The Conserved Family of the Pyridoxal Phosphate-Binding Protein (PLPBP) and Its Cyanobacterial Paradigm PipY
Abstract
:1. Introduction
2. Structural and Functional Features of PLPBPs
2.1. PLP Is Solvent-Exposed in PLPBP Structures
2.2. Dimerization of Just Some PLPBP Family Members?
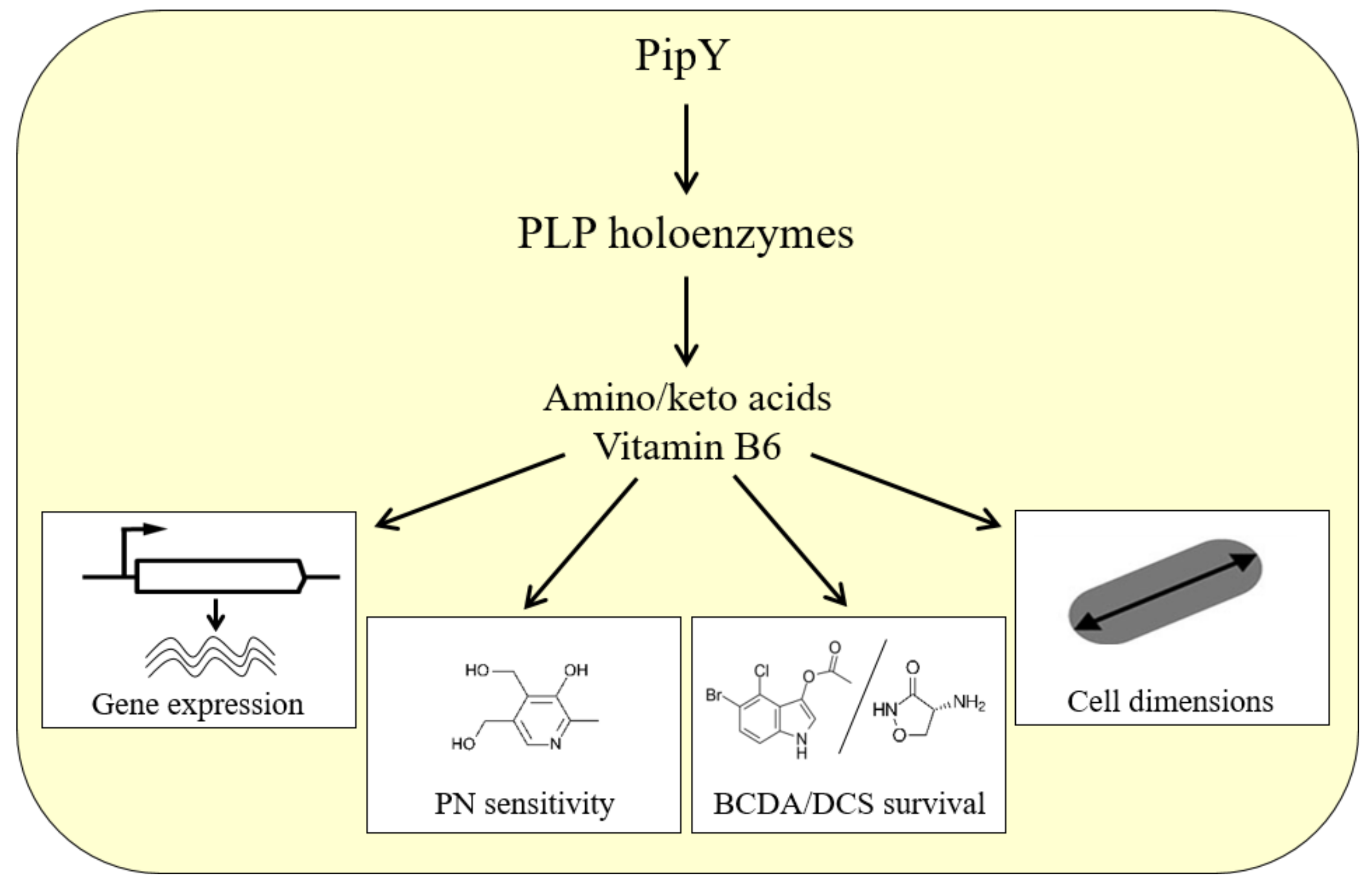
3. PLPBP-Related Phenotypes
3.1. Null Mutations, Heterologous Complementation, and Animal Disease Models Support Universal Functions of PLPBP Family Members
3.2. PLPBP Mutations Cause B6-Dependent Epilepsy in Humans
| Amino Acid Change in HuPLPHP | Molecular Mechanism: Effect on PLPBP Protein | |||||||
|---|---|---|---|---|---|---|---|---|
| Amino acid | Conserv. Score 1 | Change | Clinical effects | Reported in | Number of patients | Observed effect | Inferred from | Ref. |
| P40 | 7 | P40L | Seizures | [44] [51] | 1 (P40L/R241Q) 1 (P40L/splicing) | ↓ thermostability | In vitro studies on rHuPLPHP | [23] |
| R41 | 4 | R41Q | Seizures Mild disease 2 | [54] [43] | 2 (homozygous;R41Q/V45D) 3 (homozygous) | ↓yield/misfolding? ↓thermostability | In vitro studies on rHuPLPHP | [36] |
| R41W | Seizures, death | [50] | 1 (homozygous) | NT | NT | NT | ||
| V45 | 9 | V45D | Seizures | [54] | 1 (R41Q/V45D) | ↓↓PLP content ↓ thermostability | In vitro studies on rHuPLPHP | [36] |
| K47 | 9 | K47A | Not reported in humans (prenatally lethal?) | lack of PLP | rEcyggSK36A | [31] | ||
| E67 | 9 | E67K | Seizures Severe disease 2 | [54] [43] | 1 (homozygous) 3 (homozygous) | Misfolding | In vitro studies on rHuPLPHP | [36] |
| Y69 | 7 | Y69C | Seizures Moderate disease 2 | [44] | 1 (homozygous) | Higher dimerization ↓PLP content | In vitro studies on rHuPLPHP | [23] |
| P87 | 1 | P87L | Seizures Severe disease 2 | [41] [44] [53] | 1(P87L/R241Q) 1 (homozygous) 1 (P87L/splicing) | ↓solubility/misfolding | In vitro studies on rHuPLPHP | [23] |
| I94 | 8 | I94F | Seizures Mild disease 2 | [43] | 1 (homozygous) | Proposed ↓ in PLP saturation | Structural modeling of HuPLPHP | [43] |
| M113 | 6 | M113T | Seizures | [51] | 1 (M113T/C15X) | NT | NT | NT |
| T116 | 7 | T116I | Seizures Severe disease 2 | [43] | 2 (1 homozygous; 1 homozygous (T116I/H275D)) | Proposed ↓ in PLP saturation | Structural modeling of HuPLPHP | [43] |
| L175 | 6 | L175P | Seizures Severe disease 2 | [41] | 1 (homozygous) | Misfolding | In vitro studies on rHuPLPHP | [23] |
| R205 | 7 | R205Q | Seizures Moderate disease 2 | [44] [54] | 1 (R205Q/null) 1 (homozygous) | ↓thermostability | In vitro studies on rHuPLPHP | [23] |
| G224 | 9 | G224A | Seizures Severe disease 2 | [43] | 1 (G224A/splicing) | Proposed ↓ in PLP saturation | Structural modeling of HuPLPHP | [43] |
| S226 | 9 | S226A | Not reported in humans (prenatally lethal?) | ↓PLP saturation | rFnS201A | [24] | ||
| R241 | 9 | R241Q | Seizures | [41] [44] [52] | 1 (P87L/R241Q) 1 (P40L/R241Q) 1(R241Q/splicing) | ↓solubility ↓thermostability ↓PLP binding | In vitro studies on rHuPLPHP In vitro studies on rSePipYR210Q | [22,23] |
| I242 | 7 | I242T | Seizures | [45] | 1 (homozygous) | NT | NT | NT |
| H275 | NA | H275D | Seizures | [43] | 1 (homozygous T116I/H275D) | Variant of uncertain significance (VUS) | Structural modeling of HuPLPHP | [43] |
3.3. Phenotypes Associated with PLPBP Excess
3.4. Synthetic Lethality between PLPBP Family Members and PLP-Holoenzymes Supports Some Functional Redundancy
4. Guilty by Association Strategies to Get Insights into PLPBP Family Functions
4.1. Genes of the PLPBP Family in Clusters and Operons from Bacteria
4.2. The Close Relationship between PipX and PipY in Cyanobacteria
4.3. PLPHP Cellular Interaction Network
5. Regulation of Gene Expression by PLPBP
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blank, C.E.; Sánchez-Baracaldo, P. Timing of Morphological and Ecological Innovations in the Cyanobacteria--a Key to Understanding the Rise in Atmospheric Oxygen. Geobiology 2010, 8, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Fu, P. Biotechnological Perspectives on Algae: A Viable Option for Next Generation Biofuels. Curr. Opin. Biotechnol. 2020, 62, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Rubin, B.E.; Wetmore, K.M.; Price, M.N.; Diamond, S.; Shultzaberger, R.K.; Lowe, L.C.; Curtin, G.; Arkin, A.P.; Deutschbauer, A.; Golden, S.S. The Essential Gene Set of a Photosynthetic Organism. Proc. Natl. Acad. Sci. USA 2015, 112, E6634–E6643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Labella, J.I.; Llop, A.; Contreras, A. The Default Cyanobacterial Linked Genome: An Interactive Platform Based on Cyanobacterial Linkage Networks to Assist Functional Genomics. FEBS Lett. 2020, 594, 1661–1674. [Google Scholar] [CrossRef]
- Forchhammer, K.; Selim, K.A. Carbon/Nitrogen Homeostasis Control in Cyanobacteria. FEMS Microbiol. Rev. 2020, 44, 33–53. [Google Scholar] [CrossRef]
- Burillo, S.; Luque, I.; Fuentes, I.; Contreras, A. Interactions between the Nitrogen Signal Transduction Protein PII and N -Acetyl Glutamate Kinase in Organisms That Perform Oxygenic Photosynthesis. J. Bacteriol. 2004, 186, 3346–3354. [Google Scholar] [CrossRef] [Green Version]
- Espinosa, J.; Forchhammer, K.; Burillo, S.; Contreras, A. Interaction Network in Cyanobacterial Nitrogen Regulation: PipX, a Protein That Interacts in a 2-Oxoglutarate Dependent Manner with PII and NtcA. Mol. Microbiol. 2006, 61, 457–469. [Google Scholar] [CrossRef]
- Herrero, A.; Muro-Pastor, A.M.; Flores, E. Nitrogen Control in Cyanobacteria. J. Bacteriol. 2001, 183, 411–425. [Google Scholar] [CrossRef] [Green Version]
- Llácer, J.L.; Espinosa, J.; Castells, M.A.; Contreras, A.; Forchhammer, K.; Rubio, V. Structural Basis for the Regulation of NtcA-Dependent Transcription by Proteins PipX and PII. Proc. Natl. Acad. Sci. USA 2010, 107, 15397–15402. [Google Scholar] [CrossRef] [Green Version]
- Forcada-Nadal, A.; Llácer, J.L.; Contreras, A.; Marco-Marín, C.; Rubio, V. The PII-NAGK-PipX-NtcA Regulatory Axis of Cyanobacteria: A Tale of Changing Partners, Allosteric Effectors and Non-Covalent Interactions. Front. Mol. Biosci. 2018, 5, 91. [Google Scholar] [CrossRef]
- Espinosa, J.; Castells, M.A.; Laichoubi, K.B.; Forchhammer, K.; Contreras, A. Effects of Spontaneous Mutations in PipX Functions and Regulatory Complexes on the Cyanobacterium Synechococcus elongatus Strain PCC 7942. Microbiology 2010, 156, 1517–1526. [Google Scholar] [CrossRef] [Green Version]
- Espinosa, J.; Forchhammer, K.; Contreras, A. Role of the Synechococcus PCC 7942 Nitrogen Regulator Protein PipX in NtcA-Controlled Processes. Microbiology 2007, 153, 711–718. [Google Scholar] [CrossRef] [Green Version]
- Laichoubi, K.B.; Espinosa, J.; Castells, M.A.; Contreras, A. Mutational Analysis of the Cyanobacterial Nitrogen Regulator PipX. PLoS ONE 2012, 7, e35845. [Google Scholar] [CrossRef] [Green Version]
- Zeth, K.; Fokinas, O.; Forchhammers, K. Structural Basis and Target-Specific Modulation of ADP Sensing by the Synechococcus elongatus PII Signaling Protein. J. Biol. Chem. 2014, 289, 8960–8972. [Google Scholar] [CrossRef] [Green Version]
- Espinosa, J.; Labella, J.I.; Cantos, R.; Contreras, A. Energy Drives the Dynamic Localization of Cyanobacterial Nitrogen Regulators during Diurnal Cycles. Env. Microbiol. 2018, 20, 1240–1252. [Google Scholar] [CrossRef] [Green Version]
- Labella, J.I.; Cantos, R.; Salinas, P.; Espinosa, J.; Contreras, A. Distinctive Features of PipX, a Unique Signaling Protein of Cyanobacteria. Life 2020, 10, 79. [Google Scholar] [CrossRef]
- Espinosa, J.; Castells, M.A.; Laichoubi, K.B.; Contreras, A. Mutations at PipX Suppress Lethality of PII -Deficient Mutants of Synechococcus elongatus PCC 7942. J. Bacteriol. 2009, 191, 4863–4869. [Google Scholar] [CrossRef] [Green Version]
- Laichoubi, K.B.; Beez, S.; Espinosa, J.; Forchhammer, K.; Contreras, A. The Nitrogen Interaction Network in Synechococcus WH5701, a Cyanobacterium with Two PipX and Two PII-like Proteins. Microbiology 2011, 157, 1220–1228. [Google Scholar] [CrossRef] [Green Version]
- Labella, J.I.; Cantos, R.; Espinosa, J.; Forcada-Nadal, A.; Rubio, V.; Contreras, A. PipY, a Member of the Conserved COG0325 Family of PLP-Binding Proteins, Expands the Cyanobacterial Nitrogen Regulatory Network. Front. Microbiol. 2017, 8, 1244. [Google Scholar] [CrossRef] [Green Version]
- Schneider, G.; Käck, H.; Lindqvist, Y. The Manifold of Vitamin B6 Dependent Enzymes. Structure 2000, 8, R1–R6. [Google Scholar] [CrossRef]
- Eswaramoorthy, S.; Gerchman, S.; Graziano, V.; Kycia, H.; Studier, F.W.; Swaminathan, S. Structure of a Yeast Hypothetical Protein Selected by a Structural Genomics Approach. Acta. Cryst. D Biol. Cryst. 2003, 59, 127–135. [Google Scholar] [CrossRef] [PubMed]
- Tremiño, L.; Forcada-Nadal, A.; Contreras, A.; Rubio, V. Studies on Cyanobacterial Protein PipY Shed Light on Structure, Potential Functions, and Vitamin B6-Dependent Epilepsy. FEBS Lett. 2017, 591, 3431–3442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tremiño, L.; Forcada-Nadal, A.; Rubio, V. Insight into Vitamin B6-Dependent Epilepsy Due to PLPBP (Previously PROSC) Missense Mutations. Hum. Mutat. 2018, 39, 1002–1013. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Chen, Y.; Wang, L.; Bai, X.; Bu, T.; Zhang, J.; Lu, M.; Ha, N.-C.; Quan, C.; Nam, K.H.; et al. Structural and Functional Analysis of the Pyridoxal Phosphate Homeostasis Protein YggS from Fusobacterium nucleatum. Molecules 2022, 27, 4781. [Google Scholar] [CrossRef]
- Cantos, R.; Labella, J.I.; Espinosa, J.; Contreras, A. The Nitrogen Regulator PipX Acts in Cis to Prevent Operon Polarity. Env. Microbiol. Rep. 2019, 11, 495–507. [Google Scholar] [CrossRef] [Green Version]
- Jansonius, J.N. Structure, Evolution and Action of Vitamin B6-Dependent Enzymes. Curr. Opin. Struct. Biol. 1998, 8, 759–769. [Google Scholar] [CrossRef]
- Eliot, A.C.; Kirsch, J.F. Pyridoxal Phosphate Enzymes: Mechanistic, Structural, and Evolutionary Considerations. Annu. Rev. Biochem. 2004, 73, 383–415. [Google Scholar] [CrossRef] [Green Version]
- Parra, M.; Stahl, S.; Hellmann, H. Vitamin B6 and Its Role in Cell Metabolism and Physiology. Cells 2018, 7, 84. [Google Scholar] [CrossRef] [Green Version]
- Palm, D.; Klein, H.W.; Schinzel, R.; Buehner, M.; Helmreich, E.J.M. The Role of Pyridoxal 5’-Phosphate in Glycogen Phosphorylase Catalysis. Biochemistry 1990, 29, 1099–1107. [Google Scholar] [CrossRef]
- Goldin, B.R.; Frieden, C. The Effect of Pyridoxal Phosphate Modification on the Catalytic and Regulatory Properties of Bovine Liver Glutamate Dehydrogenase. J. Biol. Chem. 1972, 247, 2139–2144. [Google Scholar] [CrossRef]
- Ito, T.; Iimori, J.; Takayama, S.; Moriyama, A.; Yamauchi, A.; Hemmi, H.; Yoshimura, T. Conserved Pyridoxal Protein That Regulates Ile and Val Metabolism. J. Bacteriol. 2013, 195, 5439–5449. [Google Scholar] [CrossRef] [Green Version]
- Shaw, J.P.; Petsko, G.A.; Ringe, D. Determination of the Structure of Alanine Racemase from Bacillus Stearothermophilus at 1. 9-Å Resolution, Biochemistry 1997, 36, 1329–1342. [Google Scholar] [CrossRef]
- Banner, D.W.; Bloomer, A.C.; Petsko, G.A.; Phillips, D.C.; Wilson, I.A. Atomic Coordinates for Triose Phosphate Isomerase from Chicken Muscle. Biochem. Biophys. Res. Commun. 1976, 72, 146–155. [Google Scholar] [CrossRef]
- Wierenga, R.K. The TIM-Barrel Fold: A Versatile Framework for Efficient Enzymes. FEBS Lett. 2001, 492, 193–198. [Google Scholar] [CrossRef] [Green Version]
- Espaillat, A.; Carrasco-López, C.; Bernardo-García, N.; Pietrosemoli, N.; Otero, L.H.; Álvarez, L.; de Pedro, M.A.; Pazos, F.; Davis, B.M.; Waldor, M.K.; et al. Structural Basis for the Broad Specificity of a New Family of Amino-Acid Racemases. Acta Cryst. D Biol. Cryst. 2014, 70, 79–90. [Google Scholar] [CrossRef] [Green Version]
- Fux, A.; Sieber, S.A. Biochemical and Proteomic Studies of Human Pyridoxal 5′-Phosphate-Binding Protein (Plpbp). ACS Chem. Biol. 2020, 15, 254–261. [Google Scholar] [CrossRef]
- Ito, T. Role of the Conserved Pyridoxal 5ʹ-Phosphate-Binding Protein YggS/PLPBP in Vitamin B6 and Amino Acid Homeostasis. Biosci. Biotechnol. Biochem. 2022, 86, 1183–1191. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Y.; Xia, L.; Chen, L.; Liao, Y.; Chen, B.; Liu, Y.; Gong, W.; Tian, Y.; Hu, B. YggS Encoding Pyridoxal 5′-Phosphate Binding Protein Is Required for Acidovorax citrulli Virulence. Front. Microbiol. 2022, 12, 783862. [Google Scholar] [CrossRef]
- Prosser, G.A.; Rodenburg, A.; Khoury, H.; de Chiara, C.; Howell, S.; Snijders, A.P.; de Carvalho, L.P.S. Glutamate Racemase Is the Primary Target of β-Chloro-D-Alanine in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2016, 60, 6091–6099. [Google Scholar] [CrossRef] [Green Version]
- Prosser, G.A.; de Carvalho, L.P.S. Metabolomics Reveal d-Alanine: D-Alanine Ligase As the Target of d-Cycloserine in Mycobacterium tuberculosis. ACS Med. Chem. Lett. 2013, 4, 1233–1237. [Google Scholar] [CrossRef]
- Darin, N.; Reid, E.; Prunetti, L.; Samuelsson, L.; Husain, R.A.; Wilson, M.; el Yacoubi, B.; Footitt, E.; Chong, W.K.; Wilson, L.C.; et al. Mutations in PROSC Disrupt Cellular Pyridoxal Phosphate Homeostasis and Cause Vitamin-B6-Dependent Epilepsy. Am. J. Hum. Genet. 2016, 99, 1325–1337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prunetti, L.; el Yacoubi, B.; Schiavon, C.R.; Kirkpatrick, E.; Huang, L.; Bailly, M.; el Badawi-Sidhu, M.; Harrison, K.; Gregory, J.F.; Fiehn, O.; et al. Evidence That COG0325 Proteins Are Involved in PLP Homeostasis. Microbiology 2016, 162, 694–706. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, D.L.; Al-Shekaili, H.H.; Tarailo-Graovac, M.; Wolf, N.I.; Ivy, A.S.; Demarest, S.; Roussel, Y.; Ciapaite, J.; van Roermund, C.W.T.; Kernohan, K.D.; et al. PLPHP Deficiency: Clinical, Genetic, Biochemical, and Mechanistic Insights. Brain 2019, 142, 542–559. [Google Scholar] [CrossRef] [PubMed]
- Plecko, B.; Zweier, M.; Begemann, A.; Mathis, D.; Schmitt, B.; Striano, P.; Baethmann, M.; Vari, M.S.; Beccaria, F.; Zara, F.; et al. Confirmation of Mutations in PROSC as a Novel Cause of Vitamin B6-Dependent Epilepsy. J. Med. Genet. 2017, 54, 809–814. [Google Scholar] [CrossRef]
- Johannsen, J.; Bierhals, T.; Deindl, P.; Hecher, L.; Hermann, K.; Hempel, M.; Kloth, K.; Denecke, J. Excessive Seizure Clusters in an Otherwise Well-Controlled Epilepsy as a Possible Hallmark of Untreated Vitamin B6-Responsive Epilepsy Due to a Homozygous PLPBP Missense Variant. J. Pediatr. Genet. 2019, 08, 222–225. [Google Scholar] [CrossRef]
- McLean, H.; Palmquist, R.; Nadauld, L.D.; Malone Jenkins, S.; Bonkowsky, J.; Filloux, F. On the Edge—A Diagnostic Odyssey. Clin. Case. Rep. 2022, 10, e05688. [Google Scholar] [CrossRef]
- Kernohan, K.D.; Hartley, T.; Naumenko, S.; Armour, C.M.; Graham, G.E.; Nikkel, S.M.; Lines, M.; Geraghty, M.T.; Richer, J.; Mears, W.; et al. Diagnostic Clarity of Exome Sequencing Following Negative Comprehensive Panel Testing in the Neonatal Intensive Care Unit. Am. J. Med. Genet. A 2018, 176, 1688–1691. [Google Scholar] [CrossRef]
- Ahmed, S.; DeBerardinis, R.J.; Ni, M.; Afroze, B. Vitamin B6-Dependent Epilepsy Due to Pyridoxal Phosphate-Binding Protein (PLPBP) Defect—First Case Report from Pakistan and Review of Literature. Ann. Med. Surg 2020, 60, 721–727. [Google Scholar] [CrossRef]
- Pal, M.; Lace, B.; Labrie, Y.; Laflamme, N.; Rioux, N.; Setty, S.T.; Dugas, M.A.; Gosselin, L.; Droit, A.; Chrestian, N.; et al. A Founder Mutation in the PLPBP Gene in Families from Saguenay-Lac-St-Jean Region Affected by a Pyridoxine-Dependent Epilepsy. JIMD Rep. 2021, 59, 32–41. [Google Scholar] [CrossRef]
- Jensen, K.V.; Frid, M.; Stödberg, T.; Barbaro, M.; Wedell, A.; Christensen, M.; Bak, M.; Ek, J.; Madsen, C.G.; Darin, N.; et al. Diagnostic Pitfalls in Vitamin B6-Dependent Epilepsy Caused by Mutations in the PLPBP Gene. JIMD Rep. 2019, 50, 1–8. [Google Scholar] [CrossRef]
- Jiao, X.; Xue, J.; Gong, P.; Wu, Y.; Zhang, Y.; Jiang, Y.; Yang, Z. Clinical and Genetic Features in Pyridoxine-Dependent Epilepsy: A Chinese Cohort Study. Dev. Med. Child. Neurol. 2020, 62, 315–321. [Google Scholar] [CrossRef]
- Heath, O.; Pitt, J.; Mandelstam, S.; Kuschel, C.; Vasudevan, A.; Donoghue, S. Early-Onset Vitamin B6-Dependent Epilepsy Due to Pathogenic PLPBP Variants in a Premature Infant: A Case Report and Review of the Literature. JIMD Rep. 2021, 58, 3–11. [Google Scholar] [CrossRef]
- Espinoza, A.C.; Wright, M.A.; Candee, M.S.; Trandafir, C.; Nelson, G.R. Child Neurology: Late-Onset Vitamin B6-Dependent Epilepsy Identified by Rapid Genome Sequencing. Neurology 2021, 96, 911–914. [Google Scholar] [CrossRef]
- Shiraku, H.; Nakashima, M.; Takeshita, S.; Khoo, C.S.; Haniffa, M.; Ch’ng, G.S.; Takada, K.; Nakajima, K.; Ohta, M.; Okanishi, T.; et al. PLPBP Mutations Cause Variable Phenotypes of Developmental and Epileptic Encephalopathy. Epilepsia Open 2018, 3, 495–502. [Google Scholar] [CrossRef] [Green Version]
- Cao, Z.; Tang, H.; Cai, Y.; Zeng, B.; Zhao, J.; Tang, X.; Lu, M.; Wang, H.; Zhu, X.; Wu, X.; et al. Natural Variation of HTH5 from Wild Rice, Oryza rufipogon Griff., Is Involved in Conferring High-Temperature Tolerance at the Heading Stage. Plant Biotechnol. J. 2022, 20, 1591–1605. [Google Scholar] [CrossRef]
- Nichols, R.J.; Sen, S.; Choo, Y.J.; Beltrao, P.; Zietek, M.; Chaba, R.; Lee, S.; Kazmierczak, K.M.; Lee, K.J.; Wong, A.; et al. Phenotypic Landscape of a Bacterial Cell. Cell 2011, 144, 143–156. [Google Scholar] [CrossRef] [Green Version]
- Ito, T.; Hori, R.; Hemmi, H.; Downs, D.M.; Yoshimura, T. Inhibition of Glycine Cleavage System by Pyridoxine 5′-Phosphate Causes Synthetic Lethality in GlyA YggS and SerA YggS in Escherichia coli. Mol. Microbiol. 2020, 113, 270–284. [Google Scholar] [CrossRef]
- Vu, H.N.; Downs, D.M. Loss of YggS (COG0325) Impacts Aspartate Metabolism in Salmonella enterica. Mol. Microbiol. 2021, 116, 1232–1240. [Google Scholar] [CrossRef]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, Encapsulated Proteomic-Sample Processing Applied to Copy-Number Estimation in Eukaryotic Cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef]
- Percudani, R.; Peracchi, A. A Genomic Overview of Pyridoxal-Phosphate-Dependent Enzymes. EMBO Rep. 2003, 4, 850–854. [Google Scholar] [CrossRef]
- Savioz, A.; Jeenes, D.J.; Kocher, H.P.; Haas, D. Comparison of ProC and Other Housekeeping Genes of Pseudomonas aeruginosa with Their Counterparts in Escherichia coli. Gene 1990, 86, 107–111. [Google Scholar] [CrossRef]
- De Wergifosse, P.; Jacques, B.; Jonniaux, J.-L.; Purnelle, B.; Goffeau, A.; Skala, J., II. Yeast Sequencing Reports. The Sequence of a 22·4 Kb DNA Fragment from the Left Arm of Yeast Chromosome II Reveals Homologues to Bacterial Proline Synthetase and Murine α-Adaptin, as well as a New Permease and a DNA-Binding Protein. Yeast 1994, 10, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Ikegawa, S.; Isomura, M.; Koshizuka, Y.; Nakamura, Y. Cloning and Characterization of Human and Mouse PROSC (Proline Synthetase Co-Transcribed) Genes. J. Hum. Genet. 1999, 44, 337–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fadda, D.; Pischedda, C.; Caldara, F.; Whalen, M.B.; Anderluzzi, D.; Domenici, E.; Massidda, O. Characterization of DivIVA and Other Genes Located in the Chromosomal Region Downstream of the Dcw Cluster in Streptococcus pneumoniae. J. Bacteriol. 2003, 185, 6209–6214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolodkin-Gal, I.; Romero, D.; Cao, S.; Clardy, J.; Kolter, R.; Losick, R. D-Amino Acids Trigger Biofilm Disassembly. Science 2010, 328, 627–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijayan, V.; Jain, I.H.; O’Shea, E.K. A High Resolution Map of a Cyanobacterial Transcriptome. Genome. Biol. 2011, 12, R47. [Google Scholar] [CrossRef] [Green Version]
- Memon, D.; Singh, A.K.; Pakrasi, H.B.; Wangikar, P.P. A Global Analysis of Adaptive Evolution of Operons in Cyanobacteria. Antonie Van Leeuwenhoek 2013, 103, 331–346. [Google Scholar] [CrossRef]
- Salinas, P.; Contreras, A. Identification and Analysis of Escherichia coli Proteins That Interact with the Histidine Kinase NtrB in a Yeast Two-Hybrid System. Mol. Genet. Genom. 2003, 269, 574–581. [Google Scholar] [CrossRef]
- Jerez, C.; Salinas, P.; Llop, A.; Cantos, R.; Espinosa, J.; Labella, J.I.; Contreras, A. Regulatory Connections Between the Cyanobacterial Factor PipX and the Ribosome Assembly GTPase EngA. Front. Microbiol. 2021, 12, 781760. [Google Scholar] [CrossRef]
- Espinosa, J.; Rodriguez-Mateos, F.; Salinas, P.; Lanza, V.F.; Dixon, R.; de la Cruz, F.; Contreras, A. PipX, the Coactivator of NtcA, Is a Global Regulator in Cyanobacteria. Proc. Natl. Acad. Sci. USA 2014, 111, E2423–E2430. [Google Scholar] [CrossRef]
- Labella, J.I.; Obrebska, A.; Espinosa, J.; Salinas, P.; Forcada-Nadal, A.; Tremiño, L.; Rubio, V.; Contreras, A. Expanding the Cyanobacterial Nitrogen Regulatory Network: The GntR-Like Regulator PlmA Interacts with the PII-PipX Complex. Front. Microbiol. 2016, 7, 1677. [Google Scholar] [CrossRef]


| Organism | Protein | PDB File | Vitamer | Ligands | Amino Acid Changes | Resolut. (Å) | Deposition Year | Ref. |
|---|---|---|---|---|---|---|---|---|
| Escherichia coli | YggS | 1W8G | PLP | Isocitrate | None | 2.00 | 2004 | – |
| 3SY1 | PLP | MES Acetate | L32V/G56S/N58H/H81N/ I83A/H102I/M165S/S202A/ M205Q/R221A hexamutant | 1.47 | 2011 | – | ||
| 7UBQ | PNP * | None | None | 2.60 | 2022 | – | ||
| 7UB4 | PLP | None | K36A/K38A/K233A/K234 | 2.40 | 2022 | – | ||
| 7UAX | None | PO4H3 | K36A/K38A | 2.07 | 2022 | – | ||
| 7U9H | None | SO4H2 | None | 2.00 | 2022 | – | ||
| 7UBP | PLP | SO4H2 | K36A/K137A | 2.30 | 2022 | – | ||
| 7UB8 | PLP | Butanediol | K38A/K137A/K233A/K234A | 2.30 | 2022 | – | ||
| 7UAU | PLP | SO4H2 | K137A | 2.10 | 2022 | – | ||
| 7UAT | PLP | PO4H3 | K36A | 2.00 | 2022 | – | ||
| 7U9C | PLP | PO4H3 | None | 2.10 | 2022 | – | ||
| Bifidobacterium adolescentis | YggS | 3CPG | PLP | Acetate | Se-Met ** | 1.71 | 2008 | – |
| Agrobacterium tumefaciens | YggS | 3R79 | PLP | Acetate Pr+3 | Se-Met ** | 1.90 | 2011 | – |
| Synechococcus elongatus | PipY | 5NLC | None | PO4H3 | None | 1.90 | 2017 | [22] |
| 5NM8 | PLP | Ca2+ | None | 1.93 | 2017 | [22] | ||
| Fusobacterium nucleatum | YggS | 7F8E | None | SO4H2 | Se-Met ** | 2.08 | 2021 | – |
| 6KZW | None | PO4H3 | T5A/N202S, Se-Met ** | 2.08 | 2019 | – | ||
| 7YGF | Structure not released | 2.08 | 2022 | [24] | ||||
| Saccharomyces cerevisiae | YBL036C | 1CT5 | PLP | None | Se-Met ** | 2.00 | 1999 | [21] |
| 1B54 | PLP | None | None | 2.10 | 1999 | [21] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tremiño, L.; Llop, A.; Rubio, V.; Contreras, A. The Conserved Family of the Pyridoxal Phosphate-Binding Protein (PLPBP) and Its Cyanobacterial Paradigm PipY. Life 2022, 12, 1622. https://doi.org/10.3390/life12101622
Tremiño L, Llop A, Rubio V, Contreras A. The Conserved Family of the Pyridoxal Phosphate-Binding Protein (PLPBP) and Its Cyanobacterial Paradigm PipY. Life. 2022; 12(10):1622. https://doi.org/10.3390/life12101622
Chicago/Turabian StyleTremiño, Lorena, Antonio Llop, Vicente Rubio, and Asunción Contreras. 2022. "The Conserved Family of the Pyridoxal Phosphate-Binding Protein (PLPBP) and Its Cyanobacterial Paradigm PipY" Life 12, no. 10: 1622. https://doi.org/10.3390/life12101622
APA StyleTremiño, L., Llop, A., Rubio, V., & Contreras, A. (2022). The Conserved Family of the Pyridoxal Phosphate-Binding Protein (PLPBP) and Its Cyanobacterial Paradigm PipY. Life, 12(10), 1622. https://doi.org/10.3390/life12101622







