Aeromonas allosaccharophila Strain AE59-TE2 Is Highly Antagonistic towards Multidrug-Resistant Human Pathogens, What Does Its Genome Tell Us?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Isolation
2.2. Antimicrobial Activity Screening
2.3. DNA Extraction, Illumina Sequencing, Data Preprocessing, and Genome Assembly
2.4. Taxonomic Identification by Molecular Methods
2.5. Virulence Potential and Antibiotic Resistance
2.6. Genome Functional Annotation and Mining
3. Results
3.1. Sample Identification and Phenotypic Characterization of Antibacterial Activity of Aeromonas Isolates
3.2. AE59-TE2 Genome Sequencing and Assembly
3.3. AE59-TE2 Species Identification and Aeromonas Taxonomy
3.4. Virulence Potential and Antimicrobial Resistance
3.5. Genome Mining of Aeromonas allosaccharophila AE59-TE2
3.6. Identification and Comparative Analysis of AE59-TE2 Bacteriocin-Related Sequences
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hemeg, H.A. Nanomaterials for alternative antibacterial therapy. Int. J. Nanomed. 2017, 12, 8211–8225. [Google Scholar] [CrossRef]
- Ayukekbong, J.A.; Ntemgwa, M.; Atabe, A.N. The threat of antimicrobial resistance in developing countries: Causes and control strategies. Antimicrob. Resist. Infect. Control 2017, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Donkor, E.S. Nosocomial Pathogens: An In-Depth Analysis of the Vectorial Potential of Cockroaches. Trop. Med. Infect. Dis. 2019, 4, 14. [Google Scholar] [CrossRef] [PubMed]
- Tamma, P.D.; Aitken, S.L.; Bonomo, R.A.; Mathers, A.J.; van Duin, D.; Clancy, C.J. Infectious Diseases Society of America Guidance on the Treatment of Extended-Spectrum β-lactamase Producing Enterobacterales (ESBL-E), Carbapenem-Resistant Enterobacterales (CRE), and Pseudomonas aeruginosa with Difficult-to-Treat Resistance (DTR-P. aeruginosa). Clin. Infect. Dis. 2020, 72, e169–e183. [Google Scholar] [CrossRef]
- Sánchez-López, E.; Gomes, D.; Esteruelas, G.; Bonilla, L.; Lopez-Machado, A.L.; Galindo, R.; Cano, A.; Espina, M.; Ettcheto, M.; Camins, A.; et al. Metal-Based Nanoparticles as Antimicrobial Agents: An Overview. Nanomaterials 2020, 10, 292. [Google Scholar] [CrossRef]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Radaic, A.; de Jesus, M.B.; Kapila, Y.L. Bacterial anti-microbial peptides and nano-sized drug delivery systems: The state of the art toward improved bacteriocins. J. Control. Release 2020, 321, 100–118. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.; Godino, A.; Príncipe, A.; Ramírez, V.L.; Quesada, J.M.; Rigo, V.; Espinosa-Urgel, M.; Morales, G.M.; Fischer, S. Characterization of the bacteriocins and the PrtR regulator in a plant-associated Pseudomonas strain. J. Biotechnol. 2020, 307, 182–192. [Google Scholar] [CrossRef]
- Zhu, J.-W.; Zhang, S.-J.; Wang, W.-G.; Jiang, H. Strategies for Discovering New Antibiotics from Bacteria in the Post-Genomic Era. Curr. Microbiol. 2020, 77, 3213–3223. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Bravo, A.; Figueras, M.J. An Update on the Genus Aeromonas: Taxonomy, Epidemiology, and Pathogenicity. Microorganisms 2020, 8, 129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbott, S.L.; Cheung, W.K.W.; Janda, J.M. The Genus Aeromonas: Biochemical Characteristics, Atypical Reactions, and Phenotypic Identification Schemes. J. Clin. Microbiol. 2003, 41, 2348–2357. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The Genus Aeromonas: Taxonomy, Pathogenicity, and Infection. Clin. Microbiol. Rev. 2010, 23, 35–73. [Google Scholar] [CrossRef]
- Valderrama, K.; Soto-Dávila, M.; Segovia, C.; Vásquez, I.; Dang, M.; Santander, J. Aeromonas salmonicida infects Atlantic salmon (Salmo salar) erythrocytes. J. Fish Dis. 2019, 42, 1601–1608. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Han, J.E.; Kwon, H.; Park, S.C.; Kim, J.H. Recent Insights into Aeromonas salmonicida and Its Bacteriophages in Aquaculture: A Comprehensive Review. J. Microbiol. Biotechnol. 2020, 30, 1443–1457. [Google Scholar] [CrossRef] [PubMed]
- Tomás, J.M. The Main Aeromonas Pathogenic Factors. ISRN Microbiol. 2012, 2012, 256261. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Chen, Y.; Yang, W.; Qiao, Z.; Zhang, X. Complete genome sequence of fish-pathogenic Aeromonas hydrophila HX-3 and a comparative analysis: Insights into virulence factors and quorum sensing. Sci. Rep. 2020, 10, 15479. [Google Scholar] [CrossRef] [PubMed]
- Greiner, M.; Anagnostopoulos, A.; Pohl, D.; Zbinden, R.; Zbinden, A. A rare case of severe gastroenteritis caused by Aeromonas hydrophila after colectomy in a patient with anti-Hu syndrome: A case report. BMC Infect. Dis. 2021, 21, 1097. [Google Scholar] [CrossRef] [PubMed]
- Von Graevenitz, A. The Role of Aeromonas in Diarrhea: A Review. Infection 2007, 35, 59–64. [Google Scholar] [CrossRef]
- Dias, C.; Ribeiro, M.; Correia-Branco, A.; Domínguez-Perles, R.; Martel, F.; Saavedra, M.J.; Simões, M. Virulence, attachment and invasion of Caco-2 cells by multidrug-resistant bacteria isolated from wild animals. Microb. Pathog. 2019, 128, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Dias, C.; Borges, A.; Saavedra, M.J.; Simões, M. Biofilm formation and multidrug-resistant Aeromonas spp. from wild animals. J. Glob. Antimicrob. Resist. 2018, 12, 227–234. [Google Scholar] [CrossRef] [PubMed]
- Moro, E.; Weiss, R.D.N.; Friedrich, R.S.; Nunes, M.P. Bacteriocin-like Substance of Aeromonas hydrophila. Mem. Do Inst. Oswaldo Cruz 1997, 92, 115–116. [Google Scholar] [CrossRef] [PubMed]
- Messi, P.; Guerrieri, E.; Bondi, M. Bacteriocin-like substance (BLS) production in Aeromonas hydrophila water isolates. FEMS Microbiol. Lett. 2003, 220, 121–125. [Google Scholar] [CrossRef]
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, Antimicrobial Peptides from Bacterial Origin: Overview of Their Biology and Their Impact against Multidrug-Resistant Bacteria. Microorganisms 2020, 8, 639. [Google Scholar] [CrossRef]
- Kaur, S.; Kaur, S. Bacteriocins as Potential Anticancer Agents. Front. Pharmacol. 2015, 6, 272. [Google Scholar] [CrossRef] [PubMed]
- Crowe, S.J.; Bottichio, L.; Shade, L.N.; Whitney, B.M.; Corral, N.; Melius, B.; Arends, K.D.; Donovan, D.; Stone, J.; Allen, K.; et al. Shiga Toxin–Producing E. coli Infections Associated with Flour. N. Engl. J. Med. 2017, 377, 2036–2043. [Google Scholar] [CrossRef]
- Hergens, M.-P.; Öhd, J.N.; Alm, E.; Askling, H.H.; Helgesson, S.; Insulander, M.; Lagerqvist, N.; Svenungsson, B.; Tihane, M.; Tolfvenstam, T.; et al. Investigation of a food-borne outbreak of gastroenteritis in a school canteen revealed a variant of sapovirus genogroup V not detected by standard PCR, Sollentuna, Sweden, 2016. Eurosurveillance 2017, 22, 30543. [Google Scholar] [CrossRef]
- Pisoschi, A.M.; Pop, A.; Georgescu, C.; Turcuş, V.; Olah, N.K.; Mathe, E. An overview of natural antimicrobials role in food. Eur. J. Med. Chem. 2018, 143, 922–935. [Google Scholar] [CrossRef]
- Schulz, S.; Stephan, A.; Hahn, S.; Bortesi, L.; Jarczowski, F.; Bettmann, U.; Paschke, A.-K.; Tusé, D.; Stahl, C.H.; Giritch, A.; et al. Broad and efficient control of major foodborne pathogenic strains of Escherichia coli by mixtures of plant-produced colicins. Proc. Natl. Acad. Sci. USA 2015, 112, E5454–E5460. [Google Scholar] [CrossRef]
- Śmiałek, J.; Nowakowski, M.; Bzowska, M.; Bocheńska, O.; Wlizło, A.; Kozik, A.; Dubin, G.; Mak, P. Structure, Biosynthesis, and Biological Activity of Succinylated Forms of Bacteriocin BacSp222. Int. J. Mol. Sci. 2021, 22, 6256. [Google Scholar] [CrossRef]
- Palú, A.P.; Gomes, L.M.; Miguel, M.A.L.; Balassiano, I.T.; Queiroz, M.L.P.; Freitas-Almeida, A.C.; de Oliveira, S.S. Antimicrobial resistance in food and clinical Aeromonas isolates. Food Microbiol. 2006, 23, 504–509. [Google Scholar] [CrossRef]
- Martin-Carnahan, A.; Joseph, S.W. Aeromonadaceae. In The Proteobacteria, Part B, Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Brenner, D.J., Krieg, N.R., Staley, J.T., Garrity, G.M., Eds.; Springer: New York, NY, USA, 2005; Volume 2, pp. 556–580. [Google Scholar]
- Tagg, J.R.; McGiven, A.R. Assay System for Bacteriocins. Appl. Microbiol. 1971, 21, 943. [Google Scholar] [CrossRef] [PubMed]
- Giambiagi-Marval, M.; Mafra, M.A.; Penido, E.G.C.; Bastos, M.C.F. Distinct groups of plasmids correlated with bacteriocin production in Staphylococcus aureus. J. Gen. Microbiol. 1990, 136, 1591–1599. [Google Scholar] [CrossRef]
- Reich, M.; Liefeld, T.; Gould, J.; Lerner, J.; Tamayo, P.; Mesirov, J.P. GenePattern 2.0. Nat. Genet. 2006, 38, 500–501. [Google Scholar] [CrossRef] [PubMed]
- Ausubel, F.M.; Brent, R.; Kingston, R.E.; Moore, D.D.; Seidman, J.G.; Smith, J.A.; Struhl, K. (Eds.) Current Protocols in Molecular Biology; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1997. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Bosi, E.; Donati, B.; Galardini, M.; Brunetti, S.; Sagot, M.-F.; Lio, P.; Crescenzi, P.; Fani, R.; Fondi, M. MeDuSa: A multidraft based scaffolder. Bioinformatics 2015, 31, 2443–2451. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Auch, A.F.; Von Jan, M.; Klenk, H.-P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef]
- Colston, S.M.; Fullmer, M.S.; Beka, L.; Lamy, B.; Gogarten, J.P.; Graf, J. Bioinformatic Genome Comparisons for Taxonomic and Phylogenetic Assignments Using Aeromonas as a Test Case. mBio 2014, 5, e02136. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. In Nucleic Acids Symposium Series; Information Retrieval Ltd.: London, UK, 1999; Volume 41, pp. 95–98. [Google Scholar]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R, Foundation for Statistical Computing: Vienna, Austria, 2022; Available online: https://www.R-project.org/ (accessed on 18 September 2022).
- Vidal Amaral, J.R.; Jucá Ramos, R.T.; Almeida Araújo, F.; Bentes Kato, R.; Figueira Aburjaile, F.; de Castro Soares, S.; Góes-Neto, A.; Matiuzzi da Costa, M.; Azevedo, V.; Brenig, B.; et al. Bacteriocin Producing Streptococcus agalactiae Strains Isolated from Bovine Mastitis in Brazil. Microorganisms 2022, 10, 588. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Jin, Q.; Chen, L.; Yang, J. VFDB 2019: A comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 2018, 47, D687–D692. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Gonzalez-Escalona, N.; Frye, J.G.; Haendiges, J.; Haft, D.H.; Hoffmann, M.; Pettengill, J.B.; Prasad, A.B.; Tillman, G.E.; et al. AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci. Rep. 2021, 11, 12728. [Google Scholar] [CrossRef]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, N.; Sasagawa, M.; Yamamoto, M.; Komaki, H.; Yoshida, Y.; Yamazaki, S.; Fujita, N. DoBISCUIT: A database of secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 2012, 41, D408–D414. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Araujo, F.; Barh, D.; Silva, A.; Guimarães, L.C.; Ramos, R.T.J. GO FEAT: A rapid web-based functional annotation tool for genomic and transcriptomic data. Sci. Rep. 2018, 8, 1794. [Google Scholar] [CrossRef] [PubMed]
- Moriya, Y.; Itoh, M.; Okuda, S.; Yoshizawa, A.C.; Kanehisa, M. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007, 35 (Suppl. S2), W182–W185. [Google Scholar] [CrossRef]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC Bioinformatics Resource Center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef]
- Grant, J.R.; Stothard, P. The CGView Server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef]
- Gibson, L.; Woodworth, J.; George, A. Probiotic activity of Aeromonas media on the Pacific oyster, Crassostrea gigas, when challenged with Vibrio tubiashii. Aquaculture 1998, 169, 111–120. [Google Scholar] [CrossRef]
- Lategan, M.J.; Gibson, L.F. Antagonistic activity of Aeromonas media strain A199 against Saprolegnia sp., an opportunistic pathogen of the eel, Anguilla australis Richardson. J. Fish Dis. 2003, 26, 147–153. [Google Scholar] [CrossRef]
- Bassetti, M.; Giacobbe, D.R.; Giamarellou, H.; Viscoli, C.; Daikos, G.; Dimopoulos, G.; DE Rosa, F.G.; Giamarellos-Bourboulis, E.; Rossolini, G.; Righi, E.; et al. Management of KPC-producing Klebsiella pneumoniae infections. Clin. Microbiol. Infect. 2018, 24, 133–144. [Google Scholar] [CrossRef]
- Nagar, V.; Shashidhar, R.; Bandekar, J.R. Characterization of Aeromonas strains isolated from Indian foods using rpoD gene sequencing and whole cell protein analysis. World J. Microbiol. Biotechnol. 2012, 29, 745–752. [Google Scholar] [CrossRef]
- Howard, S.P.; Garland, W.J.; Green, M.J.; Buckley, J.T. Nucleotide sequence of the gene for the hole-forming toxin aerolysin of Aeromonas hydrophila. J. Bacteriol. 1987, 169, 2869–2871. [Google Scholar] [CrossRef]
- Suarez, G.; Khajanchi, B.K.; Sierra, J.C.; Erova, T.E.; Sha, J.; Chopra, A.K. Actin cross-linking domain of Aeromonas hydrophila repeat in toxin A (RtxA) induces host cell rounding and apoptosis. Gene 2012, 506, 369–376. [Google Scholar] [CrossRef] [PubMed]
- Ast, V.M.; Schoenhofen, I.C.; Langen, G.R.; Stratilo, C.W.; Chamberlain, M.D.; Howard, S.P. Expression of the ExeAB complex of Aeromonas hydrophila is required for the localization and assembly of the ExeD secretion port multimer. Mol. Microbiol. 2002, 44, 217–231. [Google Scholar] [CrossRef]
- Rangrez, A.Y.; Dayananda, K.M.; Atanur, S.; Joshi, R.; Patole, M.S.; Shouche, Y.S. Detection of Conjugation Related Type Four Secretion Machinery in Aeromonas culicicola. PLoS ONE 2006, 1, e115. [Google Scholar] [CrossRef]
- Suarez, G.; Sierra, J.C.; Kirtley, M.L.; Chopra, A.K. Role of Hcp, a type 6 secretion system effector, of Aeromonas hydrophila in modulating activation of host immune cells. Microbiology 2010, 156, 3678–3688. [Google Scholar] [CrossRef]
- Beaz-Hidalgo, R.; Figueras, M.J. Aeromonas spp. whole genomes and virulence factors implicated in fish disease. J. Fish Dis. 2013, 36, 371–388. [Google Scholar] [CrossRef]
- Bergh, P.V.; Frey, J. Aeromonas salmonicida subsp. salmonicida in the light of its type-three secretion system. Microb. Biotechnol. 2013, 7, 381–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dallaire-Dufresne, S.; Tanaka, K.H.; Trudel, M.V.; Lafaille, A.; Charette, S.J. Virulence, genomic features, and plasticity of Aeromonas salmonicida subsp. salmonicida, the causative agent of fish furunculosis. Vet. Microbiol. 2014, 169, 1–7. [Google Scholar] [CrossRef]
- Jamaluddin, N.; Stuckey, D.C.; Ariff, A.B.; Wong, F.W.F. Novel approaches to purifying bacteriocin: A review. Crit. Rev. Food Sci. Nutr. 2017, 58, 2453–2465. [Google Scholar] [CrossRef] [PubMed]
- Fath, M.J.; Mahanty, H.K.; Kolter, R. Characterization of a purF operon mutation which affects colicin V production. J. Bacteriol. 1989, 171, 3158–3161. [Google Scholar] [CrossRef]
- Gilson, L.; Mahanty, H.K.; Kolter, R. Four plasmid genes are required for colicin V synthesis, export, and immunity. J. Bacteriol. 1987, 169, 2466–2470. [Google Scholar] [CrossRef]
- Lenneman, E.M.; Barney, B.M.; Kuleshov, K.V.; Vodop’Ianov, S.O.; Dedkov, V.G.; Markelov, M.L.; Kermanov, A.V.; Kruglikov, V.D.; Vodop’Ianov, A.S.; Pisanov, R.V.; et al. Draft Genome Sequences of the Alga-Degrading Bacteria Aeromonas hydrophila Strain AD9 and Pseudomonas pseudoalcaligenes Strain AD6. Genome Announc. 2014, 2, e00624-14. [Google Scholar] [CrossRef] [PubMed]
- Pavan, M.E.; Pavan, E.E.; López, N.I.; Levin, L.; Pettinari, M.J. Living in an Extremely Polluted Environment: Clues from the Genome of Melanin-Producing Aeromonas salmonicida subsp. pectinolytica 34melT. Appl. Environ. Microbiol. 2015, 81, 5235–5248. [Google Scholar] [CrossRef]
- Chen, Y.; Simmonds, R.S.; Young, J.K.; Timkovich, R. Solution structure of the recombinant target recognition domain of zoocin A. Proteins: Struct. Funct. Bioinform. 2013, 81, 722–727. [Google Scholar] [CrossRef]
- Butler, A.R.; Gandecha, A.R.; Cundliffe, E. Influence of ancillary genes, encoding aspects of methionine metabolism, on tylosin biosynthesis in Streptomyces fradiae. J. Antibiot. 2001, 54, 642–649. [Google Scholar] [CrossRef]
- Bannantine, J.P.; Lingle, C.K.; Adam, P.R.; Ramyar, K.X.; McWhorter, W.J.; Stabel, J.R.; Picking, W.D.; Geisbrecht, B.V. NlpC/P60 domain-containing proteins of M ycobacterium avium subspecies paratuberculosis that differentially bind and hydrolyze peptidoglycan. Protein Sci. 2016, 25, 840–851. [Google Scholar] [CrossRef]
- Gargis, S.R.; Gargis, A.S.; Heath, H.E.; Heath, L.S.; LeBlanc, P.A.; Senn, M.M.; Berger-Bächi, B.; Simmonds, R.S.; Sloan, G.L. Zif, the Zoocin A Immunity Factor, Is a FemABX-Like Immunity Protein with a Novel Mode of Action. Appl. Environ. Microbiol. 2009, 75, 6205–6210. [Google Scholar] [CrossRef] [Green Version]
- Sharp, C.; Bray, J.; Housden, N.G.; Maiden, M.C.J.; Kleanthous, C. Diversity and distribution of nuclease bacteriocins in bacterial genomes revealed using Hidden Markov Models. PLoS Comput. Biol. 2017, 13, e1005652. [Google Scholar] [CrossRef]
- Schatz, A.; Bugie, E.; Waksman, S.A.; Hanssen, A.D.; Patel, R.; Osmon, D.R. The Classic: Streptomycin, a Substance Exhibiting Antibiotic Activity against Gram-Positive and Gram-Negative Bacteria. Clin. Orthop. Relat. Res. 2005, 437, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Gomes-Pepe, E.S.; Schuch, V.; de Macedo Lemos, E.G. Biotechnology of polyketides: New breath of life for the novel antibiotic genetic pathways discovery through metagenomics. Braz. J. Microbiol. 2013, 44, 1007–1034. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Devine, R.; Hutchings, M.I.; Wilkinson, B. A role for antibiotic biosynthesis monooxygenase domain proteins in fidelity control during aromatic polyketide biosynthesis. Nat. Commun. 2019, 10, 3611. [Google Scholar] [CrossRef] [Green Version]
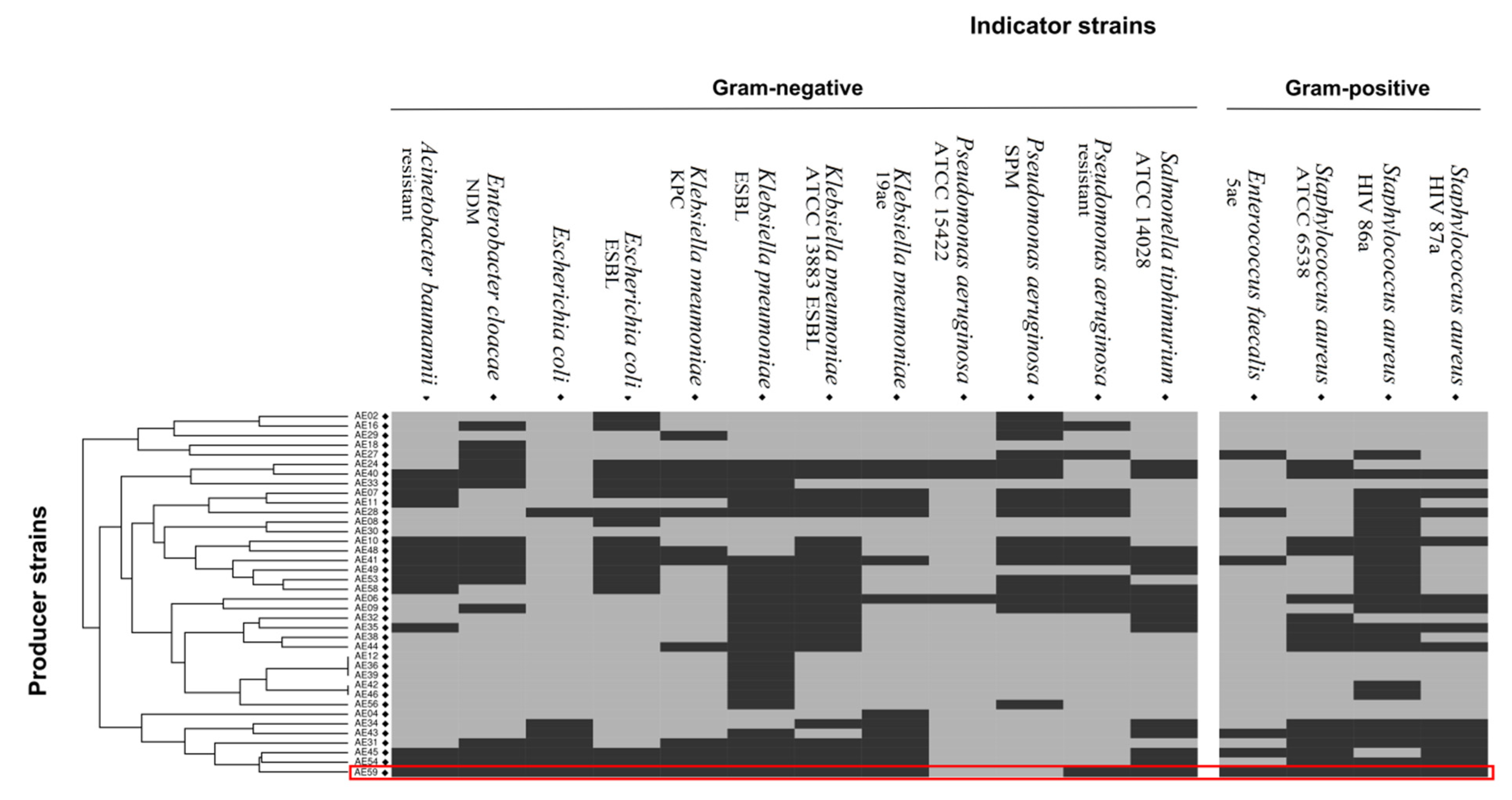



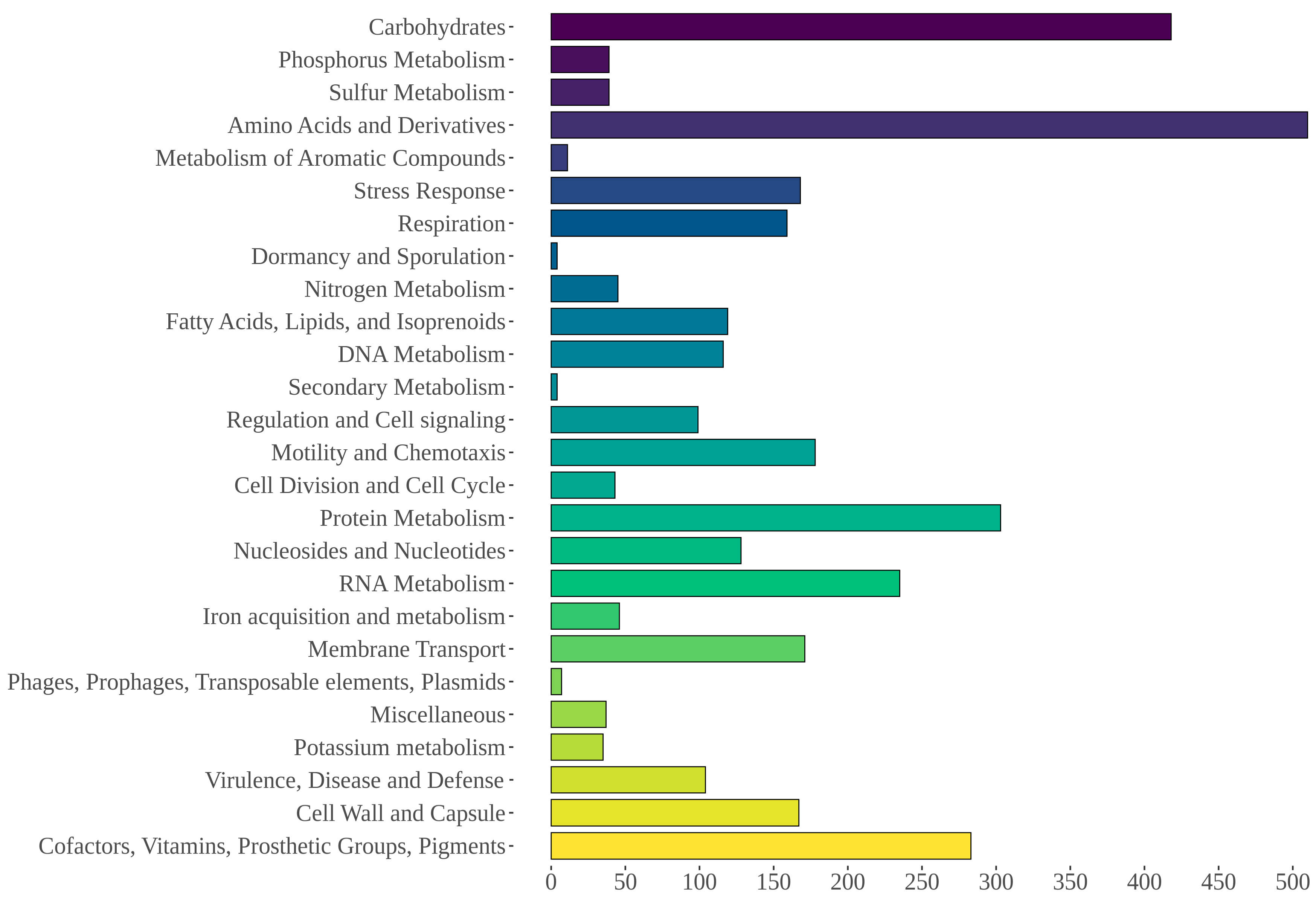
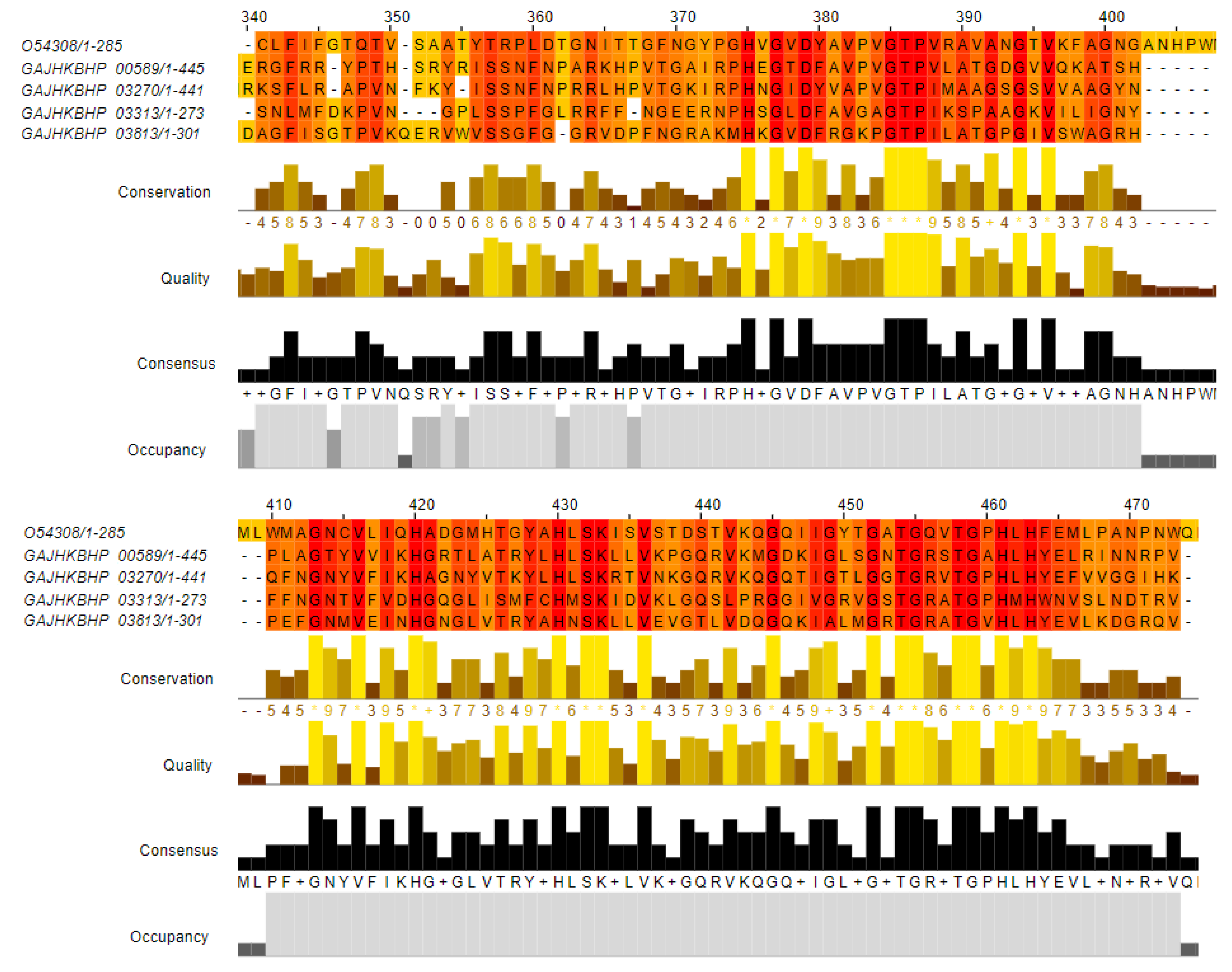
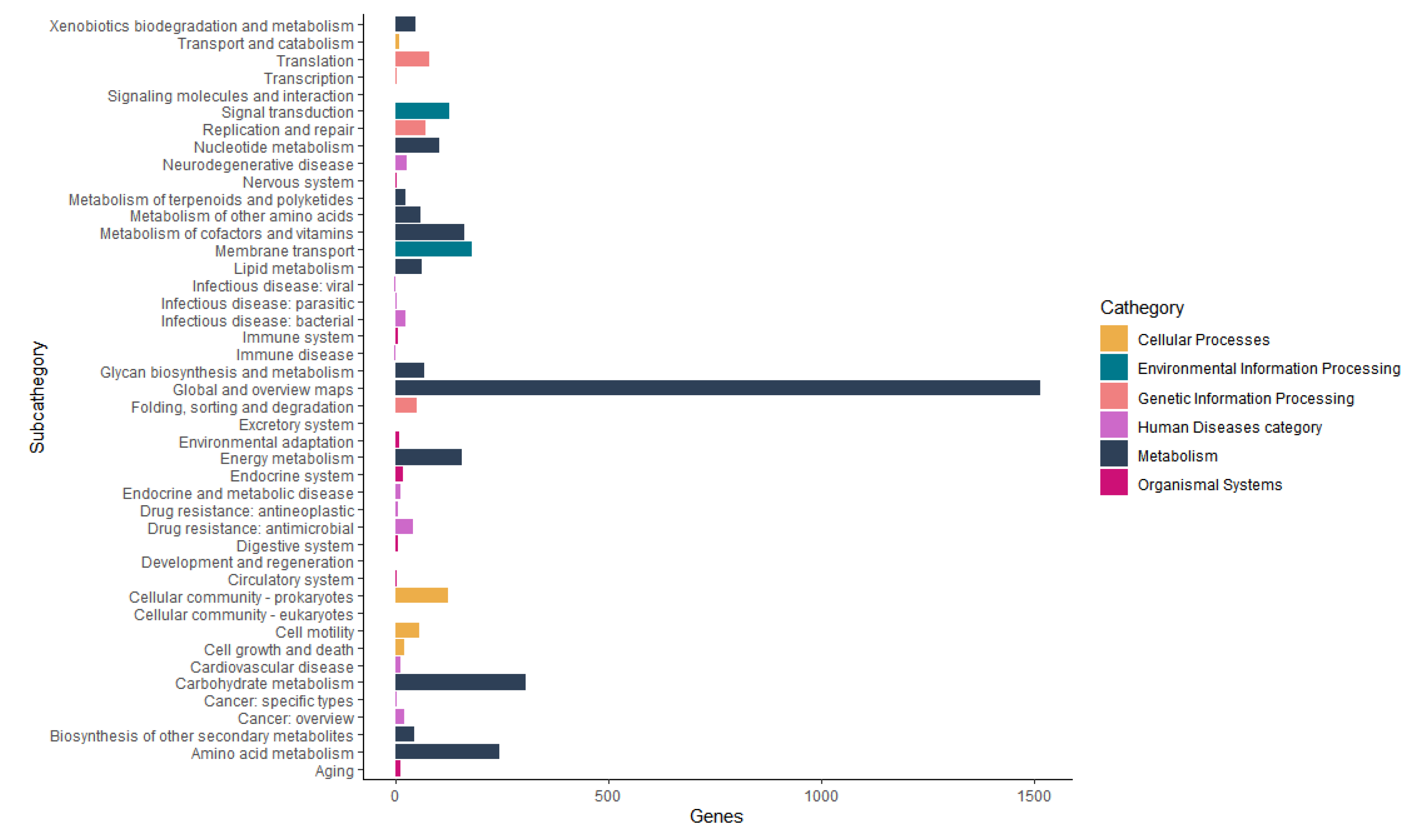
| Statistics | De Novo Assembly | Scaffolding 1 | Scaffolding 2 |
|---|---|---|---|
| Number of sequences | 109 | 62 | 51 |
| Largest contig (bp) | 473,819 | 3,152,962 | 4,498,261 |
| Average length (bp) | 41,433.55 | 72,878.34 | 88,608.96 |
| Total length (bp) | 4,516,257 | 4,518,457 | 4,519,057 |
| % GC | 58.65 | 58.63 | 58.62 |
| N50 | 263,685 | 3,152,962 | 4,498,261 |
| Subsystem | Putative Function | Gene ID |
|---|---|---|
| Colicin V and Bacteriocin Production Cluster | DedA protein | peg.679 |
| peg.2991 | ||
| peg.1442 | ||
| Amidophosphoribosyltransferase (EC 2.4.2.14) | peg.851 | |
| Colicin V production protein | peg.850 | |
| DedD protein | peg.934 | |
| Folylpolyglutamate synthase (EC 6.3.2.17); Dihydrofolate synthase (EC 6.3.2.12) | peg.933 | |
| Acetyl-coenzyme A carboxyl transferase beta chain (EC 6.4.1.2) | peg.932 | |
| tRNA pseudouridine synthase A (EC 4.2.1.70) | peg.931 | |
| Tolerance to colicin E2 | Conserved uncharacterized protein CreA | peg.2597 |
| peg.2682 | ||
| Two-component response regulator CreB | peg.2016 | |
| Two-component response regulator CreC | peg.2017 | |
| Inner membrane protein CreD | peg.2020 |
| Result of BLASTp AE59-TE2 (Amino Acid by Prokka Annotation) X Bacteriocins DB | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sequence from AE59 (Query) | % Identity | Subject Acc.Ver | Alignment Length | Mismatches | Gap Opens | q. Start | q. End | s. Start | s. End | E-Value | Bit Score |
| GAJHKBHP_00589 | 46.73% | Zoocin A (Uniprot Acc. Number: O54308) | 92 | 42 | 2 | 319 | 403 | 22 | 113 | 2.08 × 10−19 | 79.0 |
| GAJHKBHP_03270 | 50.00% | 94 | 38 | 2 | 301 | 386 | 45 | 137 | 2.81 × 10−20 | 82.4 | |
| GAJHKBHP_03313 | 34.21% | 114 | 58 | 3 | 171 | 268 | 45 | 157 | 8.07 × 10−17 | 70.5 | |
| GAJHKBHP_03813 | 45.0% | 100 | 47 | 3 | 192 | 284 | 39 | 137 | 6.67 × 10−17 | 71.2 | |
| Result of BLASTp AE59 (Prokka Annotation) X DoBiscuit Database | ||||||
|---|---|---|---|---|---|---|
| Sequence from AE59 (Query) | % Identity | Subject | Classified | Relative | PROKKA Gene | PROKKA Product |
| GAJHKBHP_00274 | 73.33% | Rifam_00640 | PKS TypeI modular | Ansamycin | rpoC | DNA-directed RNA polymerase subunit beta |
| GAJHKBHP_02296 | 63.01% | Rubra_00090 | PKS TypeI modular | Rubradirin | infA | Translation initiation factor IF-1 |
| GAJHKBHP_00363 | 62.65% | A4092_00490 | NRPS/PKS TypeIII | Glycopeptide(teicoplanin-type) | thiC | Phosphomethylpyrimidine synthase |
| GAJHKBHP_01234 | 61.41% | Salino_00470 | PKS TypeI modular | Salinomycin/Polyether | fadH | 2% 2C4-dienoyl-CoA reductase |
| GAJHKBHP_02296 | 61.19% | Rubra_00040 | PKS TypeI modular | Rubradirin/Ansamycin | infA | Translation initiation factor IF-1 |
| GAJHKBHP_03276 | 60.16% | Polk_00010 | PKS TypeI iterative | Tetracyclic quinone glycoside | metK | S-adenosylmethionine synthase |
| PKS TypeII | Polyketomycin | |||||
| Biosynthesis of Secondary Metabolites | Pathway Modules |
|---|---|
| Macrolide biosynthesis | M00773 Tylosin biosynthesis |
| M00934 Mycinamicin biosynthesis | |
| M00774 Erythromycin biosynthesis | |
| M00775 Oleandomycin biosynthesis | |
| M00776 Pikromycin/methymycin biosynthesis | |
| M00777 Avermectin biosynthesis | |
| Type II polyketide biosynthesis | M00778 Type II polyketide backbone biosynthesis |
| M00779 Dihydrokalafungin biosynthesis | |
| M00780 Tetracycline/oxytetracycline biosynthesis | |
| M00823 Chlortetracycline biosynthesis | |
| M00781 Nogalavinone/aklavinone biosynthesis | |
| M00782 Mithramycin biosynthesis | |
| M00783 Tetracenomycin C/8-demethyltetracenomycin C biosynthesis | |
| M00784 Elloramycin biosynthesis | |
| Biosynthesis of beta-lactams | M00672 Penicillin biosynthesis |
| M00673 Cephamycin C biosynthesis | |
| M00675 Carbapenem-3-carboxylate biosynthesis | |
| M00736 Nocardicin A biosynthesis | |
| M00674 Clavaminate biosynthesis | |
| Biosynthesis of other antibiotics | M00877 Kanosamine biosynthesis glucose 6-phosphate => kanosamine |
| M00889 Puromycin biosynthesis | |
| M00815 Validamycin A biosynthesis | |
| M00904 Dapdiamides biosynthesis | |
| M00785 Cycloserine biosynthesis | |
| M00787 Bacilysin biosynthesis | |
| M00848 Aurachin biosynthesis | |
| M00788 Terpentecin biosynthesis | |
| M00819 Pentalenolactone biosynthesis | |
| M00903 Fosfomycin biosynthesis | |
| M00890 Roseoflavin biosynthesis |
| Results of Searches for Keywords: “Endopeptidase, Endonuclease, Polyketide, Antibiotic, Colicin and Microcin” | |||
|---|---|---|---|
| Keyword | Annotation Tool | Sequence ID | Product |
| Endonuclease | PATRIC | peg.3314 | Endonuclease I precursor/deoxyribonuclease I activity |
| PATRIC | peg.2963 | Endonuclease IV/deoxyribonuclease IV (phage-T4-induced) activity | |
| PATRIC | peg.2394 | Predicted ATP-dependent endonuclease of the OLD family, YbjD subgroup | |
| PATRIC | peg.2112 | DNA/RNA endonuclease G | |
| PATRIC | peg.1965 | Endonuclease III/DNA-(apurinic or apyrimidinic site) lyase activity | |
| PATRIC | peg.1792 | Protein containing HNH endonuclease domain | |
| PATRIC | peg.1779 | Esterase ybfF | |
| PATRIC | peg.1125 | Flap endonuclease Xni | |
| PATRIC | peg.512 | DNA mismatch repair endonuclease MutH | |
| PROKKA | GAJHKBHP_01017 | putative DNA endonuclease SmrA | |
| PROKKA | GAJHKBHP_01117 | Flap endonuclease Xni-YgdG | |
| PROKKA | GAJHKBHP_01958 | Endonuclease III-Nth | |
| PROKKA | GAJHKBHP_02669 | Endonuclease MutS2 | |
| PROKKA | GAJHKBHP_02932 | Endonuclease 4-Nfo | |
| Endopeptidase | PATRIC | peg.384 | Murein-DD-endopeptidase |
| PATRIC | peg.577 | Murein DD-endopeptidase MepM | |
| PATRIC | peg.2124 | Probable endopeptidase NlpC | |
| PATRIC | peg.2264 | Penicillin-insensitive murein endopeptidase | |
| PATRIC | peg.3967 | Murein-DD-endopeptidase (EC 3.4.99.-) | |
| PROKKA | GAJHKBHP_00392 | D-alanyl-D-alanine endopeptidase-PbpG_1 | |
| PROKKA | GAJHKBHP_00589 | Murein DD-endopeptidase MepM | |
| PROKKA | GAJHKBHP_02126 | Oligoendopeptidase F% 2C plasmid-PepF1 | |
| PROKKA | GAJHKBHP_02243 | Penicillin-insensitive murein endopeptidase-MepA_1 | |
| PROKKA | GAJHKBHP_03259 | Neutral endopeptidase-PepO | |
| PROKKA | GAJHKBHP_03270 | Murein DD-endopeptidase MepM | |
| PROKKA | GAJHKBHP_03927 | D-alanyl-D-alanine endopeptidase-PbpG_2 | |
| Colicin | PATRIC | peg.1689 | Colicin I receptor precursor |
| PATRIC | peg.2329 | Colicin V production protein | |
| PROKKA | GAJHKBHP_00287 | Colicin I receptor-CirA_1 | |
| PROKKA | GAJHKBHP_01798 | Colicin I receptor-CirA_2 | |
| PROKKA | GAJHKBHP_01806 | Colicin I receptor-CirA_3 | |
| PROKKA | GAJHKBHP_02310 | Colicin V production protein-CvpA | |
| Antibiotic | PATRIC | peg.705 | Antibiotic biosynthesis monooxygenase |
| PROKKA | GAJHKBHP_03466 | Phenazine antibiotic resistance protein EhpR | |
| Microcin | PATRIC | peg.2417 | Microcin C7 immunity MccF-like protein |
| PROKKA | GAJHKBHP_02397 | Microcin C7 self-immunity protein MccF | |
| Polyketide | PATRIC | peg.1909 | Polyketide synthase modules and related proteins |
| PATRIC | peg.2168 | Polyketide synthase modules and related proteins | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silva, S.d.; Guedes, F.A.d.F.; Amaral, J.R.V.; Ribeiro, J.R.d.A.; Souza, Y.P.A.d.; Freitas-Almeida, Â.C.d.; Thompson, F.L.; Ramos, R.T.J.; Whiteley, A.S.; Macrae, A.; et al. Aeromonas allosaccharophila Strain AE59-TE2 Is Highly Antagonistic towards Multidrug-Resistant Human Pathogens, What Does Its Genome Tell Us? Life 2022, 12, 1492. https://doi.org/10.3390/life12101492
Silva Sd, Guedes FAdF, Amaral JRV, Ribeiro JRdA, Souza YPAd, Freitas-Almeida ÂCd, Thompson FL, Ramos RTJ, Whiteley AS, Macrae A, et al. Aeromonas allosaccharophila Strain AE59-TE2 Is Highly Antagonistic towards Multidrug-Resistant Human Pathogens, What Does Its Genome Tell Us? Life. 2022; 12(10):1492. https://doi.org/10.3390/life12101492
Chicago/Turabian StyleSilva, Sheila da, Fernanda Alves de Freitas Guedes, João Ricardo Vidal Amaral, José Roberto de Assis Ribeiro, Yuri Pinheiro Alves de Souza, Ângela Correa de Freitas-Almeida, Fabiano Lopes Thompson, Rommel Thiago Jucá Ramos, Andrew Steven Whiteley, Andrew Macrae, and et al. 2022. "Aeromonas allosaccharophila Strain AE59-TE2 Is Highly Antagonistic towards Multidrug-Resistant Human Pathogens, What Does Its Genome Tell Us?" Life 12, no. 10: 1492. https://doi.org/10.3390/life12101492
APA StyleSilva, S. d., Guedes, F. A. d. F., Amaral, J. R. V., Ribeiro, J. R. d. A., Souza, Y. P. A. d., Freitas-Almeida, Â. C. d., Thompson, F. L., Ramos, R. T. J., Whiteley, A. S., Macrae, A., & Oliveira, S. S. d. (2022). Aeromonas allosaccharophila Strain AE59-TE2 Is Highly Antagonistic towards Multidrug-Resistant Human Pathogens, What Does Its Genome Tell Us? Life, 12(10), 1492. https://doi.org/10.3390/life12101492











