AntiGan: An Epinutraceutical Bioproduct with Antitumor Properties in Cultured Cell Lines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Preparation of AntiGan (E-Congerine-10423) and Treatments
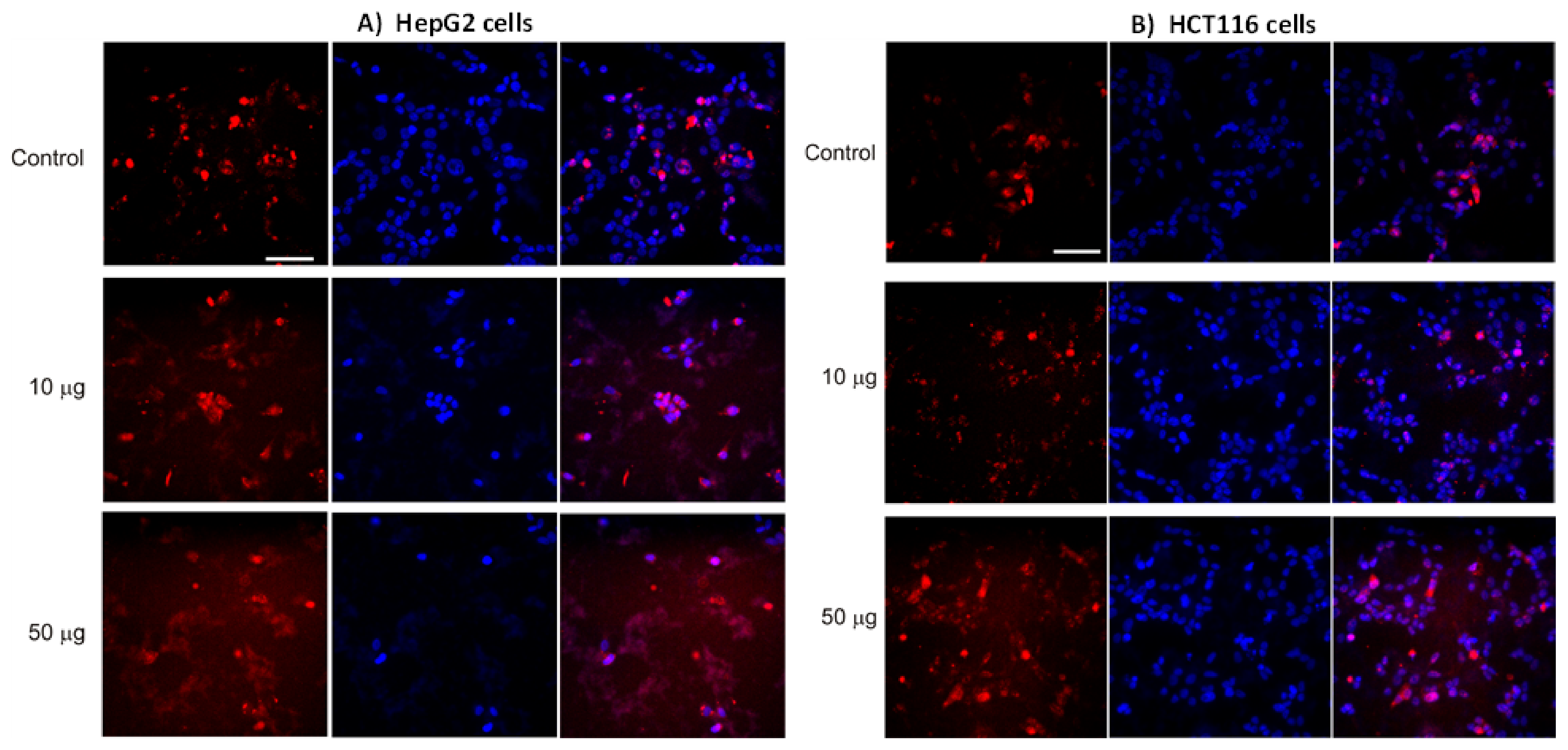
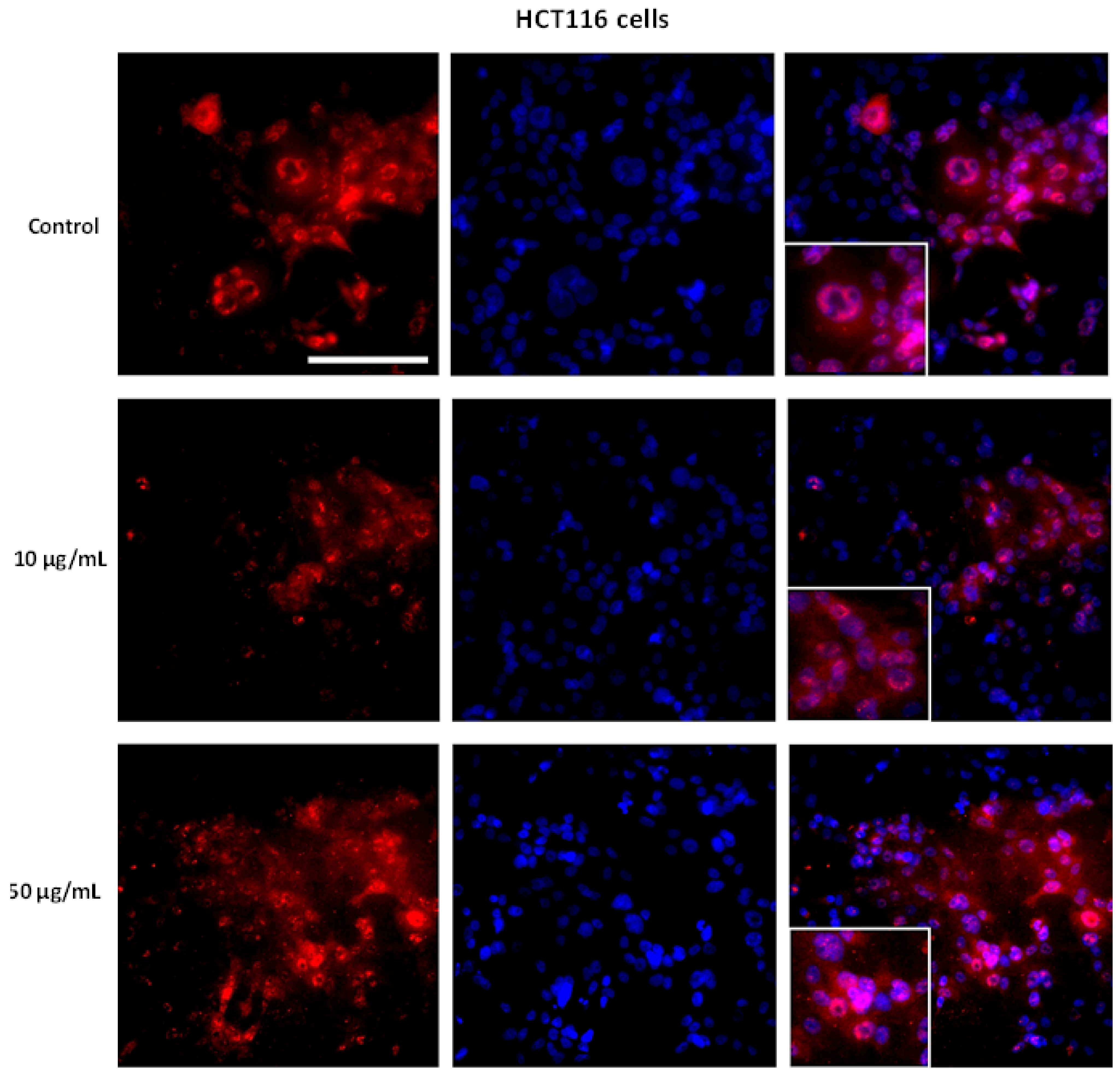
2.3. Immunofluorescence
2.4. Cell Viability Assay
2.5. DNA Extraction
2.6. Quantification of Global DNA Methylation (5mC)
2.7. RNA Extraction
2.8. Quantitative Real-Time PCR (qPCR)
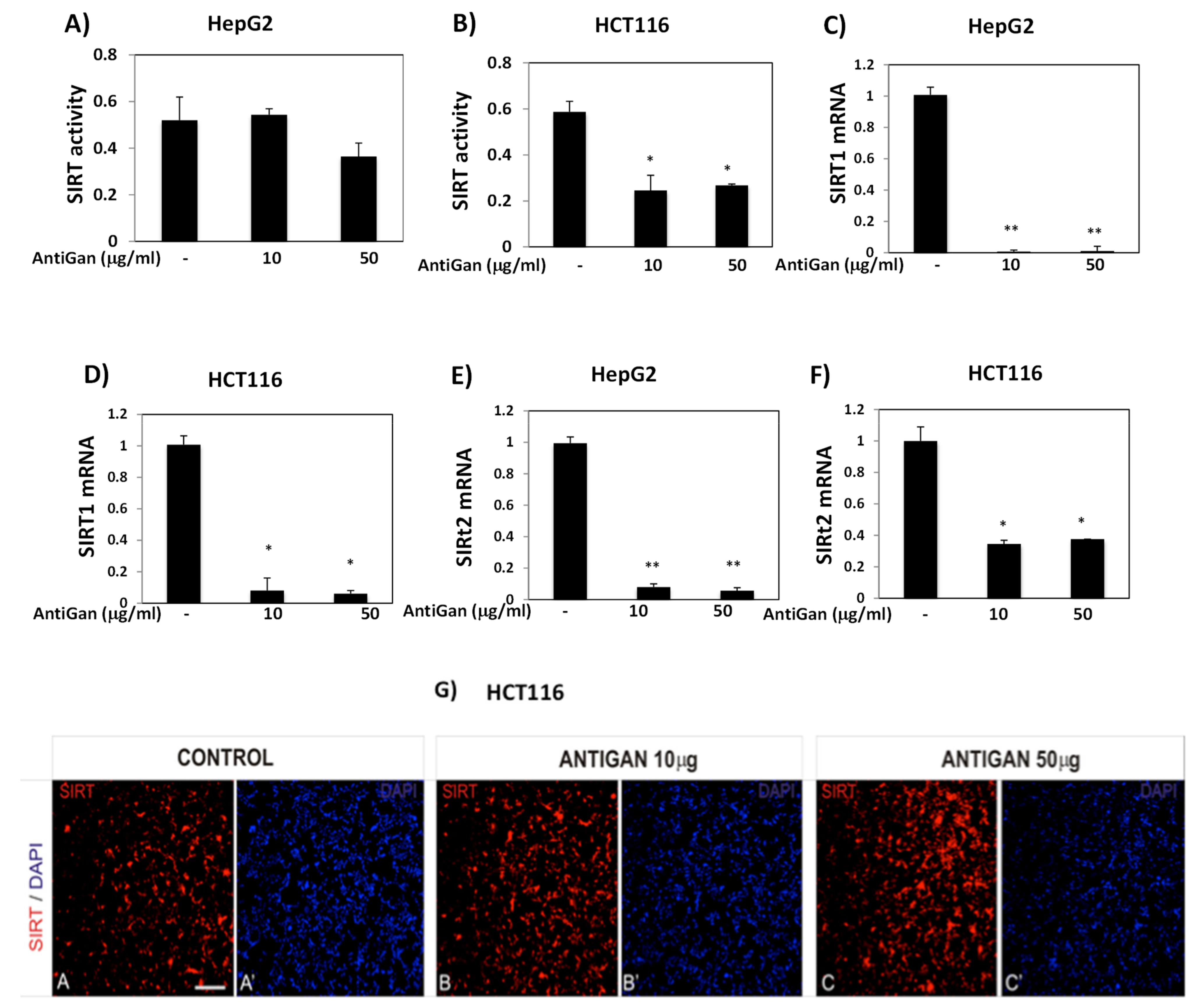
2.9. Sirtuin Activity Assay
2.9.1. Nuclear Protein Extraction
2.9.2. Quantification of SIRT Activity
2.10. Statistical Analysis
3. Results
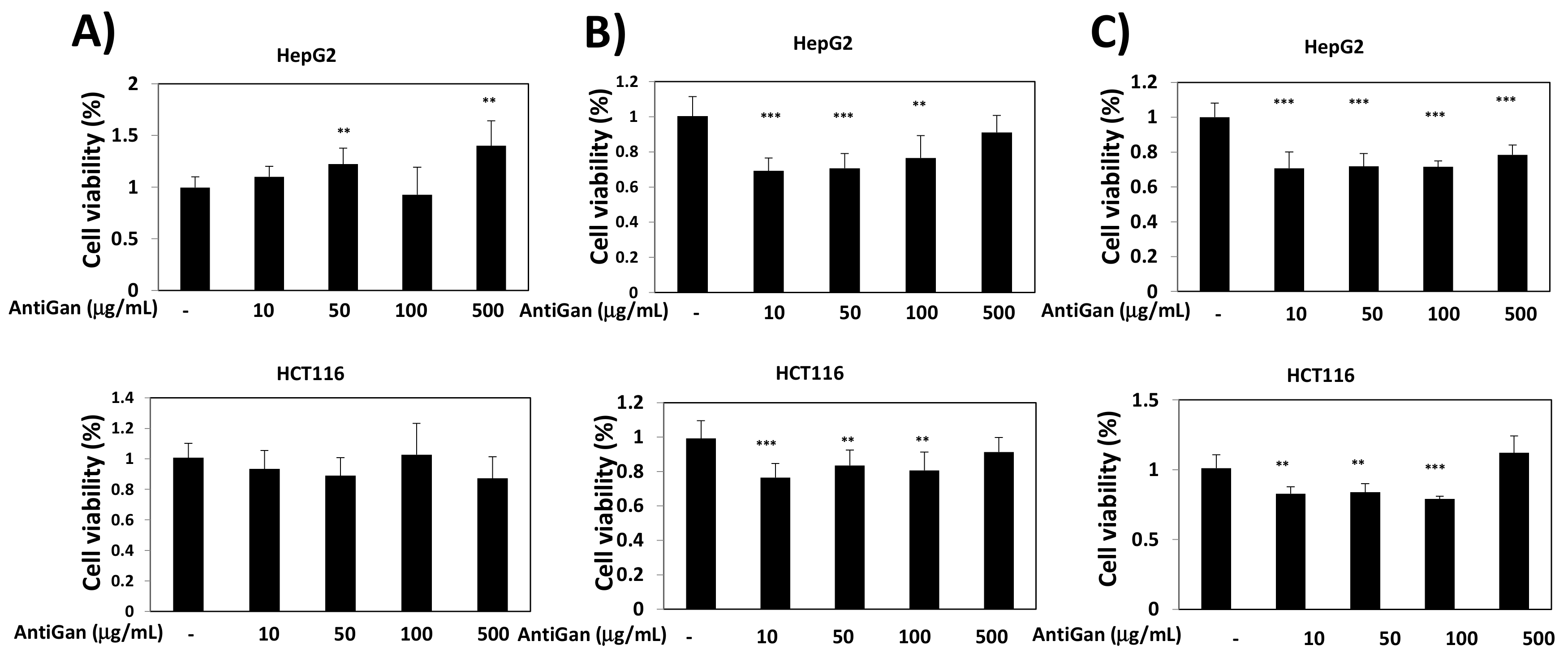
3.1. Effect of AntiGan on Cell Viability of Human Cancer Cell Lines
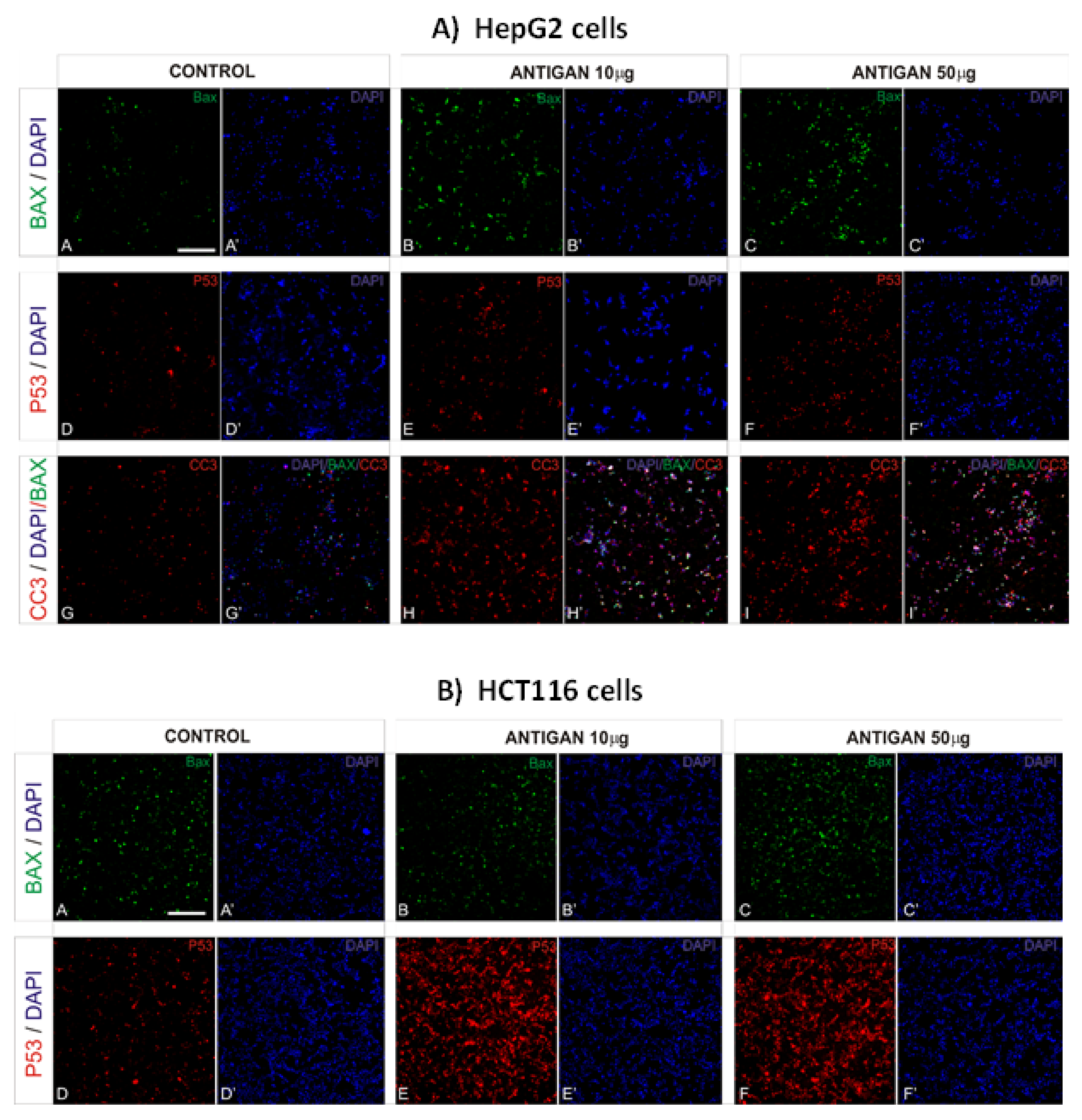
3.2. AntiGan Treatment Induces Apoptosis in Human Cancer Cell Lines
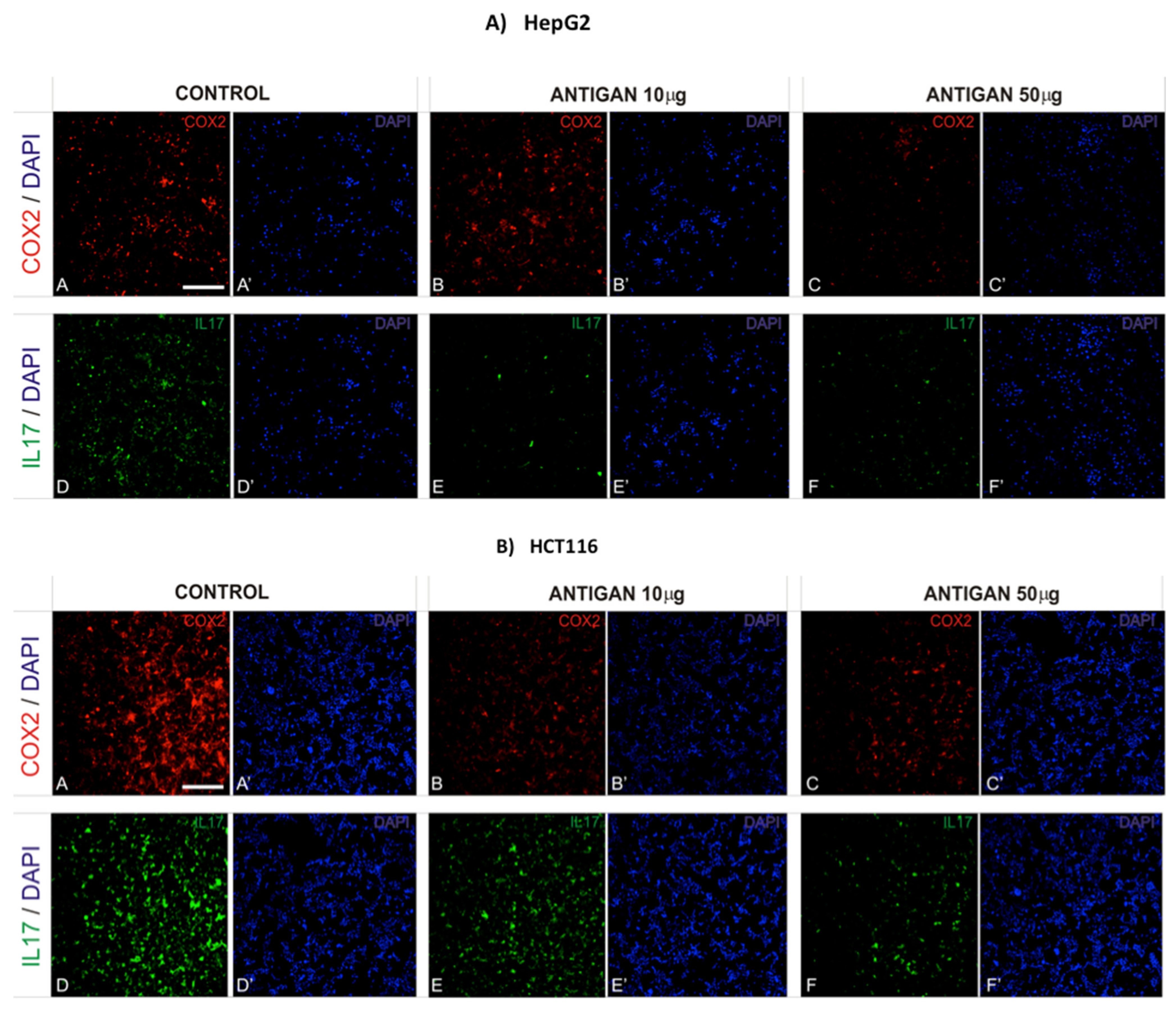
3.3. AntiGan Regulates Cyclooxygenase-2 (COX-2) and Interleukin-17 (IL-17) Expression
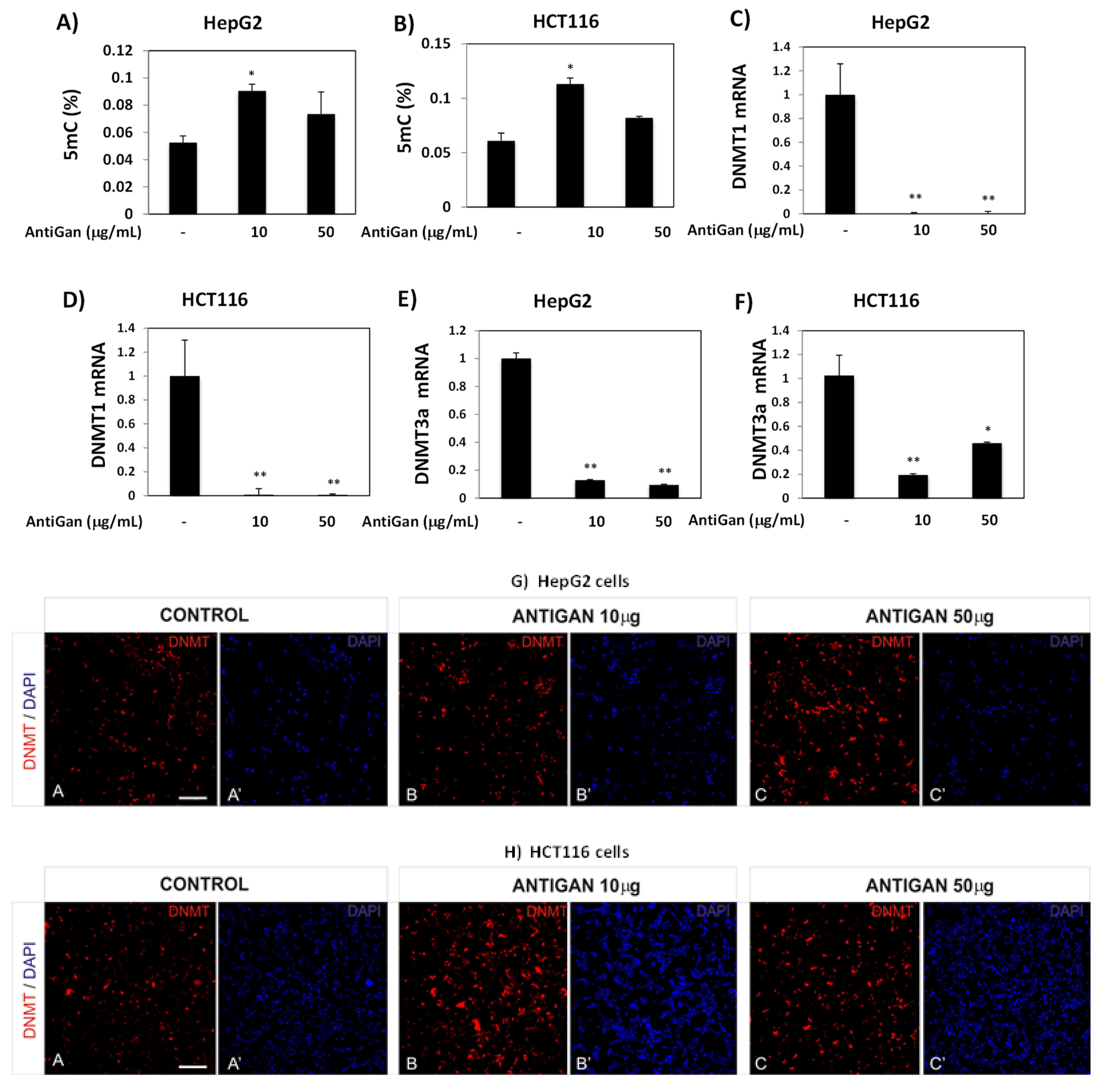
3.4. AntiGan Regulates DNA Methylation in HepG2 and HCT116 Cells
3.5. AntiGan Regulates Sirtuin Expression and Activity in HepG2 and HCT116 Cells
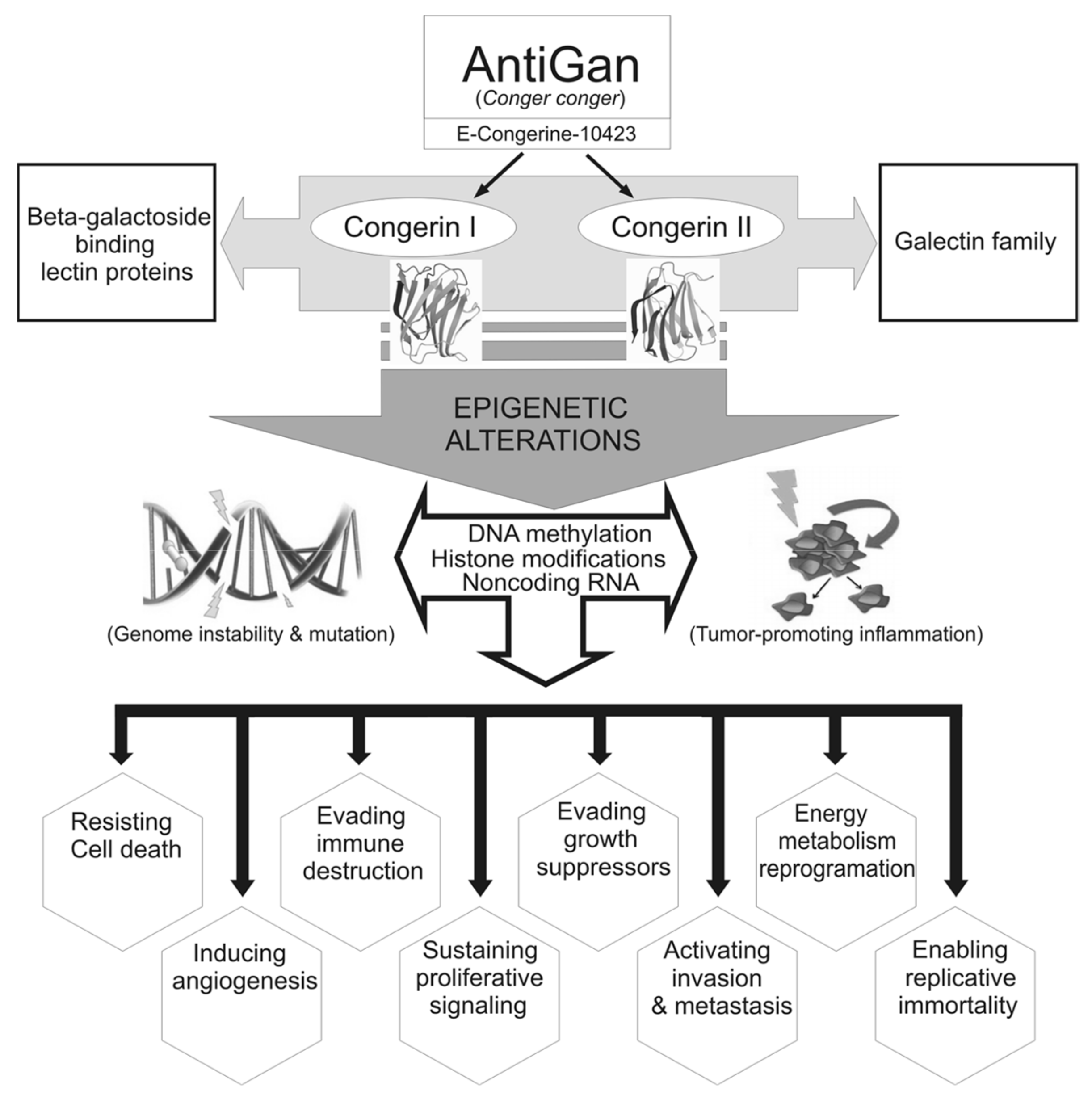
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. The Hallmaks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Rahib, L.; Wehner, M.; Matrisian, L.M.; Nead, K.T. Estimated Projection of US Cancer Incidence and Death to 2040. JAMA Netw. Open 2021, 4, e214708. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Xu, P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021, 14, 101174. [Google Scholar] [CrossRef] [PubMed]
- Singal, A.G.; Lampertico, P.; Nahon, P. Epidemiology and surveillance for hepatocellular carcinoma: New trends. J. Heptol. 2020, 72, 250–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cacabelos, R.; Tellado, I.; Cacabelos, P. The epigenetic machinery in the life cycle and pharmacoepigenetics. Pharmacoepigenetics 2019, 10, 1–100. [Google Scholar]
- Lu, Y.; Chan, Y.-T.; Tan, H.-Y.; Wang, N.; Feng, Y. Epigenetic regulation in human cancer: The potential role of epidrug in cancer therapy. Mol. Cancer 2020, 19, 1–16. [Google Scholar] [CrossRef]
- Esteller, M. Epigenetics in cancer. N. Eng. J. Med. 2008, 358, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. The essentials of DNA methylation. Cell 1992, 70, 5–8. [Google Scholar] [CrossRef]
- Hermann, A.; Gowher, H.; Jeltsch, A. Biochemistry and biology of mammalian DNA methyltransferases. Cell. Mol. Life Sci. 2004, 61, 2571–2587. [Google Scholar] [CrossRef]
- Esteller, M. Cancer epigenomics:DNA methylomes and histone-modifications maps. Nat. Rev. Genet. 2007, 8, 286–298. [Google Scholar] [CrossRef]
- Elmallah, M.I.Y.; Micheau, O. Epigenetic regulation of TRAIL signaling: Implications for cancer therapy. Cancers 2019, 11, 850. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Wei, X.; Wu, Q.; Xu, Z.; Gu, D.; Jin, Y.; Shen, Y.; Huang, H.; Fan, H.; Chen, J. Clinical significance of the expression of DNA methyltransferase proteins in gastric cancer. Mol. Med. Rep. 2011, 4, 1139–1143. [Google Scholar]
- Lin, C.H.; Hsieh, S.Y.; Sheen, I.S.; Lee, W.C.; Chen, T.C.; Shyu, W.C.; Liaw, Y.F. Genome-wide hypomethylation in hepatocellulat carcinogenesis. Cancer Res. 2001, 61, 4238–4243. [Google Scholar]
- Pappalardo, X.G.; Barra, V. Losing DNA methylation at repetitive elements and breaking bad. Epigenet. Chromatin. 2021, 14, 1–21. [Google Scholar] [CrossRef]
- Takai, D. Hypomethylation of LINE1 Retrotransposon in human ¡hepatocellular carcinoma, but not in surrounding liver cirrhosis. Jpn. J. Clin. Oncol. 2002, 30, 306–309. [Google Scholar] [CrossRef] [Green Version]
- Verdone, L.; Caserta, M.; Di Mauro, E. Role of histone acetylation in the control of gene expression. Biochem. Cell. Biol. 2005, 83, 344–353. [Google Scholar] [CrossRef]
- Carafa, V.; Rotili, D.; Forgione, M.; Cuomo, F.; Serretiello, E.; Hailu, G.-S.; Jarho, E.; Lahtela-Kakkonen, M.; Mai, A.; Altucci, L. Sirtuin functions and modulation: From chemistry to the clinic. Clin. Epig. 2016, 8, 1–21. [Google Scholar] [CrossRef]
- Michan, S.; Sinclair, D. Sirtuins in mammals: Insights into their biological function. Biochem. J. 2007, 404, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Cacabelos, R. Epigenomic networking in Drug Development: From Pathogenic Mechanisms to Pharmacogenomics. Drug. Dev. Res. 2014, 75, 348–365. [Google Scholar] [CrossRef]
- Nakamura, O.; Matsuoka, H.; Ogawa, T.; Muramoto, K.; Kamiya, H.; Watanabe, T. Opsonic effect of congerin, a mucosal galectin of the Japanese conger, Conger myriaster (Brevoort). Fish Shellfish Immunol. 2006, 20, 433–435. [Google Scholar] [CrossRef]
- Jayasinghe, C.D.; Udalamaththa, A.; Imbulana, I.B.P.S.; Suetake, I. Dietary phytochemicals as Epi-drugs: Role in modulating the epigenetic mechanisms of human diseases. Int. J. Curr. Pharm. Rev. Res. 2015, 7, 50–58. [Google Scholar]
- Carrera, I.; Martinez, O.; Cacabelos, R. Neuroprotection with natural antioxidants and nutraceiticals in the context of brain cell degeneration: The epigenetic connection. Curr. Top. Med. Chem. 2019, 19, 2999–3011. [Google Scholar] [CrossRef]
- Cacabelos, R. The role of epigenetic biomarkers in disease pathogenesis. Pharmacoepigenetics 2019, 10, 139–189. [Google Scholar]
- Carlos-Reyes, A.; López-González, J.S.; Meneses-Flores, M.; Gallardo-Rincón, D.; Ruíz-García, E.; Marchat, L.A.; Astudillo-de la Vega, H.; Hernandez de la Cruz, O.N.; López-Camarillo, C. Dietary compounds as epigeneting modulating agents in cancer. Front Genet. 2019, 10, 79. [Google Scholar] [CrossRef] [Green Version]
- Calvani, M.; Pasha, A.; Favre, C. Nutraceutical Boom in cancer: Inside the labyrinth of reactive oxygen species. Int. J. Mol. Sci. 2020, 21, 1936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozyerli-Goknar, E.; Bagci-Onder, T. Epigenetic deregulation of apoptosis in cancer. Cancers 2021, 13, 3210. [Google Scholar] [CrossRef]
- Santini, A.; Tenore, G.C.; Novellino, E. Nutraceuticals: A paradigm of proactive medicine. Eur. J. Pharm. Sci. 2017, 96, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Ranzato, E.; Martinotti, S.; Calabrese, C.; Calabrese, G. Role of nutraceuticals in cancer therapy. J. Food Res. 2014, 3, 18. [Google Scholar] [CrossRef] [Green Version]
- Divella, R.; Daniele, A.; Savino, E.; Paradiso, A. Anticancer effects of nutraceuticals in the mediterranean diet: An epigenetic diet model. Cancer Genom. Prot. 2020, 17, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, O.; Inaga, I.; Suzuki, S.; Tsutsui, S.; Muramoto, K.; Kamiya, H.; Watanabe, T. Possible immune functions of congerine, a mucosal galectin, in the intstinal lumen of Japanese conger eel. Fish Shelfish Immunol. 2007, 23, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Corzo, L.; Fernandez-Novoa, L.; Carrera, I.; Martinez, O.; Rodriguez, S.; Alejo, R.; Cacabelos, R. Nutrition, health and disease: Role of selected marine and vegetal nutraceuticals. Nutrients 2020, 12, 747. [Google Scholar] [CrossRef] [Green Version]
- Kamiya, H.; Muramoto, K.; Goto, R. Purification and properties of agglutinins from conger eel, Conger Myriaster (Brevoort), skin mucus. Dev. Comp. Immunol. 1988, 12, 309–318. [Google Scholar] [CrossRef]
- Ogawa, T.; Watanabe, M.; Naganuma, T.; Muramoto, K. Diversified carbohydrate-binding lectins from marine resources. J. Amino Acids 2011, 2011, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Lombardi, V.R.M.; Carrera, I.; Cacabelos, R. In vitro and in vivo cytotoxic effect of AntiGan against tumor cells. Exp. Therap. Med. 2018, 15, 2547–2556. [Google Scholar] [CrossRef]
- Lombardi, V.R.M.; Carrera, I.; Corzo, L.; Cacabelos, R. Role of lipofishins in prevention of inflammation and colon cancer. Semin. Cancer Biol. 2019, 56, 175–184. [Google Scholar] [CrossRef]
- Martinez-Iglesias, O.; Carrera, I.; Carrila, J.C.; Fernandez-Novoa, L.; Cacabelos, N.; Cacabelos, R. DNA methylation in Neurodegenerative and Cerebrovascular Disorders. Int. J. Mol. Sci. 2020, 21, 2220. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Iglesias, O.; Naidoo, V.; Carril, J.C.; Carrera, I.; Corzo, L.; Rodriguez, S.; Alejo, R.; Cacabelos, N.; Cacabelos, R. AtreMorine treatment regulates DNA methylation in neurodegenrative disorders: Epigenetic and pharmacogenetic studies. Curr. Pharm. Pers. Med. 2020, 17, 159–171. [Google Scholar]
- Cacabelos, R.; Carrera, I.; Martinez, O.; Alejo, R.; Fernandez-Novoa, L.; Cacabelos, P.; Corzo, L.; Rodríguez, S.; Alcaraz, S.; Nebril, L.; et al. Atremorine in Parkinson’s disease: From dopaminergic neuroprotection to pharmacogenomics. Med. Res. Rev. 2021, 41, 2841–2886. [Google Scholar] [CrossRef]
- Natalino, R.J.M.; Antoneli, C.B.G.; Ribeiro, K.; Campos, A.H.; Soares, F.A. Immunohistochemistry of apoptosis-related proteins in retinoblastoma. Pathol. Res. Pract. 2016, 212, 1144–1150. [Google Scholar] [CrossRef]
- Fernandez-Albarral, J.A.; Salazar, J.J.; de Hoz, R.; Marco, E.M.; Martín-Sanchez, B.; Flores-Salguero, E.; Salobrar-Garcia, E.; Lopez-Cuenca, I.; Barrios-Sabador, V.; Avilé-Trigueros, M.; et al. Retinal Molecular Changes Are Associated with Neuroinflammation and Loss of RGCs in an Experimental Model of Glaucoma. Int. J. Mol. Sci. 2021, 22, 2066. [Google Scholar] [CrossRef]
- Kobayashi, K.; Tomita, H.; Shimizu, M.; Tanaka, T.; Suzuki, N.; Miyazaki, T.; Hara, A. p53 Expression as a Diagnostic Biomarker in Ulcerative Colitis-Associated Cancer. Int. J. Mol. Sci. 2017, 18, 1284. [Google Scholar] [CrossRef] [PubMed]
- Eang, R.; Song, F.; Li, S.; Wu, B.; Gu, Y.; Yuan, Y. Salvianolic acid A attenuates CCl4-induced liver fibrosis by regulating the PI3K/AKT/mTOR, Bcl-2/Bax and caspase-3/cleaved caspase-3 signaling pathways. Drug Des. Devel. Ther. 2019, 13, 1889–1900. [Google Scholar]
- Sun, M.; Sheng, Y.; Zhu, Y. Ginkgolide B alleviates the inflammatory response and attenuates the activation of LPS-induced BV2 cells in vitro and in vivo. Exp. Ther. Med. 2021, 21, 586. [Google Scholar] [CrossRef] [PubMed]
- Samal, S.; Patnaik, A.; Sahu, F.; Purkait, S. Altered expression of epigenetic modifiers EZH2, H3K27me3, and DNA methyltransferases in meningiomas - Prognostic biomarkers for routine practice. Folia Neuropathol. 2020, 58, 133–142. [Google Scholar] [CrossRef]
- Hong, W.G.; Pyo, J.-S. The clinicopathological significance of SIRT1 expression in colon cancer: An immunohistochemical study and meta-analysis. Pathol. Res. Pract. 2018, 214, 1550–1555. [Google Scholar] [CrossRef]
- Wang, M.L.; Medrano, J.F. Real-time PCR for mRNA quantitation. BioTechniques 2005, 39, 75–85. [Google Scholar] [CrossRef]
- Aden, D.P.; Fogel, A.; Plotkin, S.; Damjanov, I.; Knowles, N.B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma derived cell line. Nature 1979, 282, 615–616. [Google Scholar] [CrossRef]
- Rajput, A.; San Martin, I.-D.; Rose, R.; Beko, A.; Levea, C.; Sharratt, E.; Mazurchuk, R.; Hoffman, R.M.; Brattain, M.G.; Wang, J. Characterization of HCT116 Human Colon cancer Cells in a Orthotopic Model. J. Surg. Res. 2007, 147, 276–281. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Lowe, S.W.; Lin, A.W. Apoptosis in cancer. Carcinogenesis 2000, 21, 485–495. [Google Scholar] [CrossRef] [Green Version]
- Porter, A.G.; Jänicke, R.U. Emerging role of caspase-3 in apoptosis. Cell Death Differ. 1999, 6, 99–104. [Google Scholar] [CrossRef]
- Goradel, N.H.; Najafi, M.; Salehi, E.; Farhood, B.; Mortezaee, K. Cyclooxygenase-2 in cancer: A review. J. Cell. Physiol. 2019, 234, 5683–5699. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, X.; Herjan, T.; Li, X. The role of interleukin-17 in tumor development and progression. J. Exp. Med. 2019, 217, e20190297. [Google Scholar] [CrossRef]
- Ehrlich, E. DNA hypomethylation in cancer cells. Epigenomics 2009, 1, 239–259. [Google Scholar] [CrossRef] [Green Version]
- Zhao, E.; Hou, J.; Ke, X.; Abbas, M.N.; Kausar, S.; Zhang, L.; Cui, H. The roles of Sirtuin family proteins in cancer progression. Cancers 2019, 11, 1949. [Google Scholar] [CrossRef] [Green Version]
- Saunders, L.R.; Verdin, E. Sirtuins: Critical regulators at the crossroads between cancer and aging. Oncogene 2007, 26, 5489–5504. [Google Scholar] [CrossRef] [Green Version]
- Sosnowska, B.; Penson, P.; Banach, M. The role of nutraceuticals in the prevention of cardiovascular disease. Cardiovasc. Diagn. Ther. 2017, 7, S21–S31. [Google Scholar] [CrossRef] [Green Version]
- Dadhania, V.P.; Trivedi, P.P.; Vikram, A.; Tripathi, D.N. Nutraceuticals against neurodegeneration: A mechanistic insight. Curr. Neuropharmacol. 2016, 14, 627–640. [Google Scholar] [CrossRef]
- Tropathi, Y.B.; Tripathi, P.; Arjmandi, B.H. Nutraceuticals and cancer management. Front. Biosci. 2005, 10, 1607–1618. [Google Scholar] [CrossRef]
- De Mejia, E.G.; Dia, V.P. The role of nutraceutical proteins and peptides in apoptosis, angiogenesis, and metastasis of cancer cells. Cancer Metastasis Rev. 2010, 29, 511–528. [Google Scholar] [CrossRef]
- Cerella, C.; Sobolewski, C.; Dicato, M.; Diederich, M. Targeting COX-2 expression by natural compounds: A promising alternative strategy to synthetic COX-2 inhibitors for cancer. Biochem. Pharmacol. 2010, 80, 1801–1815. [Google Scholar] [CrossRef]
- Fabre, J.; Giustiniani, J.; Garbar, C.; Antonicelli, F.; Merrouche, Y.; Bensussan, A.; Bagot, M.; Al-Dacak, R. Targeting the tumor microenvironment: The protumor effects of IL-17 realted to cancer type. Int. J. Mol. Sci. 2016, 17, 1433. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.P.; Yan, J.; Xu, J.; Pang, X.-H.; Chen, M.-S.; Li, L.; Wu, C.; Li, S.-P.; Zheng, L. Increased intratumoral IL-17-producing cells correlate with por sirvival in hepatocellular carcinoma patients. J. Hepatol. 2009, 50, 980–989. [Google Scholar] [CrossRef]
- Sharma, S.V.; Lee, D.Y.; Li, B.; Quinlan, M.P.; Takahashi, F.; Maheswaran, S.; McDermott, U.; Aziziab, N.; Zou, L.; Fischbach, M.A.; et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010, 141, 69–80. [Google Scholar] [CrossRef] [Green Version]
- El-Khoury, V.; Breuzard, G.; Fourré, N.; Dufer, J. The histone deacetylase inhibitor trichostatin A downregulates human MDR1 (ABCB1) gene expression by a transcription-dependent mechanism in a drug-resistant small cell lung carcinoma cell line model. Br. J. Cancer 2007, 97, 562–573. [Google Scholar] [CrossRef]
- Kumar, S.; Kushwaha, P.P.; Gupta, S. Emerging targets in cancer drug resistance. Cancer Drug Resist. 2019, 2, 161–177. [Google Scholar] [CrossRef] [Green Version]
- Giri, A.K.; Aittokallio, T. DNMT inhibitors increase methylation in the cancer genome. Front. Pharmacol. 2019, 10, 385. [Google Scholar] [CrossRef] [Green Version]
- Carafa, V.; Altucci, L.; Nrbbioso, A. Dual tumor suppressor and tumor promoter action of sirtuins in determining malignant phenotype. Front. Pharmacol. 2019, 10, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luos, J.; Nikolaev, A.Y.; Imai, S.; Chen, D.; Su, F.; Shiloh, A.; Guarente, L.; Gu, W. Negative control of p53 by Sir2a promotes cell survival under stress. Cell 2001, 107, 137–148. [Google Scholar]
- Hu, J.; Jing, H.; Lin, H. Sirtuin inhibitors as anticancer agents. Future Med. Chem. 2014, 6, 945–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.; Fang, D. The roles of SIRT1 in cancer. Genes Cancer 2013, 4, 97–104. [Google Scholar] [CrossRef] [Green Version]
- O’Hagan, H.M.; Wang, W.; Sen, S.; Shields, C.D.; Lee, S.S.; Zhang, Y.W.; Clements, E.G.; Cai, Y.; Neste, L.V.; Easwaran, H.; et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG islands. Cancer Cell 2011, 42, 210–223. [Google Scholar]
- Yang, T.; Zhou, R.; Yu, S.; Cui, Z.; Hu, P.; Liu, J.; Qiao, Q.; Zhang, J. Cytoplasmatic SIRT1 inhibits cell migration and invasion by impeding epithelial-mesenchymal transition in ovarian carcinoma. Mol. Cell. Biochem. 2019, 459, 157–169. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Chan, A.W.H.; To, K.-F.; Chen, W.; Zhang, Z.; Ren, J.; Song, C.; Cheung, Y.-S.; Lai, P.B.S.; Cheng, S.-H.; et al. SIRT2 overexpression in hepatocellular carcinoma mediates epithelial to mesenchymal transition by protein kinase B/glycogen synthase kinase-3beta/beta-catenin signaling. Hepatology 2013, 57, 2287–2298. [Google Scholar] [CrossRef]
- Cheon, M.G.; Kim, W.; Choi, M.; Kim, J.-E. AK-1, a specific SIRT2 inhibitor, induces cell cycle arrest by downregulation Snail in HCT116 human colon carcinoma cells. Cancer Lett. 2015, 356, 637–645. [Google Scholar] [CrossRef]
- Farooqi, A.S.; Hong, J.Y.; Cao, J.; Lu, X.; Price, R.; Zhao, Q.; Kosciuk, T.; Yang, M.; Bai, Y.J.; Lin, H. Novel lysine-based thioureas as mechanism-based inhibitors of sirtuin 2 (SIRT2) whith anticancer activities in a colorectal cancer murine model. J. Med. Chem. 2019, 62, 3131–3141. [Google Scholar] [CrossRef]
- Suzuki, T.; Khan, M.N.A.; Sawada, H.; Imai, E.; Itoh, Y.; Yamatsuta, K.; Tokuda, N.; Takeuchi, J.; Seko, T.; Nakagawa, H.; et al. Design, synthesis and biological activity of a novel series of human sirtuin-2-selective inhibitors. J. Med. Chem. 2012, 6, 5760–5773. [Google Scholar] [CrossRef]
- Kim, M.J.; Kang, Y.J.; Sung, B.; Jang, J.Y.; Ahn, Y.R.; Oh, H.J.; Choi, H.; Choi, I.; Im, E.; Moon, H.R.; et al. Novel SIRT inhibitor, MHY2256, induces cell cycle arrest, apoptosis, and autophagic cell death in HCT116 human colorectal cancer cells. Biomol. Ther. 2020, 28, 561–568. [Google Scholar] [CrossRef]
- Hontecillas-Prieto, L.; Flores-Campos, R.; Silver, A.; de Alava, E.; Hajji, N.; Garcia-Dominguez, D.J. Synergistic enhancement of cancer therapy using HDAC inhibitors: Opportunity for clinical trials. Front. Genet. 2020, 11, 578011. [Google Scholar] [CrossRef]
| COMPOSITION (%) | MINERALS per 100 g | |||
| PROTEINS | 75–85 | PHOSPHORUS | 100 mg | |
| LIPIDS | 0.5–1.5 | CALCIUM | 164 mg | |
| CARBOHYDRATES | 0.4 | MAGNESIUM | 134 mg | |
| AMINO ACIDS: % IN PROTEINS | IRON | 14 mg | ||
| GLUTAMIC ACID | 13.71 | ZINC | 3 mg | |
| ASPARTIC ACID | 8.26 | VITAMINS per 100 g | ||
| LYSINE | 7.80 | VITAMIN B3 | 760 mg | |
| LEUCINE | 6.49 | VITAMIN B1 | 0.2 mg | |
| ARGININE | 5.37 | VITAMIN B2 | 0.08 mg | |
| ALANINE | 4.76 | VITAMIN D | 0.25 mcg | |
| VALINE | 3.82 | LIPIDS | ||
| THREONINE | 3.76 | SATURATED FATTY ACIDS: % IN LIPIDS | ||
| ISOLEUCINE | 3.70 | PALMITIC | 21.8 | |
| SERINE | 3.35 | STEARIC | 8.60 | |
| PHENYLALANINE | 3.21 | MYRISTIC | 4.60 | |
| GLYCINE | 3.10 | POLYUNSATURATED FATTY ACIDS: % IN LIPIDS | ||
| PROLINE | 2.90 | OLEIC | 23.20 | |
| TYROSINE | 2.71 | PALMITOLEIC | 5.50 | |
| METHIONINE | 2.29 | GADOLEIC | 4.90 | |
| HISTIDINE | 2.09 | MONOSATURATED FATTY ACIDS: % IN LIPIDS | ||
| CYSTEINE | 0.94 | LINOLEIC | 12.00 | |
| TRYPTOPHAN | 0.84 | DHA | 6.30 | |
| LINOLENIC | 2.00 | |||
| EPA | 1.80 | |||
| Antibody | Species | Clonality | Supplier | Product Number | Ref. |
|---|---|---|---|---|---|
| Bax | Mouse | Monoclonal | Thermo Fisher | MA5-14003 | [39] |
| IL-17 | Mouse | Monoclonal | Thermo Fisher | 12-7177-81 | [40] |
| p53 | Rabbit | Monoclonal | Thermo Fisher | MA5-14516 | [41] |
| Cleaved caspase-3 | Rabbit | Polyclonal | Cell Signaling | 9664 | [42] |
| Cox-2 | Rabbit | Monoclonal | Vector Labs. | VP-RM02 | [43] |
| Dnmt3a | Rabbit | Polyclonal | Abcam | ab2850 | [44] |
| Sirt1 | Rabbit | Monoclonal | Abcam | ab32441 | [45] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Iglesias, O.; Carrera, I.; Naidoo, V.; Cacabelos, R. AntiGan: An Epinutraceutical Bioproduct with Antitumor Properties in Cultured Cell Lines. Life 2022, 12, 97. https://doi.org/10.3390/life12010097
Martínez-Iglesias O, Carrera I, Naidoo V, Cacabelos R. AntiGan: An Epinutraceutical Bioproduct with Antitumor Properties in Cultured Cell Lines. Life. 2022; 12(1):97. https://doi.org/10.3390/life12010097
Chicago/Turabian StyleMartínez-Iglesias, Olaia, Ivan Carrera, Vinogran Naidoo, and Ramón Cacabelos. 2022. "AntiGan: An Epinutraceutical Bioproduct with Antitumor Properties in Cultured Cell Lines" Life 12, no. 1: 97. https://doi.org/10.3390/life12010097
APA StyleMartínez-Iglesias, O., Carrera, I., Naidoo, V., & Cacabelos, R. (2022). AntiGan: An Epinutraceutical Bioproduct with Antitumor Properties in Cultured Cell Lines. Life, 12(1), 97. https://doi.org/10.3390/life12010097









