Abstract
Garcinia mangostana (Clusiaceae) is a rich pool of metabolites with diversified bioactivities. A new xanthone, garcixanthone E (1), and a new benzophenone, rhamnoside, as well as garcimangophenone C (9) together with garcinone E (2), α-mangostin (3), γ-mangostin (4), garcinone C (5), garcixanthone C (6), gartanin (7), and 2,4,6,3′,5′-pentahydroxybenzophenone (8) were purified from G. mangostana EtOAc extract. Their structural verification was accomplished utilizing assorted spectral tools and relating to the literature. The in vitro cytotoxic potential versus MCF-7, A549, and HCT-116 cell lines demonstrated the moderate potential of 1 (IC50s 8.5, 5.4, and 5.7 µM, respectively) in comparison to doxorubicin (IC50s 0.18, 0.6 and 0.2 µM, respectively) using a sulforhodamine B (SRB) assay. Additionally, 1 and 9 had AAI (α-amylase inhibition) with IC50s 17.8 and 12.9 µM, respectively, compared to acarbose (IC50 6.7 µM). Further, their AAI mechanisms were inspected utilizing molecular-docking evaluation by employing the crystal structure of the human α-amylase (PDB-ID: 5EOF). Compound 9 possessed a reasonable docking score of −7.746 kcal/mol compared with the native ligand 7JR which had a docking score of −9.932 kcal/mol. These results could further provide new insight into the potential usage of G. mangostana as a functional food for regulating postprandial hyperglycemia via suppressing AA.
1. Introduction
Garcinia mangostana (Clusiaceae, formerly Guttiferae) is among the most prevailing tropical fruits in southeast Asian regions. It has been widely consumed due to its high nutritional benefits, sweet unique taste, and pleasant aroma [1,2]. This plant has been famed in Chinese/Ayurvedic remedies for hundreds of years for treating various ailments, including cystitis, eczema, dysentery, gonorrhea, hyperkeratosis, gleet, psoriasis, and menstrual disorders [1,3,4]. Further, G. mangostana is known to possess diverse bioactivities, as it has anti-inflammatory, antimicrobial, antitumor, antimycobacterial, antioxidant, photoprotective, antimalarial, antiviral, and antileptospiral capacities [5,6,7]. Our former investigations of G. mangostana disclosed the characterization of xanthones as main metabolites, in addition to flavonoids, benzophenones, and phenolics [7,8,9]. Xanthones are a class of metabolites that are widely reported from lichens, plants, and fungi [10,11]. They are commonly produced by Polygalaceae, Gentianaceae, Guttiferae, Clusiaceae, and Moraceae plants [10,11]. They have an oxygenated tricyclic ring structure with diverse attached functional groups, such as methoxy, phenolic OH, and a dihydrofuran ring. These metabolites displayed various biological properties, including cytotoxic, antidiabetic, antioxidant, antileishmanial, antimicrobial, antimalarial, antitumor, antiHIV, antiquorum sensing, antihypertensive, anti-inflammatory, and larvicidal. Additionally, benzophenones are classes of metabolites reported from fungi and plants. It was stated that nearly 77% of benzophenones are separated from Clusiaceae plants [12]. These metabolites displayed various bioactivities, including antiHIV, antifungal, antiviral, antioxidant, and antimicrobial [12].
Cancer is one of the major serious illnesses that has a high, unacceptable mortality rate and incidence [13]. In total, 19.3 million new cancer cases and ≈10 million deaths because of cancer worldwide were estimated in 2020 [14]. Breast cancer with 2.3 million new cases (11.7%) has transcended lung cancer (11.4%) as the most frequently pinpointed cancer, followed by colorectal, prostate, and stomach (10.0%, 7.3%, and 5.6%, respectively) cancers. On the other hand, lung cancer with 1.8 million deaths (18%) continued to be the dominant reason of cancer death, following colorectal (9.4%) and breast (6.9%) cancers [14].
So far, the majority of anticancer agents have failed to accomplish the expected results. Therefore, there is an intensive research reorientation towards the discovery of new chemopreventive agents from natural sources [15,16]. Many natural metabolites are known to have chemoprotective potential towards various types of cancers worldwide [15]. These metabolites are widely found in fruits, vegetables, and plants [17]. It is a fact that consuming vegetables and fruits lowers carcinogenesis incidence [16]. Fruits and vegetables contain vitamins, fiber, minerals, and various bioactive metabolites, such as flavonoids, carotenoids, sterols, and phenolics, and all of them could be responsible for this protective potential [18].
The objective of this work was to discover new AAIs (α-amylase inhibitors) and anticancer agents from G. mangostana pericarps.
2. Material and Methods
2.1. General Experimental Procedures
A UV spectrum was accomplished utilizing a Hitachi-300 spectrometer (Hitachi High-Technologies Corporation/Kyoto/Japan). An ESIMS was performed with a LCQ-DECA mass spectrometer (Thermo_Finnigan/Bremen/Germany). An HRESIMS was executed utilizing a Micromass_Qtof2 spectrometer (Bruker/Rheinstetten/Germany). NMR spectra were determined on BRUKER_AVANCE600 equipment (BioSpin-Bruker/Billerica/MA/USA). IR data were estimated with an Infrared-400 Shimadzu spectrophotometer (Shimadzu/Kyoto/Japan). A chromatographic investigation was carried out on SiO2 60 (0.04–0.063 mm)/Sephadex LH-20 (0.25–0.1 mm)/RP-18 (0.04–0.063 mm) (Merck/Darmstadt/Germany). Precoated SiO260_F254 TLC plates (0.2 mm, Merck/Darmstadt/Germany) were employed for TLC examination. The metabolites’ purification and detection were carried out by employing a LiChrolut_RP-18 6 mL solid-phase extraction tube and UV inspection at λmax 366 and 255 nm and then spraying with H2SO4: p-anisaldehyde and a 110 °C heating.
2.2. Plant Material
G. mangostana fruits were secured in December 2019 from a Saudi local market. Its attestation was accomplished as earlier stated [6] and a voucher specimen (no. GM_1424) was kept in the herbarium at the Faculty of Pharmacy, KAU.
2.3. Extraction and Isolation
At room temperature, the dried pericarps (520 g) were extracted with MeOH (3 L × 5) until exhausting [7]. The combined concentrated methanol extract (GMT, 24 g) was suspended in distilled H2O (150 mL) and partitioned among n-hexane/EtOAc (500 mL × 6, each) to afford 2.7, 6.5, and 12.9 g, respectively, of n-hexane, EtOAc, and aqueous fractions. The EtOAC (6.5 g) fraction was chromatographed on SiO2CC (silica gel column chromatography) (300 g × 100 × 5 cm, EtOAC/n-hexane 5/95–0/100) to obtain four main subfractions: GME-1 (25/75), GME-2 (50/50), GME-3 (75/25), and GME-4 (100%EtOAc). The subfraction GME-2 (1.39 g) SiO2 CC (150 g × 50 × 3 cm, EtOAc/n-hexane gradient) produced 5 fractions of GME-2A: GME-2E. GME-2A (174 mg) SiO2 CC (30 g × 50 × 2 cm), EtOAC/n-hexane (10/90–30/70) provided 1, which was purified on a RP-18 LiChrolut extraction tube (acetonitrile/H2O70/30– 20/80) to give a light yellow powder of 1 (9.6 mg). The GME-2B (295 mg) fraction was handled as GME-2A to result in 2 (22.4 mg). Additionally, GME-2C (478 mg) was managed as GME-2B to yield 3 and 4, and their RP-18 column (100 g, 50 × 3 cm, H2O/MeOH gradient) produced 3 (29.4 mg) and 4 (13.7 mg). GME-3 (1.92 g) SiO2 CC (150 g × 50 × 3 cm, MeOH/CHCl3 gradient) resulted in 7 fractions: GME3A–GME3G. GME3B (320 mg) was handled on SiO2 CC (40 g, 50 × 2 cm, MeOH/CHCl3 gradient) following this RP-18 LiChrolut extraction tube (acetonitrile/H2O: gradient) and resulted in 5 (12.6 mg). The GME3C–GME3E (746 mg) fractions were combined relying on TLC and were submitted to Sephadex LH-20 (50 g, 50 × 3 cm, MeOH) to produce 6, 7, and 8, and their further purifying on RP-18 (100 g, 100 × 3 cm, H2O/MeOH gradient) yielded 6 (9.2 mg), 7 (11.6 mg), and 8 (14.8 mg). The Sephadex LH-20 of GME-4 (100% EtOAc, 1.17 g) employing MeOH produced 9, which was handled on an RP-18 column (H2O/MeOH (6:4–3:7) to obtain 9 (7.4 mg).
Spectral Data
Garcixanthone E (1)
Light-yellow powder. IR νmax (KBr): 2942, 3439, 1648, 1585, 1458 cm−1; UV (λmax, MeOH) (log ε): 237 (4.36), 269 (4.23), 322 (3.89), 386 (3.45) nm; HRESIMS m/z: 441.1908 (calc. for 441.1913, C25H29O7 [M+H]+); NMR spectral data (Table 1).

Table 1.
NMR data of garcixanthone E (1) and garcimangophenone C (9) (600 and 125 MHz).
Garcimangophenone C (9)
Brown powder. IR νmax (KBr): 3365, 2985, 1638, and 1605 cm−1; UV (λmax, MeOH) (log ε): 310 (3.96), 282 (4.15), 213 (4.49) nm; HRESIMS m/z: 393.1180 (calcd for 393.1186 for C19H21O9 [M+H]+); NMR spectral data (Table 1).
2.4. In Vitro Cytotoxic Assay
The new compounds (1 and 9) were examined for cytotoxic potential towards human MCF-7 (breast cancer), HCT-116 (colorectal carcinoma), and A549 (lung cancer) cell lines using a sulforhodamine B assay (SRB) as previously stated [8].
2.5. a-Amylase Inhibitory Assay
The AAI potential of 1 and 9 was assessed utilizing Enz-Chek® Ultra-Amylase Assay Kits as formerly stated [7].
2.6. Molecular Docking Evaluation
2.6.1. Protein Preparation
To perform the docking studies, the crystal structure of the alpha amylase (PDB ID: 5E0F) was imported from the available online protein databank. Before docking, the protein was prepared by employing the Schrödinger suite protein preparation wizard tool [19]. The hydrogen and heavy atoms were subjected to optimization by restrained minimization. Additionally, missed H atoms were added, and the correct charges were assigned using the OPSL4 force field. H2O molecules from HET groups beyond 5 Å were removed.
2.6.2. Ligand Preparation
Lig Prep was used to convert the compounds from 2D to 3D structures [20]. Strained minimization was carried out by employing the OPLS4 force field, the optimization of H-bonds was accomplished at pH 7.0 utilizing PROPKA, and water molecules beyond 3 Å were removed from the HET groups, Additionally, at 7.0 ± 2.0 pH, the metals’ HET cofactors and states were generated.
2.6.3. Receptor Grid Generation and Docking
By using Glide, both ligands docking and grid generation were accomplished [21]. The grid box was defined by selecting the cocrystalized peptide inhibitor of 5E0F, and the binding region was specified using Glide’s Receptor-Grid-Generation tool. The generated grid was utilized for the prepared ligands docking using Glide software. The selected protocol was SP (standard precision). The default 0.25 potential charge cutoff and 1.0 radii scaling factor (vdW) were set. Compounds 1 and 9, in addition to the cocrystalized ligand, 5,7-dihydroxy-4-oxo-2-(3,4,5-trihydroxyphenyl)-4H-chromen-3-yl-6-deoxy-2-O-{6-O-[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]-beta-D-glucopyranosyl}-alpha-L-mannopyranoside (code:5JZ) and acarbose were redocked using the XP (extraprecision) protocol. All other settings were retained as the default.
3. Results and Discussion
3.1. Metabolites Purification and Structural Determination of 1 and 9
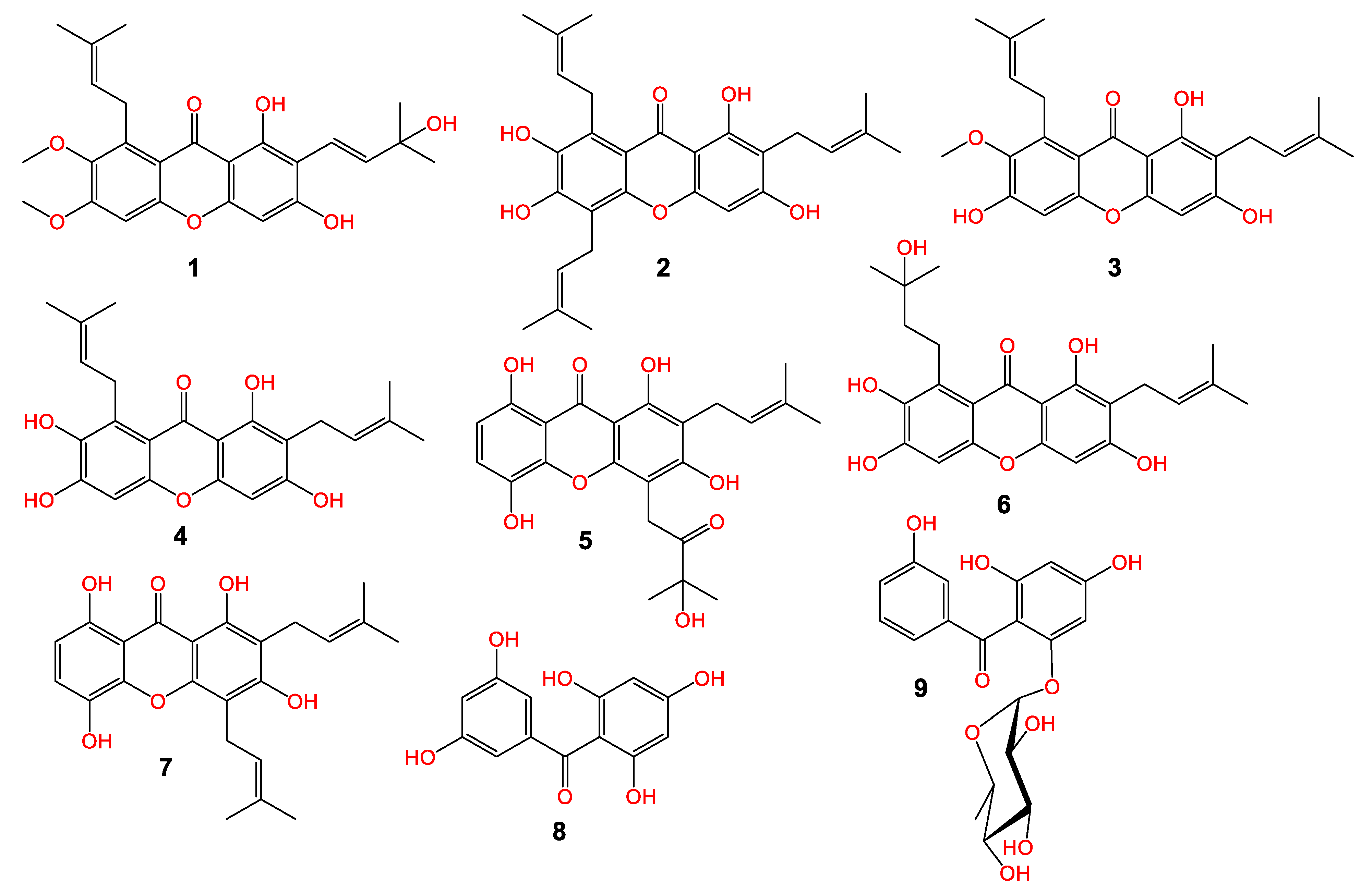
The MeOH extract of pericarps was partitioned among EtOAc and n-hexane. The EtOAC fraction was chromatographed utilizing SiO2 and CC to afford a new xanthone; garcixanthone E (1) and a new benzophenone rhamnoside; garcimangophenone C (9), along with garcinone E (2) [22], α-mangostin (3) [23], γ-mangostin (4) [24], garcinone C (5) [22], garcixanthone C (6) [9], gartanin (7) [25], and 2,4,6,3′,5′-pentahydroxybenzophenone (8) [26] (Figure 1). The former metabolites’ identification was achieved by comparing their spectral data to the earlier published ones and was proven via coTLC along authentic samples.
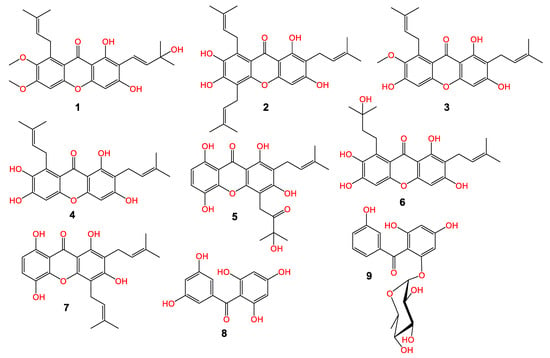
Figure 1.
Chemical structure of new metabolites: garcixanthone E (1) and garcimangophenone C (9) and known (2–5 and 6–8) metabolites.
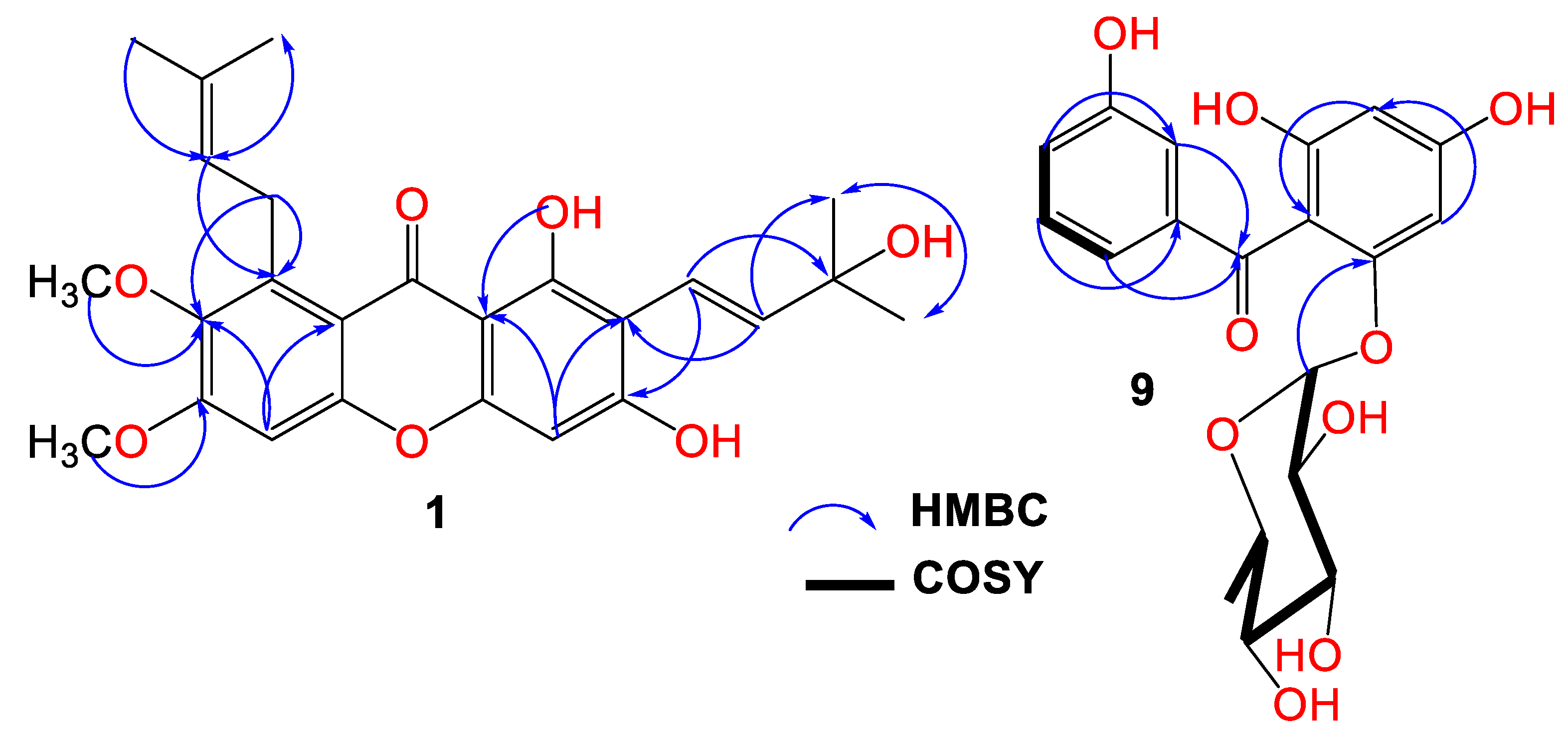
Compound 1 was purified as light-yellow powder with a C25H28O7 molecular formula, relying on the observed HRESIMS pseudomolecular peak at m/z 441.1908 [M+H]+ (calc. for 441.1913). This formula revealed 12 unsaturation degrees. The IR bands at 2942, 3439, 1648, 1458, and 1585 cm−1 characterized the presence of C-H aliphatic, OH phenolic, chelated carbonyl, C=C aromatic, and C–O functionalities, respectively [4]. It displayed UV absorptions for an oxygenated xanthone at 237, 269, 322, and 386 nm [9]. The HSQC and 13C exhibited 25 carbons: five methines, one methylene, six methyls, and twelve quaternary carbons, including an oxygen-linked aliphatic (δC 77.7, C-3′), five oxygenated-aromatic, and a carbonyl (δC 182.2, C-9) carbons (Figures S1–S4). The 1H revealed two singlets at δH 6.23 (H-4) and 6.88 (H-5) for two pentasubstituted phenyl moieties (Table 1). These signals related the carbons at δC 94.2 and 97.7 in the HSQC. The HMBC peaks of H-4/C-8b, C-2, C-3, and C-4a and H-5/C-8a, C-6, C-7, and C-4b affirmed these moieties. Additionally, a signal at δH 13.71 for chelated phenolic OH was observed. Its locality at C-1 was secured by C-1 (δC 157.9) and C-8b (δC 103.5) HMBC crosspeaks. The 1H and 13C spectra possessed disubstituted double bond signals at δH 5.55 (H-2′)/127.1 (C-2′), 6.72 (H-1′)/δC 115.8 (C-1′), two methyls at δH 1.46 (H-5′, 4′)/δC 28.5 (C-5′, 4′), and an oxygenated quaternary at δC 77.7 (C-3′), characterizing a 3-hydroxy-3-methylbut-1-enyl subunit. This was assured by the HMBC relations of H-1′/C-4′/C-2′/C-3′/C-5′, H-2′/C-4′/C-3′/C-5′, H-4′/C-2′/C-5′/C-3′, and H-5′/C-2′/C-4′/C-3′ (Figure 2).
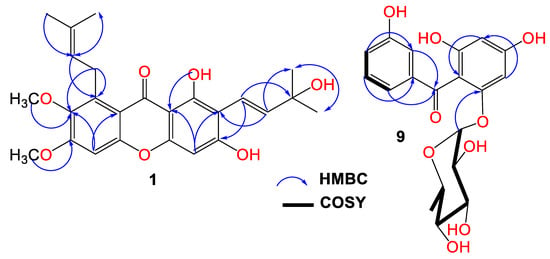
Figure 2.
Important HMBC crosspeaks of 1 and 9.
Its connectivity at C-2 was established on the basis of H-1′/C-2/C-3, H-2′/C-1/C-2 HMBC crosspeaks. Further, the presence of the 3-methylbut-2-enyl subunit was evidenced by the noticed signals at δH 4.07 (H-1″)/δC 26.5 (C-1″), 5.25 (H-2″)/123.1 (C-2″), 132.2 (C-3″), 1.68 (H-4″)/25.6 (C-4′), and 1.82 (H-5″)/18.1 (C-5″). The H-2″/C-8 and H-1″/C-8/C-7/C-8a crosspeaks in the HMBC secured the location of the 3-methylbut-2-enyl subunit at C-8. Two methoxys at δH 3.75/δC 61.5 and δH 3.81/δC 59.9 were present. Their placements at C-7 and C-6 were asserted by the HMBC crosspeaks of 7-OCH3/C-7 (δC 142.6) and 6-OCH3/C-6 (δC 151.9). Therefore, 1 was designated as garcixanthone E (1,3-dihyroxy-6,7-dimethoxy-2-(3-hydroxy-3-methylbut-1-enyl)-8-(3-methylbut-2-enyl)-xanthone).
Compound 9 was obtained as a brown powder. Its IR spectrum revealed bands at 3365, 1638, and 1605 cm−1, which signalized the existence of hydroxyl, carbonyl, and C=C functionalities in 9. Additionally, it had UV bands at 310, 282, and 213 nm [7,8]. The HRESIMS demonstrated a pseudomolecular peak at m/z 393.1180 (calcd for 393.1186 for C19H21O9), corresponding to the molecular formula C19H20O9, which required 10 unsaturation degrees for two benzene, one carbonyl, and hexose moieties. Further, the HRESIMS 246.0535 [M+H-hexose moiety]+ fragment peak indicated that 9 possessed a hexose sugar. In the HSQC and 13C, nineteen carbons were noticed, consisting of one methyl, eleven methines, and seven quaternary carbons for the oxygen-bonded aromatics at δC 163.8 (C4), 161.7 (C2), 159.5 (C6), and 158.2 (C3′), as well as one carbonyl (δC 197.5, C7) carbon. The NMR spectrum revealed signals at 6.95 (H-4′), 7.11 (H-2′), 7.17 (H-6′), and 7.19 (H-5′), having HSQC crosspeaks with carbons at 120.4, 116.9, 121.4, and 130.2, respectively (Figures S5–S8). The 1H-1H COSY relations of H-4′/H-2′ and H-6′, H-2/H-4′, H-6′/H-2′, and H-4′ featured the existence of a disubstituted benzene ring (substructure A). This was ensured by the observed HMBC crosspeaks of H-2′/C-1′/C-3′/C-4′/C-6′, H-4′/C-4′/C-2′/C-6′, and H-5′ and H-6′/C-1′/C-2′/C-3′/C-4′. In the HSQC, the carbons at δC 98.1 and 95.7 correlated to the metacoupled protons at δH 6.06 (H-3) and 6.21 (H-5), respectively, characterizing a substituted phloroglucinol moiety (substructure B). The HMBC crosspeaks of H-3/C-1/C-2/C-4/C-5 and H-5/C-1/C-2/C-4/C-3/C-6 emphasized this assignment. The connection between the two substructures through the carbonyl carbon was proved utilizing HMBC relations of H-5, H-3, H-2′, and H-6′/C-7 (δC 197.5). The 1H and 13C signals at δH 5.36 (d, J = 1.2 Hz, H-1″)/103.7 (C-1″) and δH 1.20 (H-6″)/δC 17.4 (C-6″), in addition to the signals at δH 3.82 (H-2′′)/72.4 (C-2″), 3.30 (H-3″)/72.6 (C-3″), 3.26 (H-4″)/73.6 (C-4″), and 3.69 (H-5″)/71.0 (C-2″) characterized a rhamnose moiety in 9, which was ensured by the COSY and HMBC correlations [27]. Its attachment at C-6 was confirmed by the HMBC crosspeak of H-1″ to C-6 (δC 159.5). Based on these data, 9 was specified and named garcimangophenone C. It is noteworthy that this was the first report of isolating benzophenone rhamnosides from G. mangostana.
3.2. Cytotoxic and AAI (Alpha-Amylase Inhibitory) Activities
The cytotoxic potential of 1 and 9 was assessed towards MCF-7, A549, and HCT-116 cell lines using a sulforhodamine B (SRB) assay. Compound 1 had moderate activity towards A549, MCF-7, and HCT-116 with IC50s 5.4, 8.5, and 5.7 µM, respectively, compared with doxorubicin (IC50s 0.18, 0.6, and 0.2 µM, respectively). Unfortunately, 9 had weak cytotoxic potential versus the tested cancer cell line. It is mentionable that GM pericarp extracts revealed a significant glucose-decreasing and insulin-sensitization capacity [28]. It also revealed the antihyperglycemic effectiveness through boosting insulin-forming β-cell populations, which referred to its antioxidative phenolic constituents [29]. Moreover, it amended β-cells and pancreatic glands impairment caused by STZ in diabetic mice via promoting insulin production and modulating the sensitivity to the decreased insulin [30]. The treatments of diabetic mice with GM xanthones remarkably amended the antioxidant and biochemical parameters, reformed the kidney and liver histological changes, and lessened the kidney tissue cellular apoptosis [31]. Further, GM xanthones and benzophenones were proved to display α-amylase and α-glucosidase inhibitory capacities; therefore, they could minimize postprandial hyperglycemia via the prohibition of glucose absorption [7].
Accordingly, the new metabolites 1 and 9 were assessed for their AAI capacity. They demonstrated AAI potential (IC50 17.8 and 12.9 µM, respectively) in comparison to acarbose (IC50 6.7 µM).
3.3. Molecular Docking Evaluation
3.3.1. Ligands and Protein Preparation
Compounds 1, 9, and 5J7 (native inhibitor of 5EOF) were prepared using LigPrep to convert 2D structures into 3D; additionally, the ionization state at a pH of 7.0 ± 2.0 and tautomeric forms were created. Using the protein preparation wizard, the human α-amylase’s protein crystal structure (PDB ID: 5E0F) was prepared, whereby the hydrogens were added, the bond orders were specified, and the het states using an Epik at pH 7.0 ± 2.0 were generated. The H-bonds were optimized at pH 7.0 employing PROPKA in sample water orientation, and the restrained minimization was performed using the OPLS4 force field.
3.3.2. Receptor Grid Generation and Molecular Docking Studies
The grid box was created all over the protein’s binding site of the minimized protein that contained the cocrystalized inhibitor utilizing the crystal structure (PDB-ID: 5E0F), and the binding area was specified by the 5J7 native inhibitor’s selection. The nonpolar atoms were located and the Van der Waals radii scaling factor was set to 1, and 0.25 was the partial charge cutoff. The ligands docking was executed utilizing the Schrödinger suite “ligand docking” tool, the protocol was SP (standard precision), and all other settings were retained in their default form. The redocking of the ligand 5J7 (5,7-dihydroxy-4-oxo-2-(3,4,5-trihydroxyphenyl)-4H-chromen-3-yl 6-deoxy-2-O-{6-O-[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]-beta-D-glucopyranosyl}-alpha-L-mannopyranoside) was performed to evaluate the docking study. Compound 9 exhibited a reasonable docking score (−7.746 kcal/mol) compared with the native inhibitor 5J7 (−9.932 kcal/mol) as shown in Table 2. For the docking validation, the native inhibitor was prepared and redocked alongside compounds 9 and 1; then, the poses of the protein compound complexes were examined, and the RMS of the native inhibitor 5J7 was found to be in an acceptable range (1.0862).

Table 2.
Results of in silico screening against human AA (PDB: 5E0F).
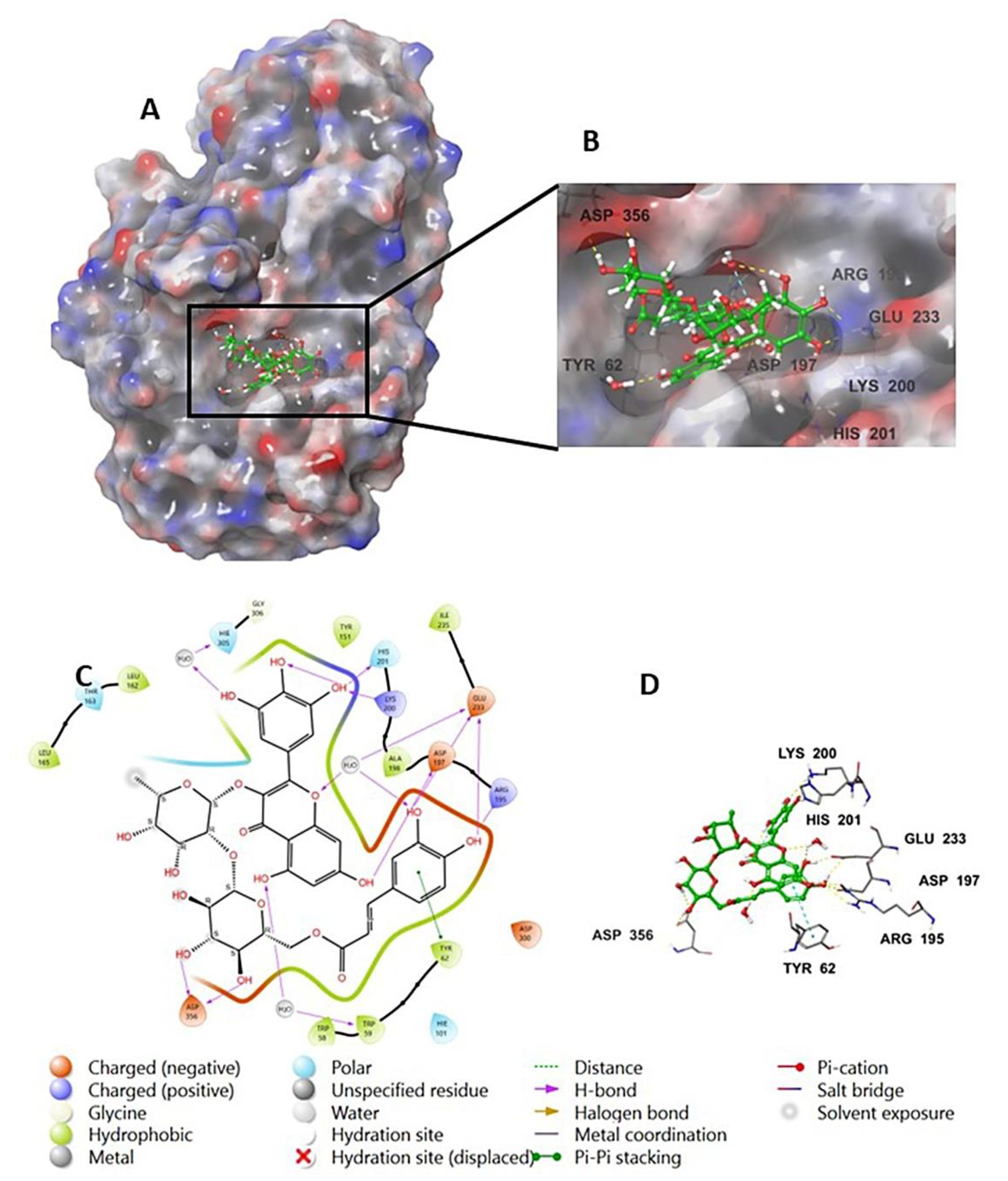
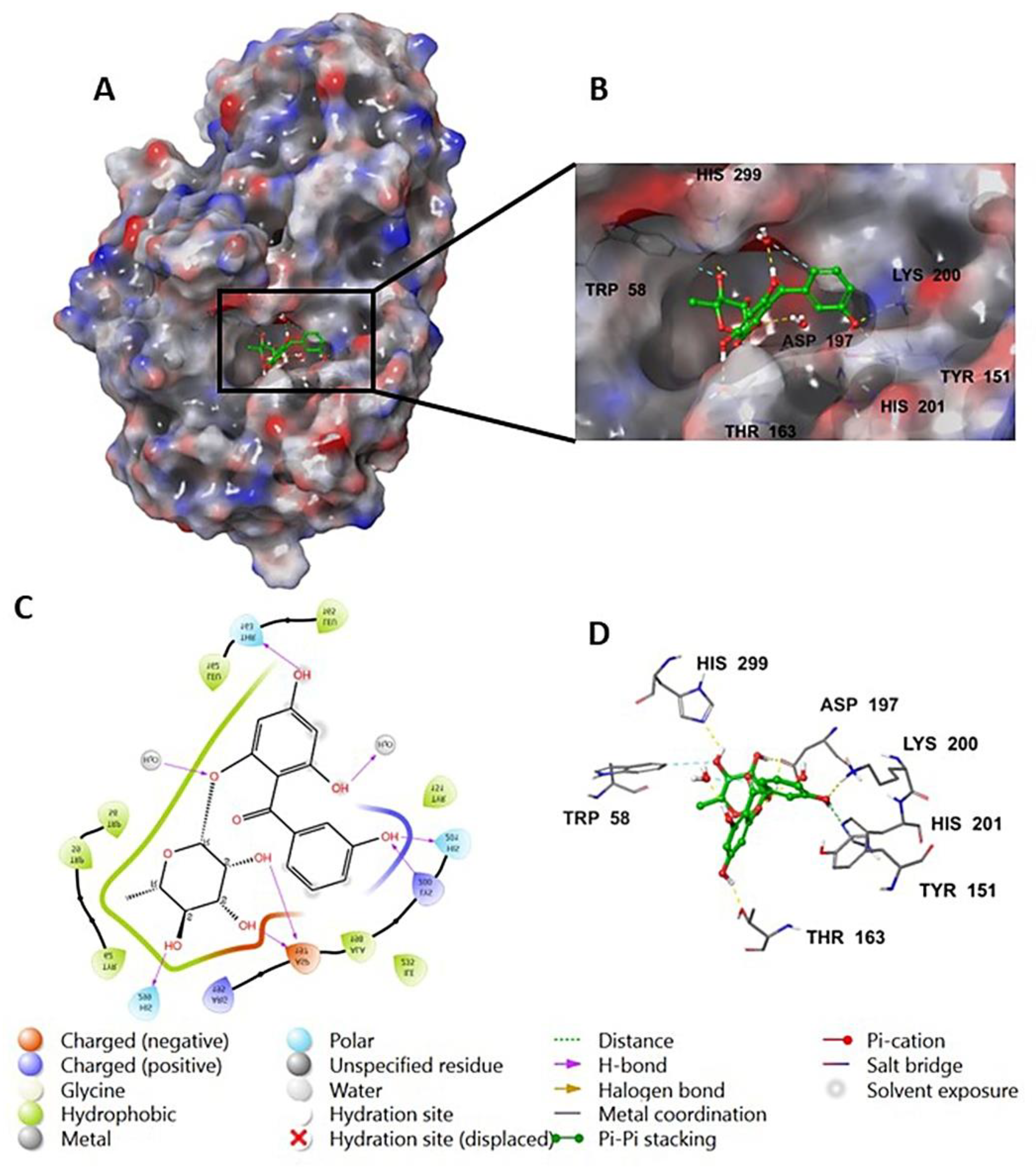
The investigation of the cocrystalized human AA with the native inhibitor 5J7 showed the formation of hydrogen bonds and hydrophobic interactions with many amino acids’ residues. Hydrogen bonds formed between 5J7 and Gln63, Asp97, Glu233, and His305 while forming a hydrophobic interaction via pi-pi stacking with the residues Tyr62 and His 29 (Figure 3). Compound 9 interacted with the AA through hydrogen bonds between its hydroxyl groups and the amino acids’ residues Thr163, Asp197, Lys200, His 201, and His 299 in addition to the aromatic hydrogen bonds’ hydroxyl groups and TRP 59 and Tyr151 (Figure 4).
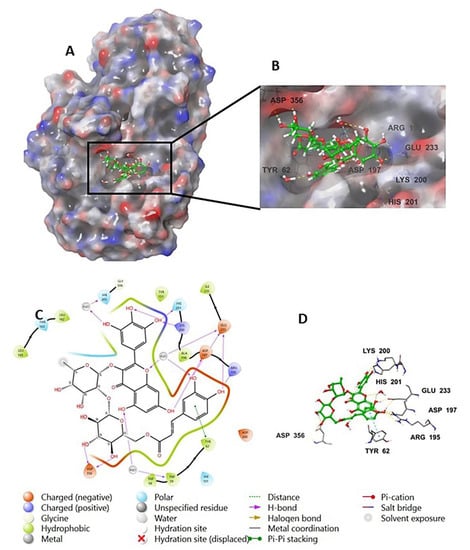
Figure 3.
Native inhibitor (5JZ) in complex with human alpha amylase PDB: 5E0F; (A) molecular surface representation with solid style and electrostatic potential color scheme (red, white, blue) (min −0.3, max +0.3); (B) close lock to the human AA binding site with 5JZ; (C) 2D representation of the binding interaction showing the important amino acids’ residues implicated in the interactions within 3 Å around the ligand; (D) 3D representation of the binding interaction, where the 5JZ was represented in green color and wire representation was applied for amino acids’ residues; hydrogen bond represented in yellow dots and aromatic hydrogen bond represented in violet dots.
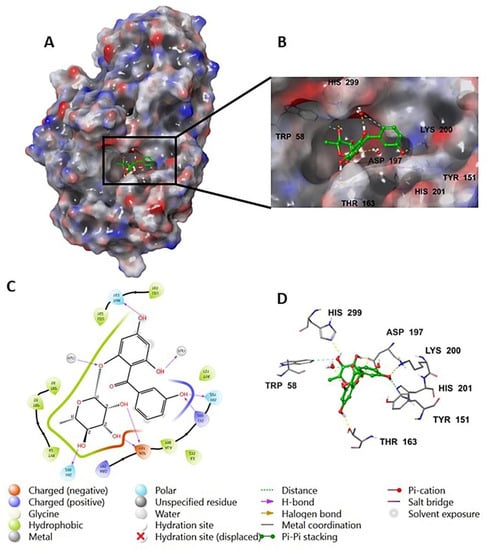
Figure 4.
Compound 9 in complex with human alpha amylase (PDB: 5E0F); (A) molecular surface representation with solid style and electrostatic potential color scheme (red, white, blue) (min −0.3, max +0.3); (B) close lock to the human AA binding site with compound 9; (C) 2D representation of the binding interaction showing the important amino acids’ residues implicated in the interactions within 3 Å around the ligand; (D) 3D representation of the binding interaction, where compound 9 was represented in green color and wire representation was applied for amino acids’ residues; hydrogen bond represented in yellow dots and aromatic hydrogen bond represented in violet dots.
4. Conclusions
G. mangostana is one of the most valuable tropical fruits and its usage as a functional product has been growing because of its bioactivities that are related to its xanthones’ content. In the current study, two new metabolites, garcixanthone E (1) and garcimangophenone C (9), along with seven known compounds were separated from G. mangostana EtOAc extract using different chromatographic tools. Their structures were assigned based on various spectral analyses, including UV, IR, MS, and NMR. Compound 1 displayed moderate in vitro cytotoxic potential versus MCF-7, A549, and HCT-116 cell lines in the SRB assay. Additionally, 1 and 9 possessed moderate AAI potential. In the molecular docking study, 9 revealed a reasonable docking score compared to the native ligand 7JR that agreed with the in vitro activity findings. These results could further prove a possible usage of G. mangostana as a functional food for treating diabetes and cancer. Certainly, more future in vivo and mechanistic studies are required to validate the activity of these interesting metabolites.
Further, to overcome the hazardous impacts and disadvantages of the conventional extraction of such metabolites by organic solvents such as MeOH, extraction using ecofriendly green solvents such as supercritical fluids, biobased solvents, and liquified gases could be applied. These solvents possess beneficial characteristics, including ease of preparation, biocompatibility, custom tunability, high selectivity, and low cost and volatility [32].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/life12111875/s1: Figure S1: 1H NMR spectrum of compound 1 (600 MHz, CDCl3); Figure S2: 13C NMR spectrum of compound 1 (150 MHz, CDCl3); Figure S3: HSQC spectrum of compound 1; Figure S4: HMBC spectrum of compound 1; Figure S5: 1H NMR spectrum of compound 9 (600 MHz, CD3DO); Figure S6: 13C NMR spectrum of compound 9 (150 MHz, CD3DO); Figure S7: HSQC spectrum of compound 9; Figure S8: HMBC spectrum of compound 9.
Author Contributions
Conceptualization, G.A.M. and S.R.M.I.; methodology, G.A.M. and S.R.M.I.; validation, G.A.M. and S.R.M.I.; investigation, G.A.M. and S.R.M.I.; resources, G.A.M.; writing—original draft preparation, G.A.M. and S.R.M.I.; writing—review and editing, G.A.M. and S.R.M.I.; visualization, G.A.M.; supervision, G.A.M.; project administration, G.A.M.; funding acquisition, G.A.M. All authors have read and agreed to the published version of the manuscript.
Funding
The Deanship of Scientific Research (DSR) at King Abdulaziz University (KAU), Jeddah, Saudi Arabia has funded this project under grant no. (G: 065-166-1443).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The Deanship of Scientific Research (DSR) at King Abdulaziz University (KAU), Jeddah, Saudi Arabia has funded this project under grant no. (G: 065-166-1443). The authors acknowledge the DSR with thanks for the technical and financial support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ovalle-Magallanes, B.; Eugenio-Pérez, D.; Pedraza-Chaverri, J. Medicinal properties of mangosteen (Garcinia mangostana L.): A comprehensive update. Food Chem. Toxicol. 2017, 109, 102–122. [Google Scholar] [CrossRef] [PubMed]
- Saraswathy, S.U.P.; Lalitha, L.C.P.; Rahim, S.; Gopinath, C.; Haleema, S.; SarojiniAmma, S.; Aboul-Enein, H.Y. A review on synthetic and pharmacological potential of compounds isolated from Garcinia mangostana Linn. Phytomedicine Plus 2022, 2, 100253. [Google Scholar] [CrossRef]
- Ghazali, S.A.I.S.M.; Lian, G.E.C.; Abd Ghani, K.D. Chemical constituent from roots of Garcinia mangostana (Linn.). Int. J. Chem. 2010, 2, 134. [Google Scholar]
- Obolskiy, D.; Pischel, I.; Siriwatanametanon, N.; Heinrich, M. Garcinia mangostana L.: A phytochemical and pharmacological review. Phytother. Res. 2009, 23, 1047–1065. [Google Scholar] [CrossRef]
- Pedraza-Chaverri, J.; Cárdenas-Rodríguez, N.; Orozco-Ibarra, M.; Pérez-Rojas, J.M. Medicinal properties of mangosteen (Garcinia mangostana). Food Chem. Toxicol. 2008, 46, 3227–3239. [Google Scholar] [CrossRef] [PubMed]
- Vien, L.C.; Chinnappan, S.; Mogana, R. Antioxidant activity of Garcinia mangostana L and alpha mangostin: A review. Res. J. Pharm. Technol. 2021, 14, 4466–4470. [Google Scholar] [CrossRef]
- Alhakamy, N.A.; Mohamed, G.A.; Fahmy, U.A.; Eid, B.G.; Ahmed, O.A.A.; Al-Rabia, M.W.; Khedr, A.I.M.; Nasrullah, M.Z.; Ibrahim, S.R.M. New Alpha-Amylase Inhibitory Metabolites from Pericarps of Garcinia Mangostana. Life 2022, 12, 384. [Google Scholar] [CrossRef]
- Mohamed, G.A.; Ibrahim, S.R. New Benzophenones and a Dihydroflavanonol from Garcinia Mangostana Pericarps and their Antioxidant and Cytotoxic Activities. Phytochem. Lett. 2020, 39, 43–48. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Abdallah, H.M.; El-Halawany, A.M.; Radwan, M.F.; Shehata, I.A.; Al-Harshany, E.M.; Zayed, M.F.; Mohamed, G.A. Garcixanthones B and C, New Xanthones from the Pericarps of Garcinia Mangostana and their Cytotoxic Activity. Phytochem. Lett. 2018, 25, 12–16. [Google Scholar] [CrossRef]
- Gul, S.; Aslam, K.; Pirzada, Q.; Rauf, A.; Khalil, A.A.; Semwal, P.; Bawazeer, S.; Al-Awthan, Y.S.; Bahattab, O.; Al-Duais, M.A.; et al. Xanthones: A class of heterocyclic compounds with anticancer potential. Curr. Top. Med. Chem. 2022, 10, 2174. [Google Scholar]
- Huang, Q.; Wang, Y.; Wu, H.; Yuan, M.; Zheng, C.; Xu, H. Xanthone glucosides: Isolation, bioactivity and synthesis. Molecules 2021, 26, 5575. [Google Scholar] [CrossRef] [PubMed]
- Surana, K.; Chaudhary, B.; Diwaker, M.; Sharma, S. Benzophenone: A Ubiquitous Scaffold in Medicinal Chemistry. MedChemComm 2018, 9, 1803–1817. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Health Estimates 2020: Deaths by Cause, Age, Sex, by Country and by Region, 2000–2019; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Cancer Statistics for the Year 2020: An Overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef]
- Dutta, S.; Mahalanobish, S.; Saha, S.; Ghosh, S.; Sil, P.C. Natural Products: An Upcoming Therapeutic Approach to Cancer. Food Chem. Toxicol. 2019, 128, 240–255. [Google Scholar] [CrossRef] [PubMed]
- Cragg, G.M.; Grothaus, P.G.; Newman, D.J. Impact of Natural Products on Developing New Anti-Cancer Agents. Chem. Rev. 2009, 109, 3012–3043. [Google Scholar] [CrossRef]
- Reddy, L.; Odhav, B.; Bhoola, K.D. Natural Products for Cancer Prevention: A Global Perspective. Pharmacol. Ther. 2003, 99, 1–13. [Google Scholar] [CrossRef]
- Rajendra, G. Mehta, Genoveva Murillo, Rajesh Naithani & Xinjian Peng. Cancer Chemoprevention by Natural Products: How Far have We Come? Pharm. Res. 2010, 27, 950–961. [Google Scholar]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and Ligand Preparation: Parameters, Protocols, and Influence on Virtual Screening Enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Schrödinger, L.L.C. Schrödinger Release 2021-4: LigPrep; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Schrödinger, L.L.C. Schrödinger Release 2021-4: Glide; Schrödinger, LLC: New York, NY, USA, 2021. [Google Scholar]
- Sen, A.K.; Sarkar, K.K.; Mazumder, P.C.; Banerji, N.; Uusvuori, R.; Hase, T.A. The Structures of Garcinones a, B and C: Three New Xanthones from Garcinia Mangostana. Phytochemistry 1982, 21, 1747–1750. [Google Scholar] [CrossRef]
- Iwo, M.I.; Soemardji, A.A.; Hanafi, M. Sunscreen Activity of A-Mangostin from the Pericarps of Garcinia Mangostana. J. Appl. Pharm. Sci. 2013, 3, 70–73. [Google Scholar]
- Govindachari, T.R.; Kalyanaraman, P.S.; Muthukumaraswamy, N.; Pai, B.R. Xanthones of Garcinia Mangostana Linn. Tetrahedron 1971, 27, 3919–3926. [Google Scholar] [CrossRef]
- Bennett, G.J.; Lee, H.; Lee, L. Synthesis of Minor Xanthones from Garcinia Mangostana. J. Nat. Prod. 1990, 53, 1463–1470. [Google Scholar] [CrossRef]
- Ishiguro, K.; Nakajima, M.; Fukumoto, H.; Isoi, K. A Xanthone Substituted with an Irregular Monoterpene in Cell Suspension Cultures of Hypericum Patulum. Phytochemistry 1995, 39, 903–905. [Google Scholar] [CrossRef]
- Tanaka, N.; Kubota, T.; Kashiwada, Y.; Takaishi, Y.; Kobayashi, J. Petiolins F—I, Benzophenone Rhamnosides from Hypericum Pseudopetiolatum Var. Kiusianum. Chem. Pharm. Bull. 2009, 57, 1171–1173. [Google Scholar] [CrossRef][Green Version]
- Mekseepralard, C.; Areebambud, C.; Suksamrarn, S.; Jariyapongskul, A. Effects of Long-Term Alpha-Mangostin Supplementation on Hyperglycemia and Insulin Resistance in Type 2 Diabetic Rats Induced by High Fat Diet and Low Dose Streptozotocin. J. Med. Assoc. Thail. Chotmaihet Thangphaet 2015, 98, 23. [Google Scholar]
- Taher, M.; Zakaria, T.; Syafiq, T.M.F.; Susanti, D.; Zakaria, Z.A. Hypoglycaemic Activity of Ethanolic Extract of Garcinia Mangostana Linn. in Normoglycaemic and Streptozotocin-Induced Diabetic Rats. BMC Complement. Altern. Med. 2016, 16, 1–12. [Google Scholar] [CrossRef]
- Husen, S.A.; Kalqutny, S.H.; Ansori, A.N.M.; Susilo, R.J.K.; Alymahdy, A.D.; Winarni, D. Antioxidant and Antidiabetic Activity of Garcinia Mangostana L. Pericarp Extract in Streptozotocin-Induced Diabetic Mice. Biosci. Res. 2017, 14, 1238–1245. [Google Scholar]
- Karim, N.; Rahman, M.A.; Changlek, S.; Tangpong, J. Short-Time Administration of Xanthone from Garcinia Mangostana Fruit Pericarp Attenuates the Hepatotoxicity and Renotoxicity of Type II Diabetes Mice. J. Am. Coll. Nutr. 2020, 39, 501–510. [Google Scholar] [CrossRef]
- Janicka, P.; Płotka-Wasylka, J.; Jatkowska, N.; Chabowska, A.; Fares, M.Y.; Andruch, V.; Kaykhaii, M.; Gębicki, J. Trends in the New Generation of Green Solvents in Extraction Processes. Curr. Opin. Green Sustain. Chem. 2022, 37, 100670. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).