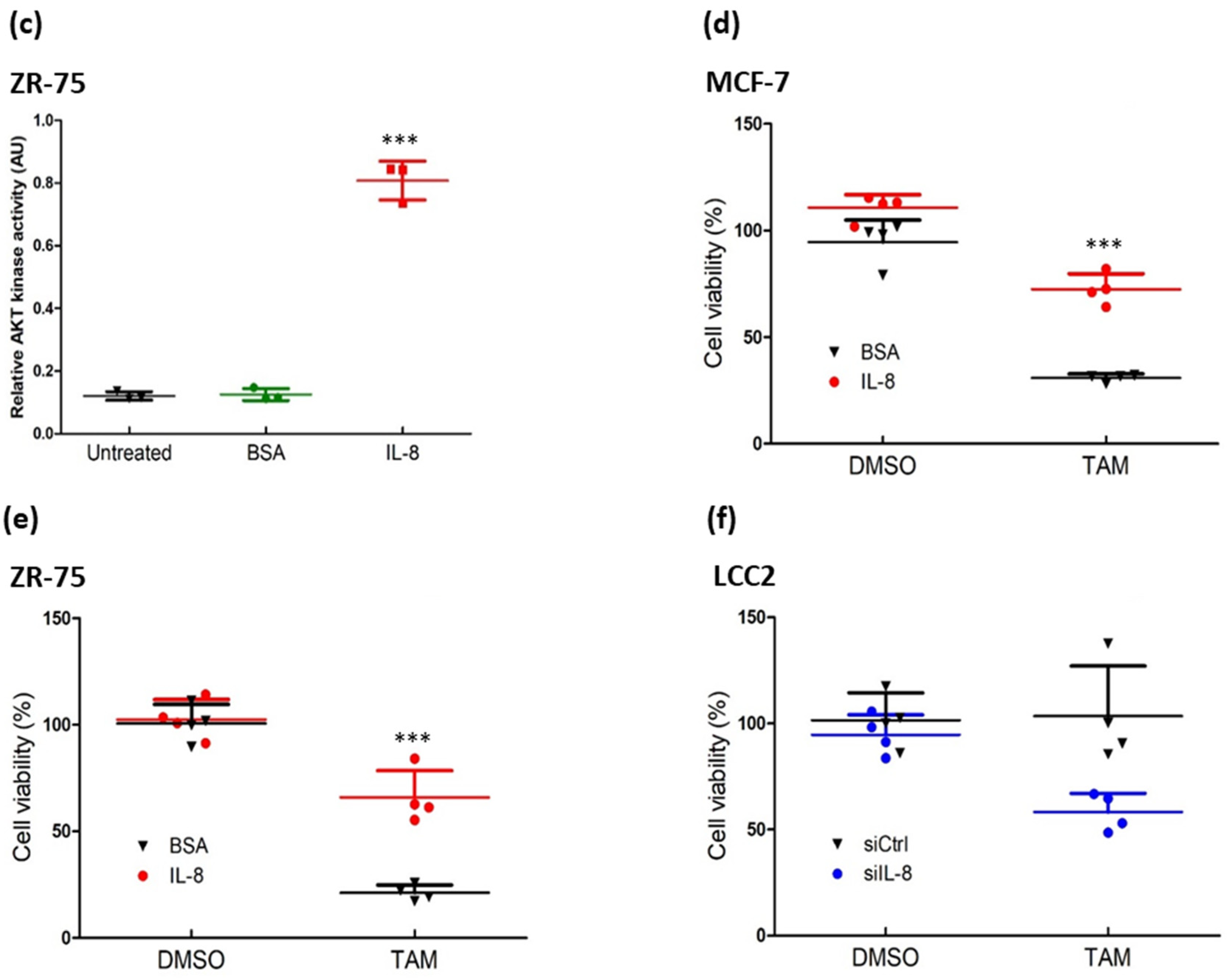
3.3. IL-8 Activated the AKT-ERK1/2 Axis to Modulate the Response to Tamoxifen
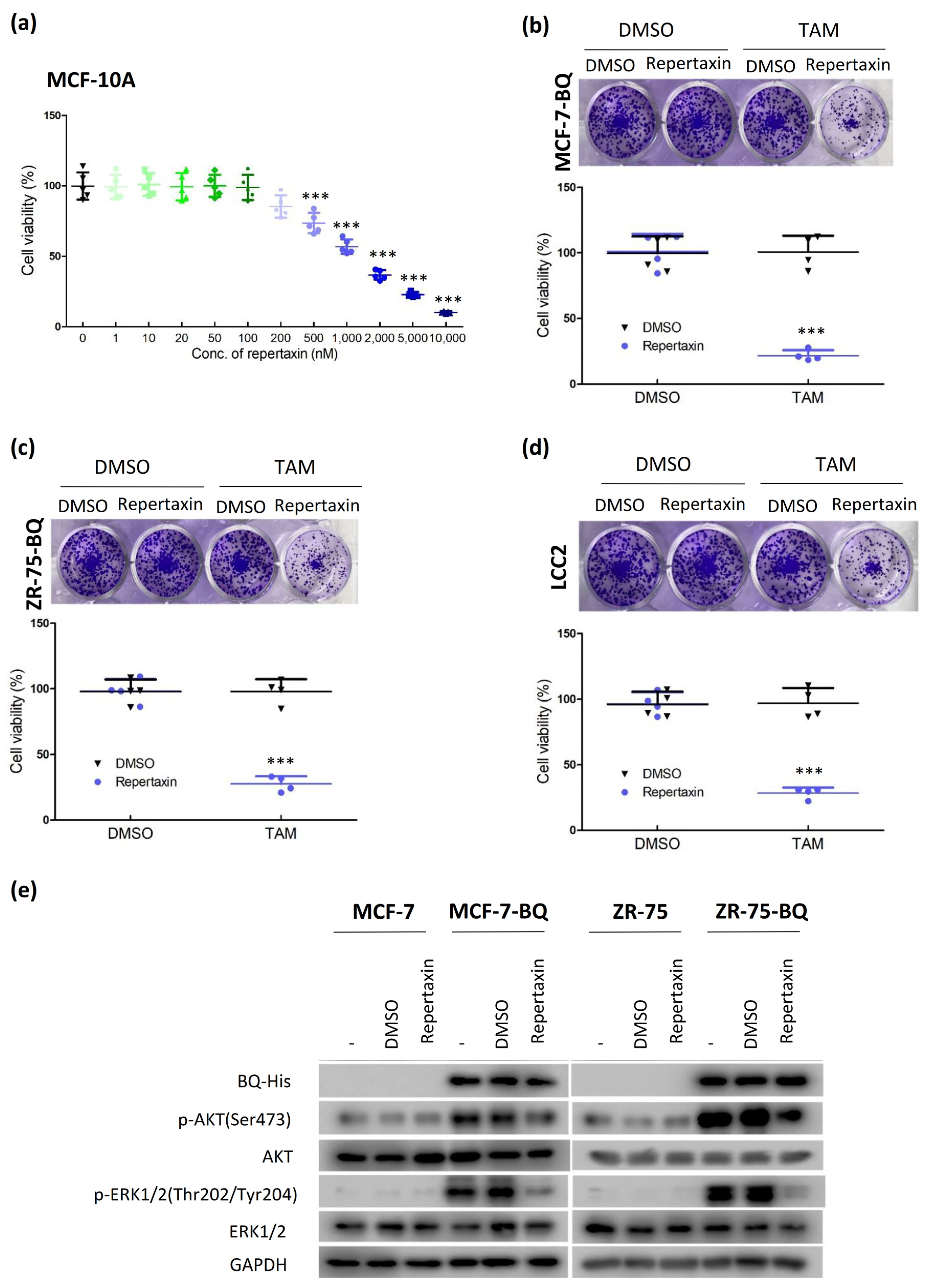
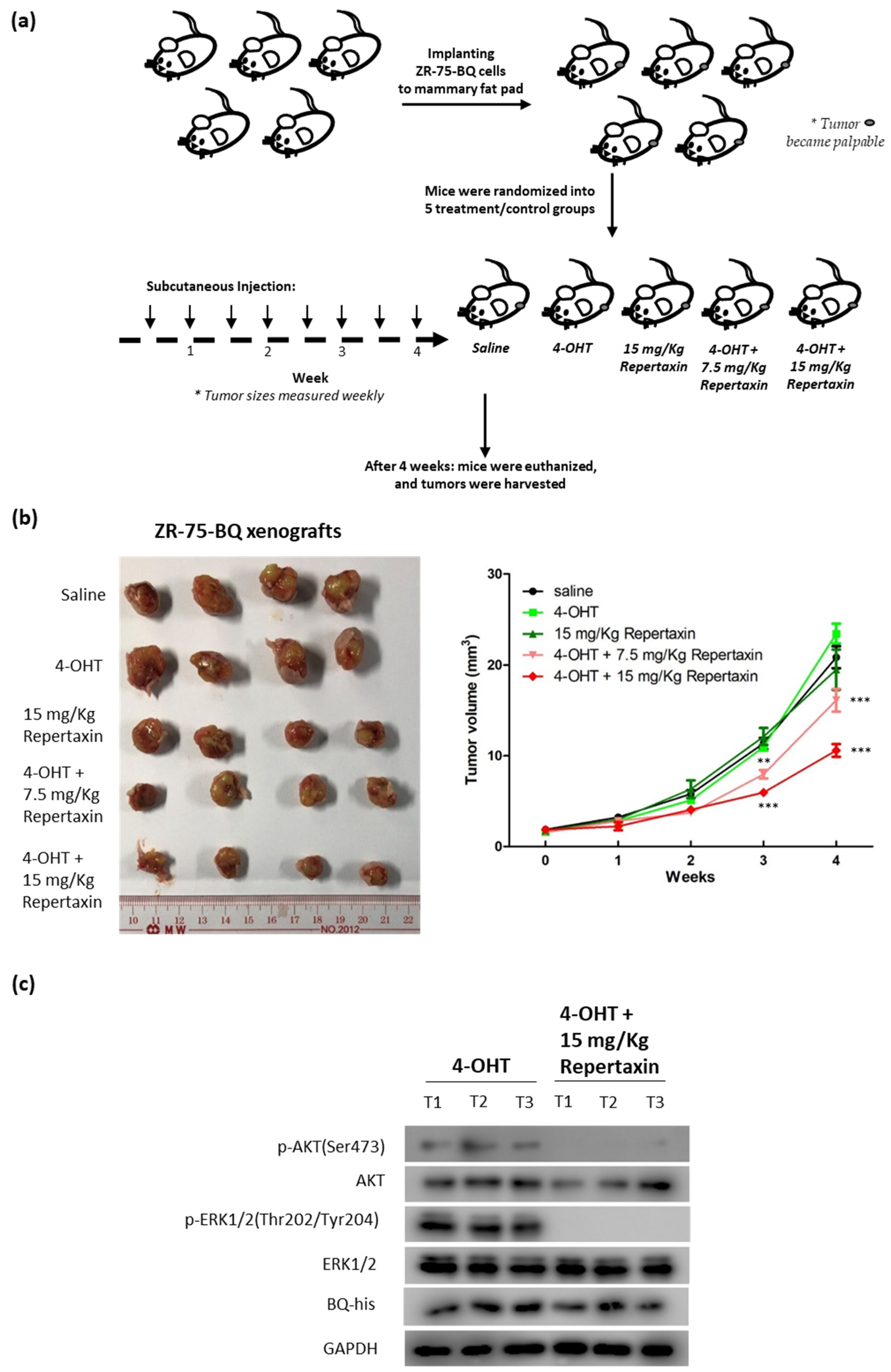
3.4. Targeting CXCR1/2 Could Reverse Tamoxifen Resistance in Breast Cancer In Vitro and In Vivo
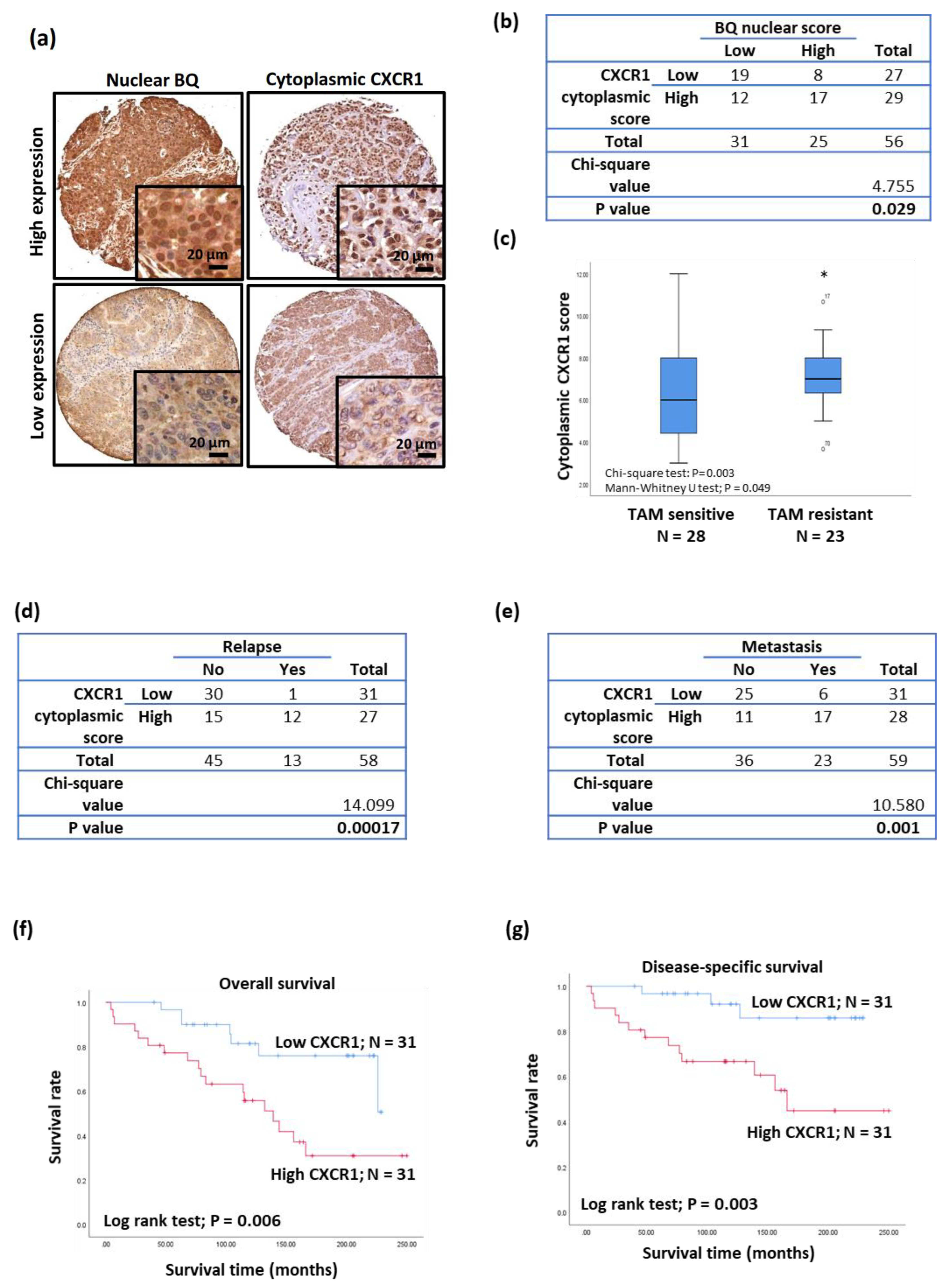
3.5. Clinical Significance of CXCR1 in Breast Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Sørlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Børresen-Dale, A.L. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef]
- Rozeboom, B.; Dey, N.; De, P. ER+ metastatic breast cancer: Past, present, and a prescription for an apoptosis-targeted future. Am. J. Cancer Res. 2019, 9, 2821–2831. [Google Scholar] [PubMed]
- Kulkoyluoglu, E.; Madak-Erdogan, Z. Nuclear and extranuclear-initiated estrogen receptor signaling crosstalk and endocrine resistance in breast cancer. Steroids 2016, 114, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Katzenellenbogen, J.A.; Mayne, C.G.; Katzenellenbogen, B.S.; Greene, G.L.; Chandarlapaty, S. Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance. Nat. Rev. Cancer 2018, 18, 377–388. [Google Scholar] [CrossRef]
- Patel, H.K.; Bihani, T. Selective estrogen receptor modulators (SERMs) and selective estrogen receptor degraders (SERDs) in cancer treatment. Pharm. Ther. 2018, 186, 1–24. [Google Scholar] [CrossRef]
- Nazarali, S.A.; Narod, S.A. Tamoxifen for women at high risk of breast cancer. Breast Cancer Targets Ther. 2014, 6, 29–36. [Google Scholar]
- Davies, C.; Godwin, J.; Gray, R.; Clarke, M.J.; Cutter, D.; Darby, S.; Peto, R. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomized trials. Lancet 2011, 378, 771–784. [Google Scholar] [PubMed]
- Yao, J.; Deng, K.; Huang, J.; Zeng, R.; Zuo, J. Progress in the Understanding of the Mechanism of Tamoxifen Resistance in Breast Cancer. Front. Pharmacol. 2020, 11, 592912. [Google Scholar] [CrossRef] [PubMed]
- Dowsett, M.; Haynes, B.P. Hormonal effects of aromatase inhibitors: Focus on premenopausal effects and interaction with tamoxifen. J. Steroid Biochem. Mol. Biol. 2003, 86, 255–263. [Google Scholar] [CrossRef]
- Chang, M. Tamoxifen Resistance in Breast Cancer. Biomol. Ther. 2012, 20, 256–267. [Google Scholar] [CrossRef]
- Osborne, C.K.; Bardou, V.; Hopp, T.A.; Chamness, G.C.; Hilsenbeck, S.G.; Fuqua, S.A.; Schiff, R. Role of the estrogen receptor co-activator AIB1 (SRC-3) and HER-2/neu in tamoxifen resistance in breast cancer. J. Natl. Cancer Inst. 2003, 95, 353–361. [Google Scholar] [CrossRef]
- Keeton, E.K.; Brown, M. Cell cycle progression stimulated by tamoxifen-bound estrogen receptor-alpha and promoter-specific effects in breast cancer cells deficient in N-CoR and SMRT. Mol. Endocrinol. 2005, 19, 1543–1554. [Google Scholar] [CrossRef]
- Zhang, L.; Gong, C.; Lau, S.L.; Yang, N.; Wong, O.G.; Cheung, A.N.; Khoo, U.S. SpliceArray Profiling of Breast Cancer Reveals a Novel Variant of NCOR2/SMRT That Is Associated with Tamoxifen Resistance and Control of ER alpha Transcriptional Activity. Cancer Res. 2013, 73, 246–255. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Man, E.P.; Tsoi, H.; Lee, T.K.; Lee, P.; Ma, S.T.; Khoo, U.S. BQ323636,1, a Novel Splice Variant to NCOR2, as a Predictor for Tamoxifen-Resistant Breast Cancer. Clin. Cancer Res. 2018, 24, 3681–3691. [Google Scholar] [CrossRef]
- Bostner, J.; Karlsson, E.; Pandiyan, M.J.; Westman, H.; Skoog, L.; Fornander, T.; Stål, O. Activation of Akt, mTOR, and the estrogen receptor as a signature to predict tamoxifen treatment benefit. Breast Cancer Res. Treat. 2013, 137, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, N.A.; Fang, J.; Huang, J.; Tian, F.; Li, C.; Xie, F. Role of PKC-ERK signaling in tamoxifen-induced apoptosis and tamoxifen resistance in human breast cancer cells. Oncol. Rep. 2012, 27, 1879–1886. [Google Scholar] [PubMed]
- Jahan, N.; Jones, C.; Rahman, R.L. Androgen receptor expression in breast cancer: Implications on prognosis and treatment, a brief review. Mol. Cell. Endocrinol. 2021, 531. [Google Scholar] [CrossRef] [PubMed]
- De Amicis, F.; Thirugnansampanthan, J.; Cui, Y.; Selever, J.; Beyer, A.; Parra, I.; Fuqua, S.A. Androgen receptor overexpression induces tamoxifen resistance in human breast cancer cells. Breast Cancer Res. Treat. 2010, 121, 1–11. [Google Scholar] [CrossRef]
- Varlakhanova, N.; Snyder, C.; Jose, S.; Hahm, J.B.; Privalsky, M.L. Estrogen Receptors Recruit SMRT and N-CoR Corepressors through Newly Recognized Contacts between the Corepressor N Terminus and the Receptor DNA Binding Domain. Mol. Cell. Biol. 2010, 30, 1434–1445. [Google Scholar] [CrossRef] [PubMed]
- Liao, G.; Chen, L.Y.; Zhang, A.; Godavarthy, A.; Xia, F.; Ghosh, J.C.; Chen, J.D. Regulation of androgen receptor activity by the nuclear receptor co-repressor SMRT. J. Biol. Chem. 2003, 278, 5052–5061. [Google Scholar] [CrossRef]
- Tsoi, H.; Man, E.P.; Chau, K.M.; Khoo, U.S. Targeting the IL-6/STAT3 Signalling Cascade to Reverse Tamoxifen Resistance in Estrogen Receptor Positive Breast Cancer. Cancers 2021, 13, 1511. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Wongvipat, J.; Choi, D.; Wang, P.; Lee, Y.S.; Zheng, D.; Sawyers, C.L. GREB1 amplifies androgen receptor output in human prostate cancer and contributes to antiandrogen resistance. Elife 2019, 8, e41913. [Google Scholar] [CrossRef]
- Hall, J.M.; McDonnell, D.P. The estrogen receptor beta-isoform (ERbeta) of the human estrogen receptor modulates ERalpha transcriptional activity and is a key regulator of the cellular response to estrogens and antiestrogens. Endocrinology 1999, 140, 5566–5578. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.K.; Simões, B.M.; Howell, S.J.; Farnie, G.; Clarke, R.B. Recent advances reveal IL-8 signaling as a potential key to targeting breast cancer stem cells. Breast Cancer Res. 2013, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Waugh, D.J.J.; Wilson, C. The Interleukin-8 Pathway in Cancer. Clin. Cancer Res. 2008, 14, 6735–6741. [Google Scholar] [CrossRef]
- Citro, A.; Cantarelli, E.; Piemonti, L. The CXCR1/2 Pathway: Involvement in Diabetes Pathophysiology and Potential Target for T1D Interventions. Curr. Diabetes Rep. 2015, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Wang, X.; Miao, W.; Wang, B.; Qiu, Y. CXCL8 promotes the invasion of human osteosarcoma cells by regulation of PI3K/Akt signaling pathway. Apmis 2017, 125, 773–780. [Google Scholar] [CrossRef]
- Liu, Q.; Li, A.; Tian, Y.; Wu, J.D.; Liu, Y.; Li, T.; Wu, K. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 2016, 31, 61–71. [Google Scholar] [CrossRef]
- Rajagopalan, L.; Rajarathnam, K. Ligand selectivity and affinity of chemokine receptor CXCR1—Role of N-terminal domain. J. Biol. Chem. 2004, 279, 30000–30008. [Google Scholar] [CrossRef]
- Milovanović, J.; Todorović-Raković, N.; Vujasinović, T.; Rabi, Z.A. Interleukin 8 in progression of hormone-dependent early breast cancer. J. Biosci. 2017, 42, 265–274. [Google Scholar] [CrossRef]
- Shi, Z.; Yang, W.M.; Chen, L.P.; Yang, D.H.; Zhou, Q.; Zhu, J.; Huang, R.P. Enhanced chemosensitization in multidrug-resistant human breast cancer cells by inhibition of IL-6 and IL-8 production. Breast Cancer Res. Treat. 2012, 135, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Jeselsohn, R.; Pereira, R.; Hollingsworth, E.F.; Creighton, C.J.; Li, F.; Schiff, R. FOXA1 overexpression mediates endocrine resistance by altering the ER transcriptome and IL-8 expression in ER-positive breast cancer. Proc. Natl. Acad. Sci. USA 2016, 113, E6600–E6609. [Google Scholar] [CrossRef]
- Sharma, I.; Singh, A.; Siraj, F.; Saxena, S. IL-8/CXCR1/2 signalling promotes tumor cell proliferation, invasion and vascular mimicry in glioblastoma. J. Biomed. Sci. 2018, 25, 1–13. [Google Scholar] [CrossRef]
- Du, L.; Han, X.G.; Tu, B.; Wang, M.Q.; Qiao, H.; Zhang, S.H.; Tang, T.T. CXCR1/Akt signaling activation induced by mesenchymal stem cell-derived IL-8 promotes osteosarcoma cell anoikis resistance and pulmonary metastasis. Cell Death Dis. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Raghuwanshi, S.K.; Su, Y.; Singh, V.; Haynes, K.; Richmond, A.; Richardson, R.M. The chemokine receptors CXCR1 and CXCR2 couple to distinct G protein-coupled receptor kinases to mediate and regulate leukocyte functions. J. Immunol. 2012, 189, 2824–2832. [Google Scholar] [CrossRef] [PubMed]
- Hasson, S.P.; Rubinek, T.; Ryvo, L.; Wolf, I. Endocrine resistance in breast cancer: Focus on the phosphatidylinositol 3-kinase/akt/mammalian target of rapamycin signaling pathway. Breast Care 2013, 8, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Cui, Y.K.; Huang, W.H.; Man, K.; Zhang, G.J. Phosphorylation of estrogen receptor alpha at serine 118 is correlated with breast cancer resistance to tamoxifen. Oncol. Lett. 2013, 6, 118–124. [Google Scholar] [CrossRef][Green Version]
- Guo, Y.; Zang, Y.; Lv, L.; Cai, F.; Qian, T.; Zhang, G.; Feng, Q. IL8 promotes proliferation and inhibition of apoptosis via STAT3/AKT/NFkappaB pathway in prostate cancer. Mol. Med. Rep. 2017, 16, 9035–9042. [Google Scholar] [CrossRef]
- Guo, F.; Long, L.; Wang, J.; Wang, Y.; Liu, Y.; Wang, L.; Luo, F. Insights on CXC chemokine receptor 2 in breast cancer: An emerging target for oncotherapy. Oncol. Lett. 2019, 18, 5699–5708. [Google Scholar] [CrossRef]
- Niemeier, L.A.; Dabbs, D.J.; Beriwal, S.; Striebel, J.M.; Bhargava, R. Androgen receptor in breast cancer: Expression in estrogen receptor-positive tumors and in estrogen receptor-negative tumors with apocrine differentiation. Mod. Pathol. 2010, 23, 205–212. [Google Scholar] [CrossRef]
- Collins, L.C.; Cole, K.S.; Marotti, J.D.; Hu, R.; Schnitt, S.J.; Tamimi, R.M. Androgen receptor expression in breast cancer in relation to molecular phenotype: Results from the Nurses’ Health Study. Mod. Pathol. 2011, 24, 924–931. [Google Scholar] [CrossRef]
- Aleskandarany, M.A.; Abduljabbar, R.; Ashankyty, I.; Elmouna, A.; Jerjees, D.; Ali, S.; Rakha, E.A. Prognostic significance of androgen receptor expression in invasive breast cancer: Transcriptomic and protein expression analysis. Breast Cancer Res. Treat. 2016, 159, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Witzel, I.; Graeser, M.; Karn, T.; Schmidt, M.; Wirtz, R.; Schütze, D.; Müller, V. Androgen receptor expression is a predictive marker in chemotherapy-treated patients with endocrine receptor-positive primary breast cancers. J. Cancer Res. Clin. Oncol. 2013, 139, 809–816. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Dawood, S.; Holmes, M.D.; Collins, L.C.; Schnitt, S.J.; Cole, K.; Tamimi, R.M. Androgen receptor expression and breast cancer survival in postmenopausal women. Clin. Cancer Res. 2011, 17, 1867–1874. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, D.R.; Bernales, S.; Jacobsen, B.M.; Cittelly, D.M.; Howe, E.N.; D’Amato, N.C.; Richer, J.K. Role of the androgen receptor in breast cancer and preclinical analysis of enzalutamide. Breast Cancer Res. 2014, 16, R7. [Google Scholar] [CrossRef]

| Clinical-Pathological Parameters | Univariate Analysis | |
|---|---|---|
| RR (95% CI) | p value | |
| Age (n = 69) | 1.682 (0.785, 3.602) | 0.181 |
| T-stage (n = 30) | 5.522 (1.226, 24.871) | 0.026 |
| Lymph-node involvement (n = 63) | 0.981 (0.438, 2.197) | 0.962 |
| Tumor-Grade (n = 68) | 1.389 (0.637, 3.027) | 0.409 |
| Histological type (n = 69) | 1.368 (0.323, 5.795) | 0.671 |
| HER2 status (n = 49) | 1.11 (0.427, 2.888) | 0.83 |
| Tumor size (n = 52) | 0.938 (0.377, 2.334) | 0.89 |
| Cases with Hi-CXCR1 cytoplasm score (n = 62) | 3.171 (1.322, 7.61) | 0.01 |
| Cases with both Hi-CXCR1 & BQ score (n = 36) | 3.205 (1.107, 9.276) | 0.032 |
| Clinical-pathological parameters | Multivariate analysis | |
| RR (95% CI) | p value | |
| T-stage (n = 26) | 8.332 (1.363, 50.943) | 0.022 |
| Cases with Hi-CXCR1 cytoplasm score (n = 26) | 3.265 (0.805, 13.247) | 0.098 |
| Clinical-pathological parameters | Multivariate analysis | |
| RR (95% CI) | p value | |
| T-stage (n = 17) | 9.31 (1.016, 85.311) | 0.048 |
| Cases with both Hi-CXCR1 & BQ score (n = 17) | 5.053 (0.796, 32.066) | 0.086 |
| Clinical-Pathological Parameters | Univariate Analysis | |
|---|---|---|
| RR (95% CI) | p value | |
| Age (n = 69) | 1.37 (0.543, 3.456) | 0.505 |
| T-stage (n = 30) | 3.695 (0.67, 20.372) | 0.133 |
| Lymph-node involvement (n = 63) | 1.373 (0.497, 3.796) | 0.541 |
| Tumor-Grade (n = 68) | 3.463 (1.139, 10.525) | 0.029 |
| Histological type (n = 69) | 0.81 (0.186, 3.527) | 0.778 |
| HER2 status (n = 49) | 1.715 (0.499, 5.893) | 0.392 |
| Tumor size (n = 52) | 1.341 (0.403, 4.46) | 0.632 |
| Cases with Hi-CXCR1 cytoplasm score (n = 62) | 5.35 (1.519, 18.84) | 0.009 |
| Cases with both Hi-CXCR1 & BQ score (n = 36) | 5.401 (1.5, 19.449) | 0.01 |
| Clinical-pathological parameters | Multivariate analysis | |
| RR (95% CI) | p value | |
| Tumor-Grade (n = 61) | 3.113 (0.879, 11.026) | 0.078 |
| Cases with Hi-CXCR1 cytoplasm score (n = 61) | 4.661 (1.313, 16.545) | 0.017 |
| Clinical-pathological parameters | Multivariate analysis | |
| RR (95% CI) | p value | |
| Tumor-Grade (n = 36) | 1.642 (0.447, 6.04) | 0.455 |
| Cases with both Hi-CXCR1 & BQ score (n = 36) | 4.86 (1.318, 17.919) | 0.018 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsoi, H.; Shi, L.; Leung, M.-H.; Man, E.P.S.; So, Z.-Q.; Chan, W.-L.; Khoo, U.-S. Overexpression of BQ323636.1 Modulated AR/IL-8/CXCR1 Axis to Confer Tamoxifen Resistance in ER-Positive Breast Cancer. Life 2022, 12, 93. https://doi.org/10.3390/life12010093
Tsoi H, Shi L, Leung M-H, Man EPS, So Z-Q, Chan W-L, Khoo U-S. Overexpression of BQ323636.1 Modulated AR/IL-8/CXCR1 Axis to Confer Tamoxifen Resistance in ER-Positive Breast Cancer. Life. 2022; 12(1):93. https://doi.org/10.3390/life12010093
Chicago/Turabian StyleTsoi, Ho, Ling Shi, Man-Hong Leung, Ellen P. S. Man, Zi-Qing So, Wing-Lok Chan, and Ui-Soon Khoo. 2022. "Overexpression of BQ323636.1 Modulated AR/IL-8/CXCR1 Axis to Confer Tamoxifen Resistance in ER-Positive Breast Cancer" Life 12, no. 1: 93. https://doi.org/10.3390/life12010093
APA StyleTsoi, H., Shi, L., Leung, M.-H., Man, E. P. S., So, Z.-Q., Chan, W.-L., & Khoo, U.-S. (2022). Overexpression of BQ323636.1 Modulated AR/IL-8/CXCR1 Axis to Confer Tamoxifen Resistance in ER-Positive Breast Cancer. Life, 12(1), 93. https://doi.org/10.3390/life12010093






