Analysis of the Potential Relationship between Aging and Pulmonary Fibrosis Based on Transcriptome
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Treatments
2.2. Histopathological, Immunohistochemical, and Immunohistofluorescent Examination
2.3. Hydroxyproline Determination and ELISA Assay
2.4. Real-Time qPCR Assay
2.5. RNA Extraction Library Construction and Sequencing
2.6. Bioinformatics Analysis
2.6.1. Sequence and Filtering of Clean Reads
2.6.2. Analysis of RNA-Seq Data and Differentially Expressed Genes
2.6.3. GO and KEGG Enrichment Analysis
2.7. Statistical Analysis
3. Results
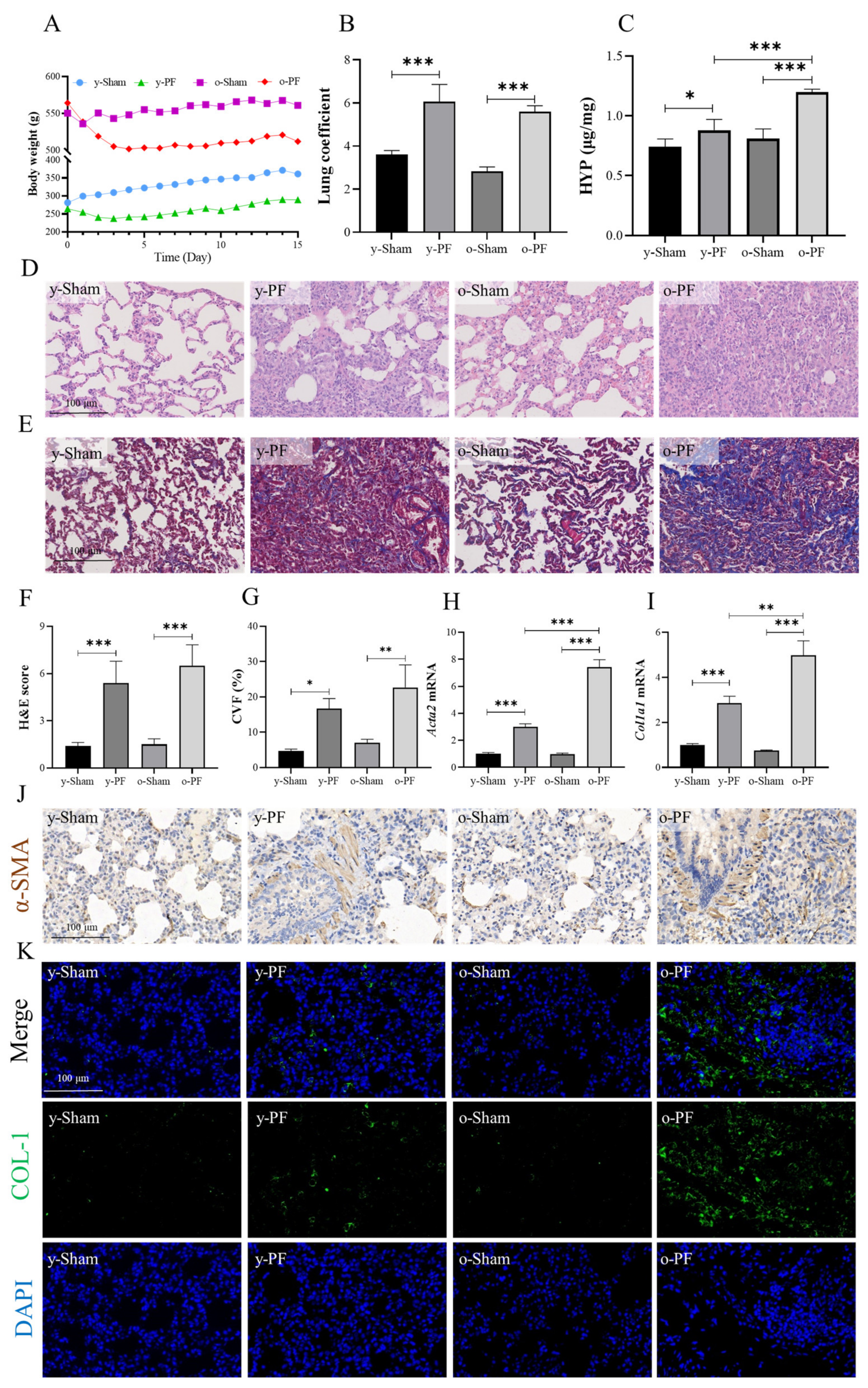
3.1. Pulmonary Fibrosis in Old Rats Is More Serious than That in Young Rats
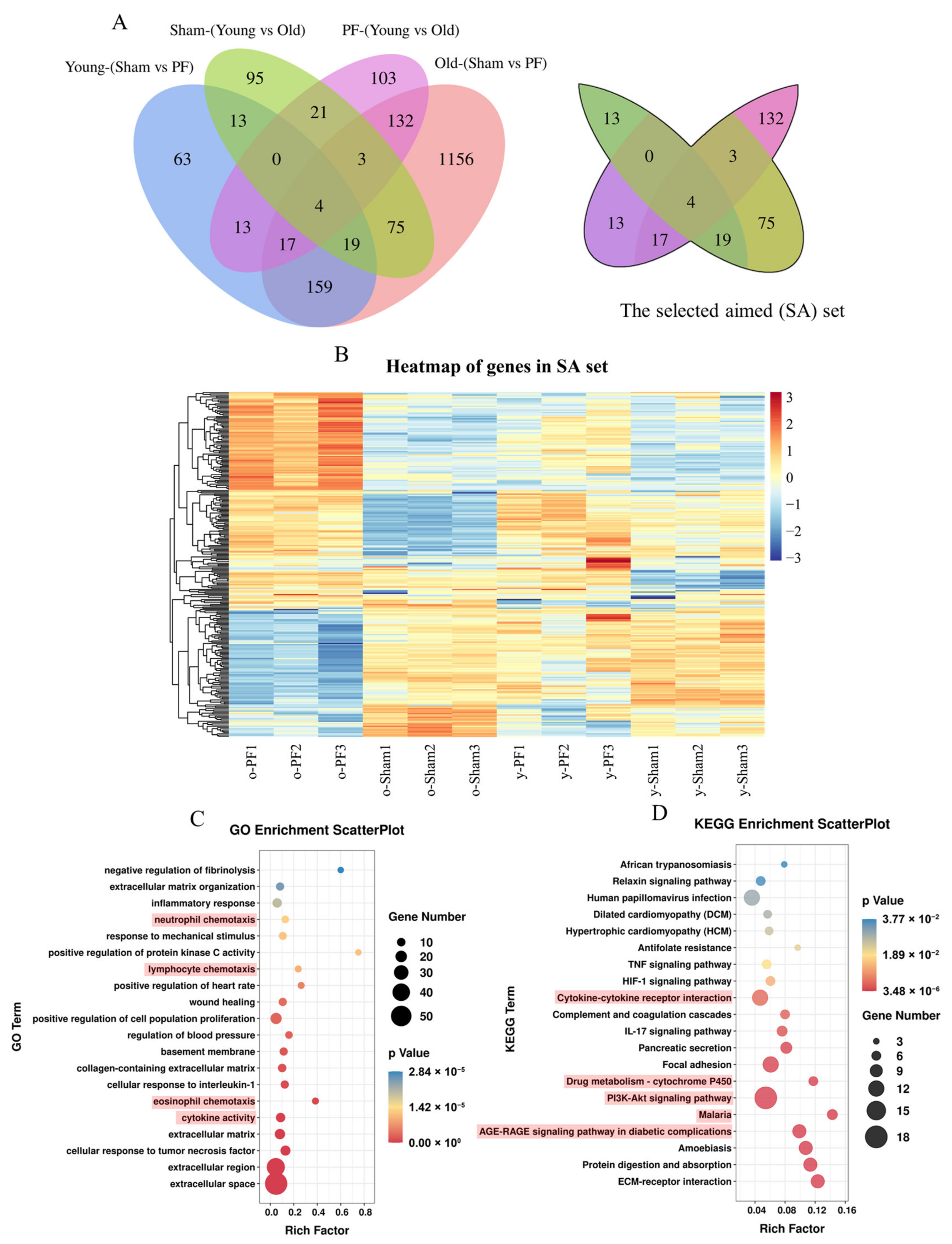
3.2. Analysis of Differentially Expressed Genes between PF Groups and Sham Groups, or Young Groups and Old Groups
3.3. Enrichment of GO and KEGG Pathways of Differentially Expressed Genes between Sham and PF Groups
3.4. Screening and Analysis of Genes Associated with Both Pulmonary Fibrosis and Aging
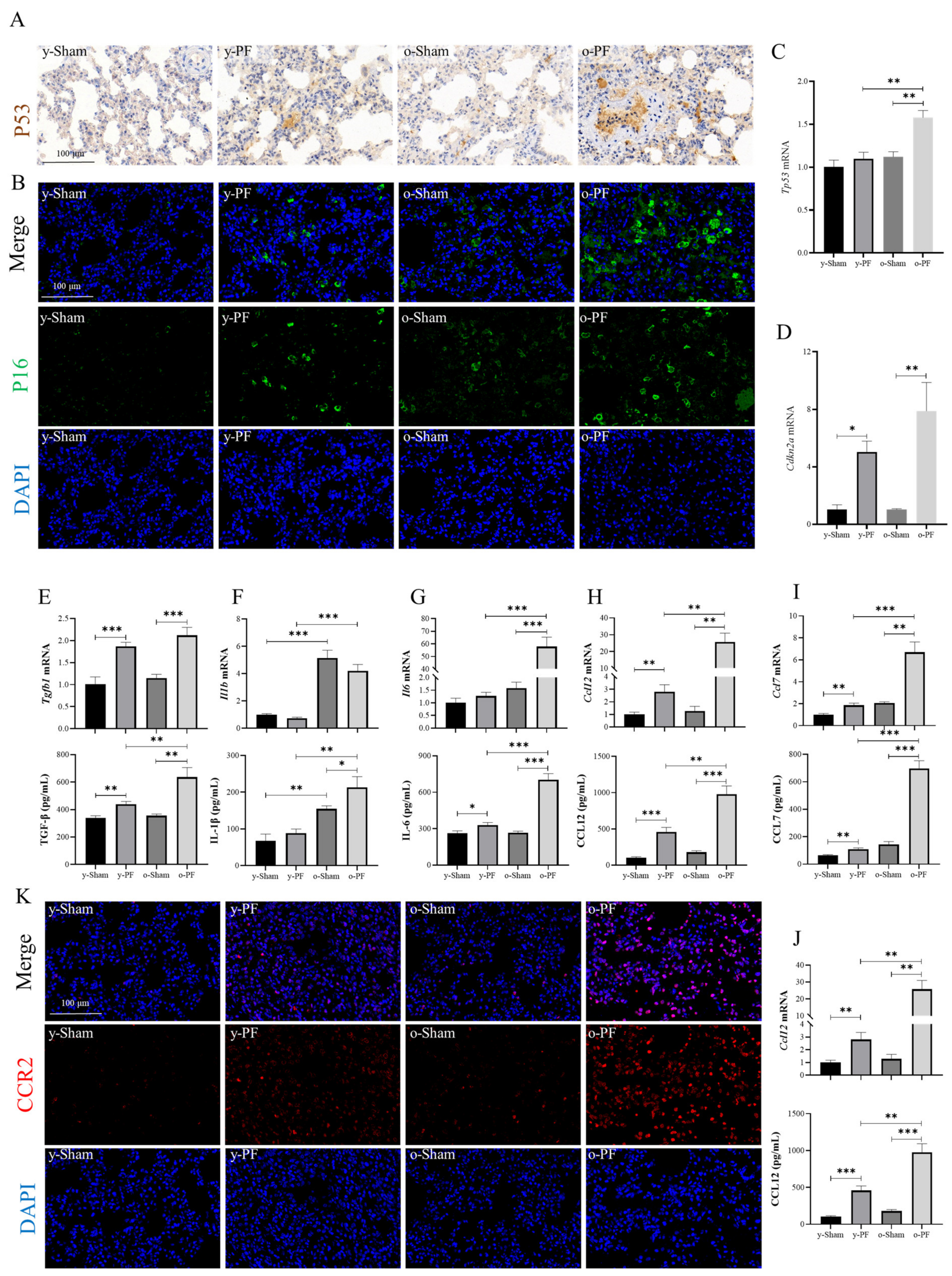
3.5. Validation of Related Indicators: Biomarkers of Senescence and Senescence-Associated Secretory Phenotype (SASP) in Aged and Young Rats
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Glass, D.S.; Grossfeld, D.; Renna, H.A.; Agarwala, P.; Spiegler, P.; Kasselman, L.J.; Glass, A.D.; DeLeon, J.; Reiss, A.B. Idiopathic pulmonary fibrosis: Molecular mechanisms and potential treatment approaches. Respir. Investig. 2020, 58, 320–335. [Google Scholar] [CrossRef]
- Martinez, F.J.; Collard, H.R.; Pardo, A.; Raghu, G.; Richeldi, L.; Selman, M.; Swigris, J.J.; Taniguchi, H.; Wells, A.U. Idiopathic pulmonary fibrosis. Nat. Rev. Dis. Prim. 2017, 3, 17074. [Google Scholar] [CrossRef] [PubMed]
- Armanios, M.Y.; Chen, J.J.; Cogan, J.D.; Alder, J.K.; Ingersoll, R.G.; Markin, C.; Lawson, W.E.; Xie, M.; Vulto, I.; Phillips, J.A., 3rd; et al. Telomerase mutations in families with idiopathic pulmonary fibrosis. N. Engl. J. Med. 2007, 356, 1317–1326. [Google Scholar] [CrossRef] [PubMed]
- Hecker, L.; Logsdon, N.J.; Kurundkar, D.; Kurundkar, A.; Bernard, K.; Hock, T.; Meldrum, E.; Sanders, Y.Y.; Thannickal, V.J. Reversal of persistent fibrosis in aging by targeting Nox4-Nrf2 redox imbalance. Sci. Transl. Med. 2014, 6, 231ra247. [Google Scholar] [CrossRef]
- Schafer, M.J.; White, T.A.; Iijima, K.; Haak, A.J.; Ligresti, G.; Atkinson, E.J.; Oberg, A.L.; Birch, J.; Salmonowicz, H.; Zhu, Y.; et al. Cellular senescence mediates fibrotic pulmonary disease. Nat. Commun. 2017, 8, 14532. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, D.L.; Kellogg, D.L., Jr.; Musi, N.; Nambiar, A.M. Cellular Senescence in Idiopathic Pulmonary Fibrosis. Curr. Mol. Biol. Rep. 2021, 7, 31–40. [Google Scholar] [CrossRef]
- Manzoni, C.; Kia, D.A.; Vandrovcova, J.; Hardy, J.; Wood, N.W.; Lewis, P.A.; Ferrari, R. Genome, transcriptome and proteome: The rise of omics data and their integration in biomedical sciences. Brief. Bioinform. 2018, 19, 286–302. [Google Scholar] [CrossRef]
- Zupanic, A.; Bernstein, H.C.; Heiland, I. Systems biology: Current status and challenges. Cell. Mol. Life Sci. 2020, 77, 379–380. [Google Scholar] [CrossRef]
- Gonzaga-Jauregui, C.; Lupski, J.R.; Gibbs, R.A. Human genome sequencing in health and disease. Annu. Rev. Med. 2012, 63, 35–61. [Google Scholar] [CrossRef]
- Venter, J.C.; Smith, H.O.; Adams, M.D. The Sequence of the Human Genome. Clin. Chem. 2015, 61, 1207–1208. [Google Scholar] [CrossRef]
- Sahraeian, S.M.E.; Mohiyuddin, M.; Sebra, R.; Tilgner, H.; Afshar, P.T.; Au, K.F.; Bani Asadi, N.; Gerstein, M.B.; Wong, W.H.; Snyder, M.P.; et al. Gaining comprehensive biological insight into the transcriptome by performing a broad-spectrum RNA-seq analysis. Nat. Commun. 2017, 8, 59. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Gene Ontology, C. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res 2021, 49, D325–D334. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef] [PubMed]
- Salama, R.; Sadaie, M.; Hoare, M.; Narita, M. Cellular senescence and its effector programs. Genes Dev. 2014, 28, 99–114. [Google Scholar] [CrossRef] [PubMed]
- Parimon, T.; Yao, C.; Stripp, B.R.; Noble, P.W.; Chen, P. Alveolar Epithelial Type II Cells as Drivers of Lung Fibrosis in Idiopathic Pulmonary Fibrosis. Int. J. Mol. Sci. 2020, 21, 2269. [Google Scholar] [CrossRef]
- Barnes, P.J.; Baker, J.; Donnelly, L.E. Cellular Senescence as a Mechanism and Target in Chronic Lung Diseases. Am. J. Respir. Crit. Care Med. 2019, 200, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Lopes-Paciencia, S.; Saint-Germain, E.; Rowell, M.C.; Ruiz, A.F.; Kalegari, P.; Ferbeyre, G. The senescence-associated secretory phenotype and its regulation. Cytokine 2019, 117, 15–22. [Google Scholar] [CrossRef]
- Popiolek-Barczyk, K.; Ciechanowska, A.; Ciapała, K.; Pawlik, K.; Oggioni, M.; Mercurio, D.; De Simoni, M.G.; Mika, J. The CCL2/CCL7/CCL12/CCR2 pathway is substantially and persistently upregulated in mice after traumatic brain injury, and CCL2 modulates the complement system in microglia. Mol. Cell. Probes 2020, 54, 101671. [Google Scholar] [CrossRef]
- Yang, J.; Agarwal, M.; Ling, S.; Teitz-Tennenbaum, S.; Zemans, R.L.; Osterholzer, J.J.; Sisson, T.H.; Kim, K.K. Diverse Injury Pathways Induce Alveolar Epithelial Cell CCL2/12, Which Promotes Lung Fibrosis. Am. J. Respir. Cell Mol. Biol. 2020, 62, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Gulati, S.; Thannickal, V.J. The Aging Lung and Idiopathic Pulmonary Fibrosis. Am. J. Med. Sci. 2019, 357, 384–389. [Google Scholar] [CrossRef]
- Agostini, C.; Gurrieri, C. Chemokine/cytokine cocktail in idiopathic pulmonary fibrosis. Proc. Am. Thorac. Soc. 2006, 3, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Kolahian, S.; Fernandez, I.E.; Eickelberg, O.; Hartl, D. Immune Mechanisms in Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 2016, 55, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Adib, R.; Montgomery, J.M.; Atherton, J.; O’Regan, L.; Richards, M.W.; Straatman, K.R.; Roth, D.; Straube, A.; Bayliss, R.; Moores, C.A.; et al. Mitotic phosphorylation by NEK6 and NEK7 reduces the microtubule affinity of EML4 to promote chromosome congression. Sci. Signal. 2019, 12, eaaw2939. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.; Ferreira, M.; Reichert, C.O.; de Almeida, L.V.; Brocardo, G.; Lage, L.; Culler, H.F.; Nukui, Y.; Bydlowski, S.P.; Pereira, J. Cell Cycle Changes, DNA Ploidy, and PTTG1 Gene Expression in HTLV-1 Patients. Front. Microbiol. 2020, 11, 1778. [Google Scholar] [CrossRef] [PubMed]
- Wasielak-Politowska, M.; Kordowitzki, P. Chromosome Segregation in the Oocyte: What Goes Wrong during Aging. Int. J. Mol. Sci. 2022, 23, 2880. [Google Scholar] [CrossRef] [PubMed]
- Holland, A.J.; Cleveland, D.W. The deubiquitinase USP44 is a tumor suppressor that protects against chromosome missegregation. J. Clin. Investig. 2012, 122, 4325–4328. [Google Scholar] [CrossRef]
- Margadant, C.; Sonnenberg, A. Integrin-TGF-beta crosstalk in fibrosis, cancer and wound healing. EMBO Rep. 2010, 11, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Munger, J.S.; Huang, X.; Kawakatsu, H.; Griffiths, M.J.; Dalton, S.L.; Wu, J.; Pittet, J.F.; Kaminski, N.; Garat, C.; Matthay, M.A.; et al. The integrin alpha v beta 6 binds and activates latent TGF beta 1: A mechanism for regulating pulmonary inflammation and fibrosis. Cell 1999, 96, 319–328. [Google Scholar] [CrossRef]
- Di Guglielmo, G.M. TSP-1 in lung fibrosis. J. Cell Commun. Signal. 2010, 4, 185–186. [Google Scholar] [CrossRef] [PubMed]
- Ezzie, M.E.; Piper, M.G.; Montague, C.; Newland, C.A.; Opalek, J.M.; Baran, C.; Ali, N.; Brigstock, D.; Lawler, J.; Marsh, C.B. Thrombospondin-1-deficient mice are not protected from bleomycin-induced pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2011, 44, 556–561. [Google Scholar] [CrossRef]
- Yang, J.; Nie, J.; Ma, X.; Wei, Y.; Peng, Y.; Wei, X. Targeting PI3K in cancer: Mechanisms and advances in clinical trials. Mol. Cancer 2019, 18, 26. [Google Scholar] [CrossRef]
- Wang, J.; Hu, K.; Cai, X.; Yang, B.; He, Q.; Wang, J.; Weng, Q. Targeting PI3K/AKT signaling for treatment of idiopathic pulmonary fibrosis. Acta Pharm. Sin. B 2022, 12, 18–32. [Google Scholar] [CrossRef] [PubMed]
- Montero, P.; Milara, J.; Roger, I.; Cortijo, J. Role of JAK/STAT in Interstitial Lung Diseases; Molecular and Cellular Mechanisms. Int. J. Mol. Sci. 2021, 22, 6211. [Google Scholar] [CrossRef] [PubMed]
- Pedroza, M.; Le, T.T.; Lewis, K.; Karmouty-Quintana, H.; To, S.; George, A.T.; Blackburn, M.R.; Tweardy, D.J.; Agarwal, S.K. STAT-3 contributes to pulmonary fibrosis through epithelial injury and fibroblast-myofibroblast differentiation. FASEB J. 2016, 30, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Milara, J.; Hernandez, G.; Ballester, B.; Morell, A.; Roger, I.; Montero, P.; Escrivá, J.; Lloris, J.M.; Molina-Molina, M.; Morcillo, E.; et al. The JAK2 pathway is activated in idiopathic pulmonary fibrosis. Respir. Res. 2018, 19, 24. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, U.; Kasembeli, M.M.; Robinson, P.; Tweardy, D.J. Targeting Janus Kinases and Signal Transducer and Activator of Transcription 3 to Treat Inflammation, Fibrosis, and Cancer: Rationale, Progress, and Caution. Pharmacol. Rev. 2020, 72, 486–526. [Google Scholar] [CrossRef]
- Waters, D.W.; Blokland, K.E.C.; Pathinayake, P.S.; Wei, L.; Schuliga, M.; Jaffar, J.; Westall, G.P.; Hansbro, P.M.; Prele, C.M.; Mutsaers, S.E.; et al. STAT3 Regulates the Onset of Oxidant-induced Senescence in Lung Fibroblasts. Am. J. Respir. Cell Mol. Biol. 2019, 61, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Senatus, L.M.; Schmidt, A.M. The AGE-RAGE Axis: Implications for Age-Associated Arterial Diseases. Front. Genet. 2017, 8, 187. [Google Scholar] [CrossRef]
- Machahua, C.; Montes-Worboys, A.; Llatjos, R.; Escobar, I.; Dorca, J.; Molina-Molina, M.; Vicens-Zygmunt, V. Increased AGE-RAGE ratio in idiopathic pulmonary fibrosis. Respir. Res. 2016, 17, 144. [Google Scholar] [CrossRef] [PubMed]
- Baek, H.; Jang, S.; Park, J.; Jang, J.; Lee, J.; Hong, S.H.; Kim, W.J.; Park, S.M.; Yang, S.R. Reduced receptor for advanced glycation end products is associated with alpha-SMA expression in patients with idiopathic pulmonary fibrosis and mice. Lab. Anim. Res. 2021, 37, 28. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Randive, R.; Stewart, J.A. Molecular mechanisms of AGE/RAGE-mediated fibrosis in the diabetic heart. World J. Diabetes 2014, 5, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, K.; Iwamoto, H.; Sakamoto, S.; Horimasu, Y.; Masuda, T.; Miyamoto, S.; Nakashima, T.; Fujitaka, K.; Hamada, H.; Hattori, N. Association of the RAGE/RAGE-ligand axis with interstitial lung disease and its acute exacerbation. Respir. Investig. 2022, 60, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Sharma, I.; Tupe, R.S.; Wallner, A.K.; Kanwar, Y.S. Contribution of myo-inositol oxygenase in AGE:RAGE-mediated renal tubulointerstitial injury in the context of diabetic nephropathy. Am. J. Physiol. Renal. Physiol. 2018, 314, F107–F121. [Google Scholar] [CrossRef]
- Kumar, V.; Fleming, T.; Terjung, S.; Gorzelanny, C.; Gebhardt, C.; Agrawal, R.; Mall, M.A.; Ranzinger, J.; Zeier, M.; Madhusudhan, T.; et al. Homeostatic nuclear RAGE-ATM interaction is essential for efficient DNA repair. Nucleic Acids Res. 2017, 45, 10595–10613. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.Y.F.; Tullis, B.; Breithaupt, K.L.; Fowers, R.; Jones, N.; Grajeda, S.; Reynolds, P.R.; Arroyo, J.A. A Role for RAGE in DNA Double Strand Breaks (DSBs) Detected in Pathological Placentas and Trophoblast Cells. Cells 2021, 10, 857. [Google Scholar] [CrossRef]
- Aghali, A.; Koloko Ngassie, M.L.; Pabelick, C.M.; Prakash, Y.S. Cellular Senescence in Aging Lungs and Diseases. Cells 2022, 11, 1781. [Google Scholar] [CrossRef] [PubMed]
- Yeo, E.J. Hypoxia and aging. Exp. Mol. Med. 2019, 51, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Tzivion, G.; Hay, N. PI3K-AKT-FoxO axis in cancer and aging. Biochim. Biophys. Acta 2011, 1813, 1925. [Google Scholar] [CrossRef] [PubMed]
- Rossner, R.; Kaeberlein, M.; Leiser, S.F. Flavin-containing monooxygenases in aging and disease: Emerging roles for ancient enzymes. J. Biol. Chem. 2017, 292, 11138–11146. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Shen, Y.; Li, W.; Li, Q.; Miao, Y.; Zhong, Y. Upregulation of flavin-containing monooxygenase 3 mimics calorie restriction to retard liver aging by inducing autophagy. Aging 2020, 12, 931–944. [Google Scholar] [CrossRef]
- Ni, C.; Chen, Y.; Xu, Y.; Zhao, J.; Li, Q.; Xiao, C.; Wu, Y.; Wang, J.; Wang, Y.; Zhong, Z.; et al. Flavin Containing Monooxygenase 2 Prevents Cardiac Fibrosis via CYP2J3-SMURF2 Axis. Circ. Res. 2022, 131, 189–206. [Google Scholar] [CrossRef] [PubMed]
- Christofides, A.; Konstantinidou, E.; Jani, C.; Boussiotis, V.A. The role of peroxisome proliferator-activated receptors (PPAR) in immune responses. Metabolism 2021, 114, 154338. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Quiles, M.; Broekema, M.F.; Kalkhoven, E. PPARgamma in Metabolism, Immunity, and Cancer: Unified and Diverse Mechanisms of Action. Front. Endocrinol. 2021, 12, 624112. [Google Scholar] [CrossRef]
- Brody, S.L.; Gunsten, S.P.; Luehmann, H.P.; Sultan, D.H.; Hoelscher, M.; Heo, G.S.; Pan, J.; Koenitzer, J.R.; Lee, E.C.; Huang, T.; et al. Chemokine Receptor 2-targeted Molecular Imaging in Pulmonary Fibrosis. A Clinical Trial. Am. J. Respir. Crit. Care Med. 2021, 203, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Das, A.M.; Seideman, J.; Griswold, D.; Afuh, C.N.; Kobayashi, T.; Abe, S.; Fang, Q.; Hashimoto, M.; Kim, H.; et al. The CC chemokine ligand 2 (CCL2) mediates fibroblast survival through IL-6. Am. J. Respir. Cell Mol. Biol. 2007, 37, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Moore, B.B.; Murray, L.; Das, A.; Wilke, C.A.; Herrygers, A.B.; Toews, G.B. The role of CCL12 in the recruitment of fibrocytes and lung fibrosis. Am. J. Respir. Cell Mol. Biol. 2006, 35, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Burgy, O.; Bellaye, P.S.; Causse, S.; Beltramo, G.; Wettstein, G.; Boutanquoi, P.M.; Goirand, F.; Garrido, C.; Bonniaud, P. Pleural inhibition of the caspase-1/IL-1beta pathway diminishes profibrotic lung toxicity of bleomycin. Respir. Res. 2016, 17, 162. [Google Scholar] [CrossRef]
- Trachalaki, A.; Tsitoura, E.; Mastrodimou, S.; Invernizzi, R.; Vasarmidi, E.; Bibaki, E.; Tzanakis, N.; Molyneaux, P.L.; Maher, T.M.; Antoniou, K. Enhanced IL-1beta Release Following NLRP3 and AIM2 Inflammasome Stimulation Is Linked to mtROS in Airway Macrophages in Pulmonary Fibrosis. Front. Immunol. 2021, 12, 661811. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, S.; Tang, X.; Xu, Y.; Song, X.; Qian, X.; Hu, Y.; Zhang, M. Analysis of the Potential Relationship between Aging and Pulmonary Fibrosis Based on Transcriptome. Life 2022, 12, 1961. https://doi.org/10.3390/life12121961
Fu S, Tang X, Xu Y, Song X, Qian X, Hu Y, Zhang M. Analysis of the Potential Relationship between Aging and Pulmonary Fibrosis Based on Transcriptome. Life. 2022; 12(12):1961. https://doi.org/10.3390/life12121961
Chicago/Turabian StyleFu, San, Xiaoyan Tang, Yiming Xu, Xianrui Song, Xiuhui Qian, Yingying Hu, and Mian Zhang. 2022. "Analysis of the Potential Relationship between Aging and Pulmonary Fibrosis Based on Transcriptome" Life 12, no. 12: 1961. https://doi.org/10.3390/life12121961
APA StyleFu, S., Tang, X., Xu, Y., Song, X., Qian, X., Hu, Y., & Zhang, M. (2022). Analysis of the Potential Relationship between Aging and Pulmonary Fibrosis Based on Transcriptome. Life, 12(12), 1961. https://doi.org/10.3390/life12121961




