Whole-Lesion CT Texture Analysis as a Quantitative Biomarker for the Identification of Homogeneous Renal Tumors
Abstract
1. Introduction
2. Materials and Methods
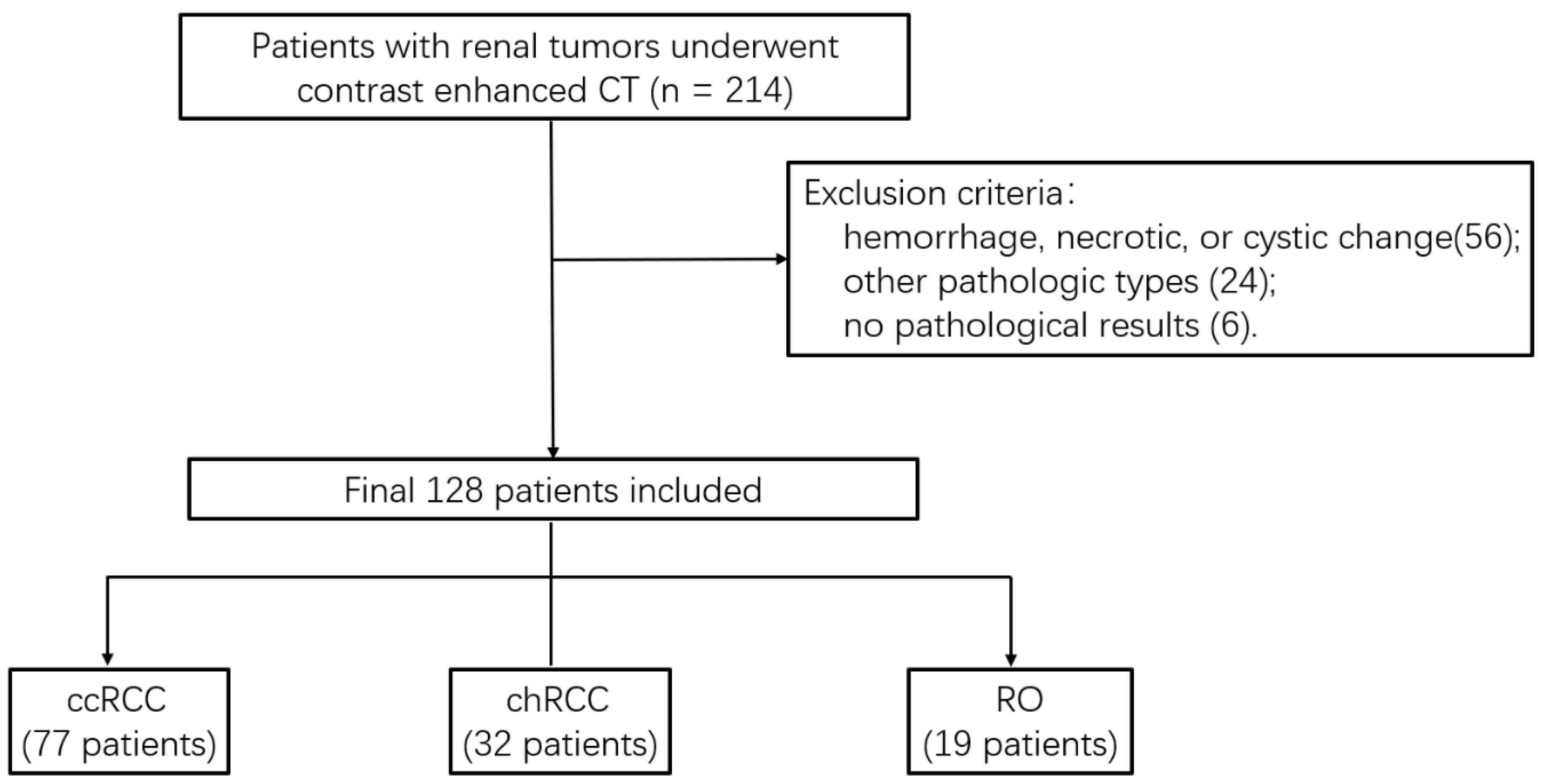
2.1. Patients
2.2. CT Examination
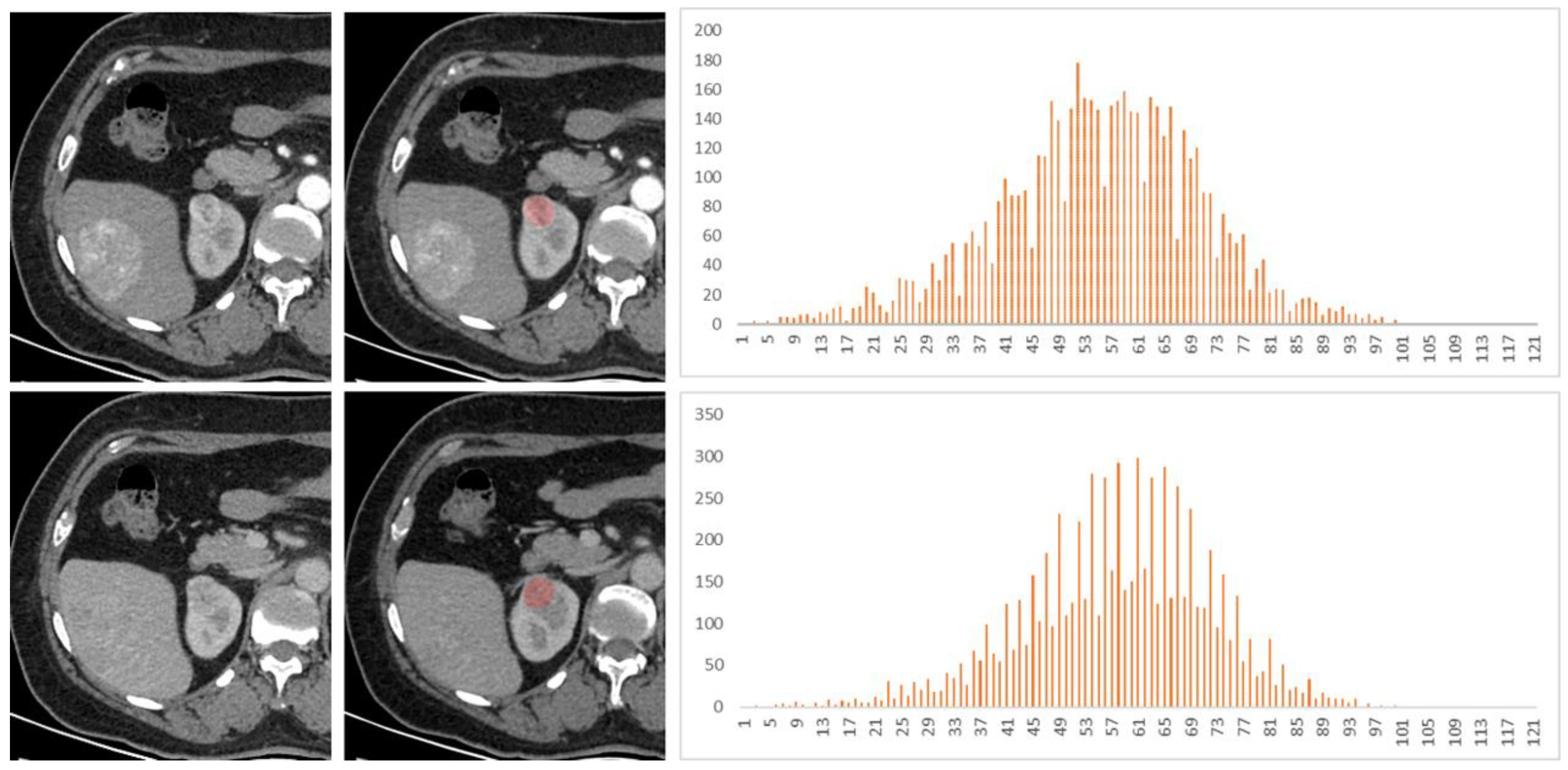
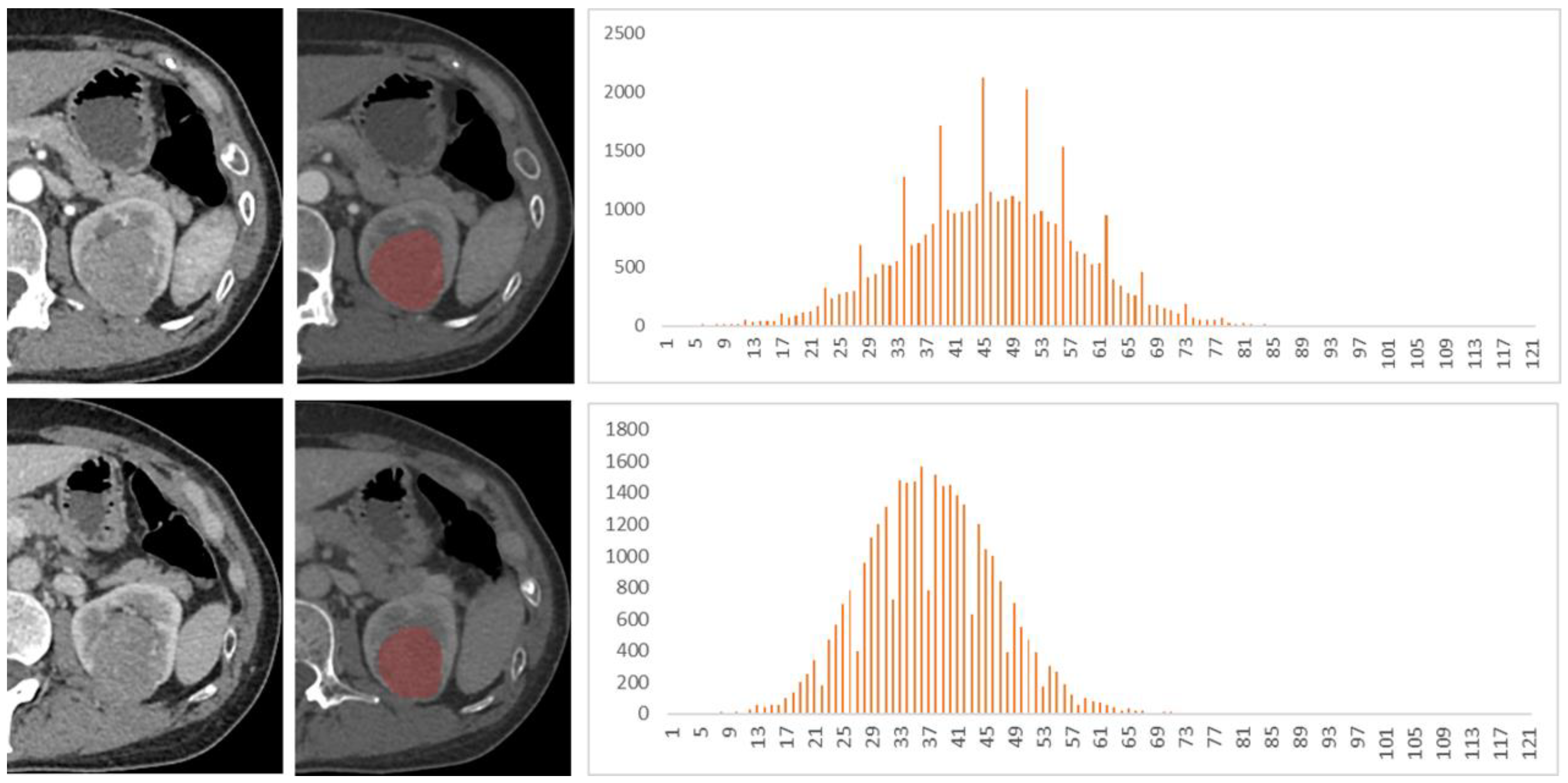
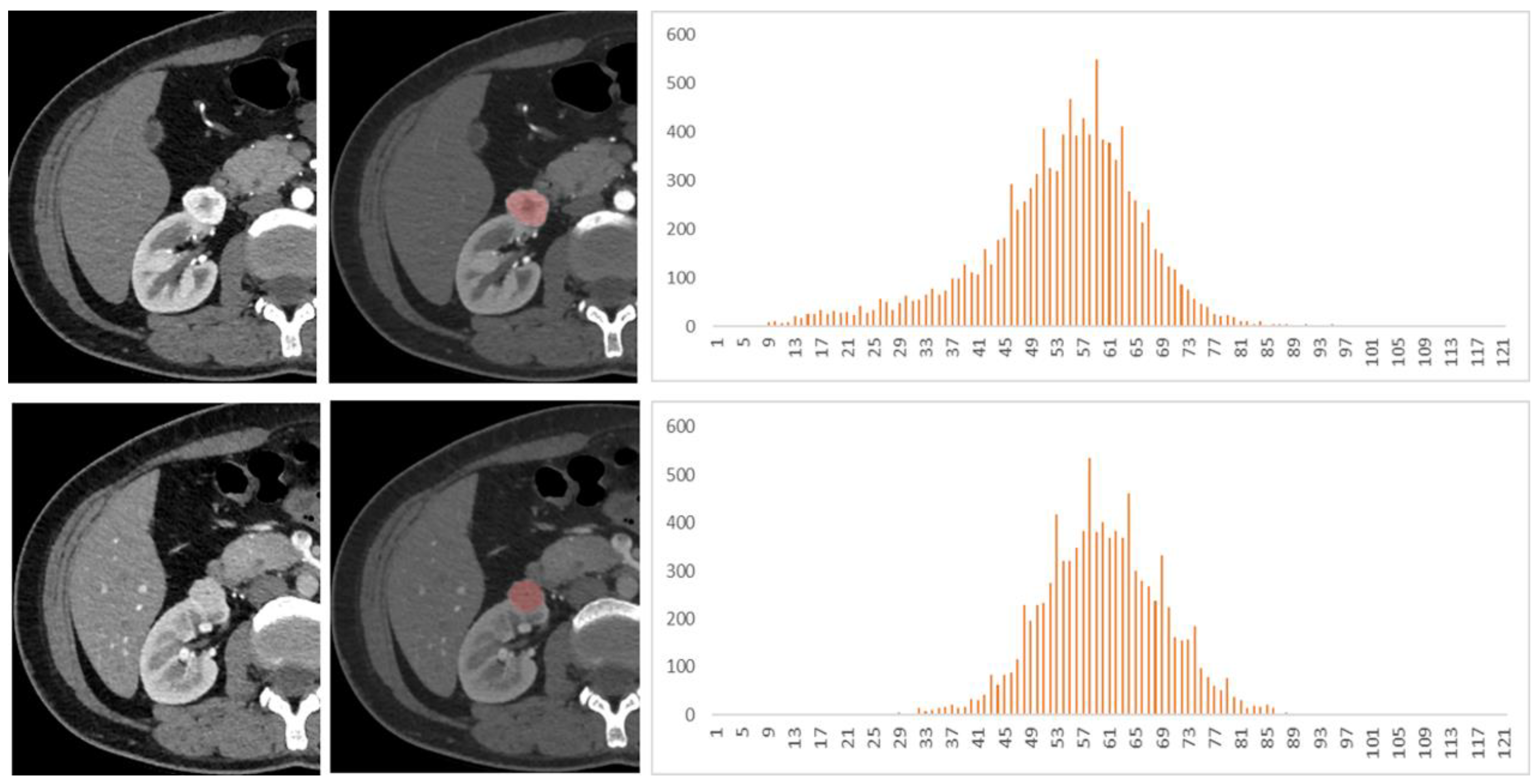
2.3. Imaging Analysis
2.4. Statistical Analysis
3. Results
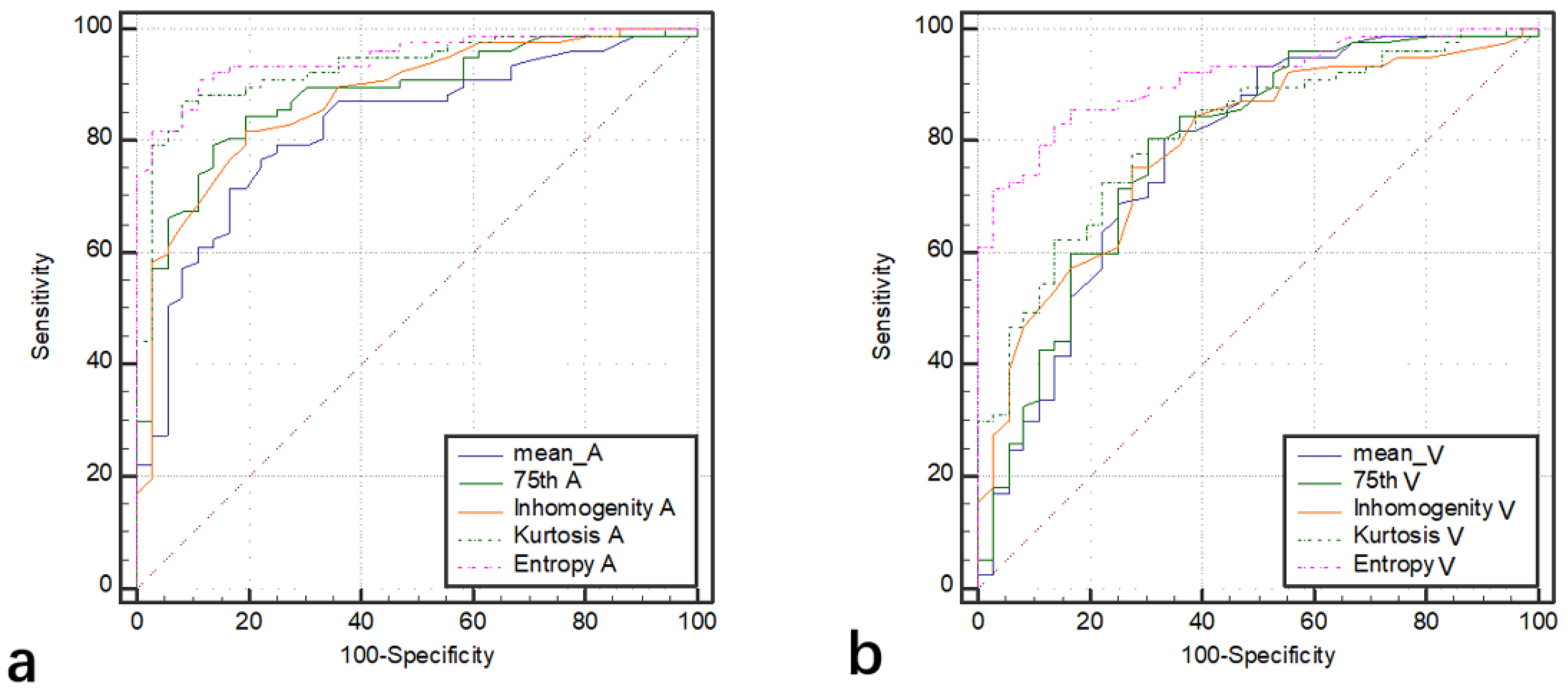
3.1. ccRCC vs. chRCC
3.2. ccRCC vs. RO
3.3. chRCC vs. RO
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Motzer, R.J.; Jonasch, E.; Agarwal, N.; Alva, A.; Baine, M.; Beckermann, K.; Carlo, M.I.; Choueiri, T.K.; Costello, B.A.; Derweesh, I.H.; et al. Kidney Cancer, Version 3.2022, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2022, 20, 71–90. [Google Scholar] [CrossRef] [PubMed]
- Shuch, B.; Amin, A.; Armstrong, A.J.; Eble, J.N.; Ficarra, V.; Lopez-Beltran, A.; Martignoni, G.; Rini, B.I.; Kutikov, A. Understanding Pathologic Variants of Renal Cell Carcinoma: Distilling Therapeutic Opportunities from Biologic Complexity. Eur. Urol. 2015, 67, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Soule, E.; Samuel, A.; Shah, S.; Cui, E.; Asare-Sawiri, M.; Sundaram, C.; Lall, C.; Sandrasegaran, K. CT texture analysis in the differentiation of major renal cell carcinoma subtypes and correlation with Fuhrman grade. Eur. Radiol. 2019, 29, 6922–6929. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.M.Y.; Shi, B.; Xue, H.D.; Ganeshan, B.; Sun, H.; Jin, Z.Y. Can quantitative CT texture analysis be used to differentiate subtypes of renal cell carcinoma? Clin. Radiol. 2019, 74, 287–294. [Google Scholar] [CrossRef]
- Minervini, A.; Di Cristofano, C.; Gacci, M.; Serni, S.; Menicagli, M.; Lanciotti, M.; Salinitri, G.; Rocca, C.D.; Lapini, A.; Nesi, G.; et al. Prognostic Role of Histological Necrosis for Nonmetastatic Clear Cell Renal Cell Carcinoma: Correlation with Pathological Features and Molecular Markers. J. Urol. 2008, 180, 1284–1289. [Google Scholar] [CrossRef]
- Sasaguri, K.; Takahashi, N. CT and MR imaging for solid renal mass characterization. Eur. J. Radiol. 2018, 99, 40–54. [Google Scholar] [CrossRef]
- Sasaguri, K.; Takahashi, N.; Gomez-Cardona, D.; Leng, S.; Schmit, G.D.; Carter, R.E.; Leibovich, B.C.; Kawashima, A. Small (<4 cm) Renal Mass: Differentiation of Oncocytoma from Renal Cell Carcinoma on Biphasic Contrast-Enhanced CT. AJR. Am. J. Roentgenol. 2015, 205, 999. [Google Scholar]
- Bektas, C.T.; Kocak, B.; Yardimci, A.H.; Turkcanoglu, M.H.; Yucetas, U.; Koca, S.B.; Erdim, C.; Kilickesmez, O. Clear Cell Renal Cell Carcinoma: Machine Learning-Based Quantitative Computed Tomography Texture Analysis for Prediction of Fuhrman Nuclear Grade. Eur. Radiol. 2019, 29, 1153–1163. [Google Scholar] [CrossRef]
- Wobker, S.E.; Williamson, S.R. Modern Pathologic Diagnosis of Renal Oncocytoma. J. Kidney Cancer Vhl 2017, 4, 1–12. [Google Scholar] [CrossRef]
- Alaghehbandan, R.; Przybycin, C.G.; Verkarre, V.; Mehra, R. Chromophobe renal cell carcinoma: Novel molecular insights and clinicopathologic updates. Asian J. Urol. 2022, 9, 1–11. [Google Scholar] [CrossRef]
- Zhong, Y.; Wang, H.; Shen, Y.; Guo, A.; Wang, J.; Kang, S.; Ma, L.; Pan, J.; Ye, H. Diffusion-weighted imaging versus contrast-enhanced MR imaging for the differentiation of renal oncocytomas and chromophobe renal cell carcinomas. Eur. Radiol. 2017, 27, 4913–4922. [Google Scholar] [CrossRef] [PubMed]
- Scrima, A.T.; Lubner, M.G.; Abel, E.J.; Havighurst, T.C.; Shapiro, D.D.; Huang, W.; Pickhardt, P.J. Texture analysis of small renal cell carcinomas at MDCT for predicting relevant histologic and protein biomarkers. Abdom. Radiol. 2019, 44, 1999–2008. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Ibarrola, R.; Hein, S.; Reis, G.; Gratzke, C.; Miernik, A. Current and future applications of machine and deep learning in urology: A review of the literature on urolithiasis, renal cell carcinoma, and bladder and prostate cancer. World J. Urol. 2019, 38, 2329–2347. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Shen, Q.; Li, Y.; Hu, Z. CT texture analysis: A potential tool for predicting the Fuhrman grade of clear-cell renal carcinoma. Cancer Imaging 2019, 19, 6. [Google Scholar] [CrossRef]
- Yu, H.; Scalera, J.; Khalid, M.; Touret, A.; Bloch, N.; Li, B.; Qureshi, M.M.; Soto, J.A.; Anderson, S.W. Texture analysis as a radiomic marker for differentiating renal tumors. Abdom. Radiol. 2017, 42, 2470–2478. [Google Scholar] [CrossRef]
- Varghese, B.A.; Chen, F.; Hwang, D.H.; Cen, S.Y.; Desai, B.; Gill, I.S.; Duddalwar, V.A. Differentiation of Predominantly Solid Enhancing Lipid-Poor Renal Cell Masses by Use of Contrast-Enhanced CT: Evaluating the Role of Texture in Tumor Subtyping. Am. J. Roentgenol. 2018, 211, W288–W296. [Google Scholar] [CrossRef]
- Raman, S.P.; Johnson, P.T.; Allaf, M.E.; Netto, G.; Fishman, E.K. Chromophobe renal cell carcinoma: Multiphase MDCT enhancement patterns and morphologic features. Am. J. Roentgenol. 2013, 201, 1268–1276. [Google Scholar] [CrossRef]
- Yang, G.; Gong, A.; Nie, P.; Yan, L.; Miao, W.; Zhao, Y.; Wu, J.; Cui, J.; Jia, Y.; Wang, Z. Contrast-Enhanced CT Texture Analysis for Distinguishing Fat-Poor Renal Angiomyolipoma from Chromophobe Renal Cell Carcinoma. Mol. Imaging 2019, 18, 1536012119883161. [Google Scholar] [CrossRef]
- Schieda, N.; Lim, R.S.; Krishna, S.; Mcinnes, M.D.F.; Flood, T.A.; Thornhill, R.E. Diagnostic Accuracy of Unenhanced CT Analysis to Differentiate Low-Grade from High-Grade Chromophobe Renal Cell Carcinoma. AJR. Am. J. Roentgenol. 2018, 210, 1079. [Google Scholar] [CrossRef]
- Hötker, A.M.; Mazaheri, Y.; Wibmer, A.; Zheng, J.; Moskowitz, C.S.; Tickoo, S.K.; Russo, P.; Hricak, H.; Akin, O. Use of DWI in the Differentiation of Renal Cortical Tumors. Am. J. Roentgenol. 2016, 206, 100. [Google Scholar] [CrossRef][Green Version]
- Lu, J.; Hu, D.; Tang, H.; Hu, X.; Shen, Y.; Li, Z.; Peng, Y.; Kamel, I. Assessment of tumor heterogeneity: Differentiation of periampullary neoplasms based on CT whole-lesion histogram analysis. Eur. J. Radiol. 2019, 115, 1–9. [Google Scholar] [CrossRef] [PubMed]
| Mean | 25th | 50th | 75th | Inhomogeneity | Skewness | Kurtosis | Entropy | ||
|---|---|---|---|---|---|---|---|---|---|
| Arterial phase | ccRCC | 127 ± 50 | 91 ± 43 | 125 ± 50 | 159 ± 54 | 0.044 ± 0.011 | −0.20 ± 0.48 | 0.70 ± 1.35 | 3.77 ± 0.21 |
| chRCC | 83 ± 24 | 61 ± 22 | 80 ± 24 | 98 ± 26 | 0.028 ± 0.007 | −0.71 ± 1.12 | 4.97 ± 6.91 | 3.25 ± 0.29 | |
| RO | 114 ± 61 | 84 ± 52 | 111 ± 61 | 135 ± 67 | 0.034 ± 0.010 | −0.30 ± 0.48 | 1.24 ± 1.50 | 3.65 ± 0.29 | |
| Venous phase | ccRCC | 140 ± 52 | 106 ± 44 | 138 ± 51 | 164 ± 56 | 0.038 ± 0.011 | −0.47 ± 0.46 | 1.40 ± 1.89 | 3.72 ± 0.26 |
| chRCC | 100 ± 46 | 77 ± 44 | 97 ± 47 | 115 ± 48 | 0.028 ± 0.007 | −0.33 ± 0.70 | 3.65 ± 2.83 | 3.22 ± 0.35 | |
| RO | 130 ± 49 | 103 ± 46 | 128 ± 49 | 148 ± 51 | 0.031 ± 0.010 | −0.70 ± 0.56 | 2.37 ± 2.53 | 3.64 ± 0.23 |
| Arterial Phase | Venous Phase | |||||
|---|---|---|---|---|---|---|
| ccRCC vs. chRCC | ccRCC vs. RO | RO vs. chRCC | ccRCC vs. chRCC | ccRCC vs. RO | RO vs. chRCC | |
| Mean | <0.001 | 0.767 | 0.080 | 0.001 | 1.000 | 0.116 |
| 25th | 0.002 | 1.000 | 0.186 | 0.006 | 1.000 | 0.113 |
| 50th | <0.001 | 0.748 | 0.070 | <0.001 | 1.000 | 0.103 |
| 75th | <0.001 | 0.202 | 0.056 | <0.001 | 0.813 | 0.106 |
| Inhomogeneity | <0.001 | 0.001 | 0.133 | <0.001 | 0.013 | 1.000 |
| Skewness | 1.000 | 1.000 | 1.000 | 1.000 | 0.363 | 0.160 |
| Kurtosis | <0.001 | 0.525 | <0.001 | <0.001 | 0.112 | 0.385 |
| Entropy | <0.001 | 0.081 | <0.001 | <0.001 | 0.634 | <0.001 |
| Arterial Phase | Venous Phase | |||||
|---|---|---|---|---|---|---|
| AUC (Sensitivity, Specificity) | Cut-Off Value | AUC (Sensitivity, Specificity) | Cut-Off Value | p | ||
| ccRCC vs. chRCC | Mean | 0.82 (71%, 83%) | 100 | 0.78 (81%, 67%) | 94 | 0.13 |
| 25th | 0.76 (65%, 78%) | 70 | 0.74 (78%, 64%) | 71 | 0.60 | |
| 50th | 0.83 (78%, 78%) | 93 | 0.78 (82%, 67%) | 91 | 0.09 | |
| 75th | 0.88 (79%, 86%) * | 119 | 0.79 (81%, 70%) * | 114 | 0.002 | |
| Inhomogeneity | 0.88 (82%, 81%) * | 0.033 | 0.79 (75%, 72%) * | 0.031 | 0.001 | |
| Kurtosis | 0.93 (87%, 92%) * | 1.31 | 0.81 (73%, 78%) * | 1.87 | 0.004 | |
| Entropy | 0.95 (91%, 89%) | 3.54 | 0.91 (83%, 86%) | 3.56 | 0.05 | |
| ccRCC vs. RO | Inhomogeneity | 0.77 (61%, 90%) | 0.039 | 0.66 (92%, 47%) | 0.025 | 0.18 |
| RO vs. chRCC | Kurtosis | 0.84 (92%, 74%) | 1.34 | - | - | - |
| Entropy | 0.85 (92%, 74%) | 3.56 | 0.84 (97%, 58%) | 3.62 | 0.92 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, X.; Li, S.; Feng, C.; Hu, D.; Li, Z.; Niu, Y. Whole-Lesion CT Texture Analysis as a Quantitative Biomarker for the Identification of Homogeneous Renal Tumors. Life 2022, 12, 2148. https://doi.org/10.3390/life12122148
Meng X, Li S, Feng C, Hu D, Li Z, Niu Y. Whole-Lesion CT Texture Analysis as a Quantitative Biomarker for the Identification of Homogeneous Renal Tumors. Life. 2022; 12(12):2148. https://doi.org/10.3390/life12122148
Chicago/Turabian StyleMeng, Xiaoyan, Shichao Li, Cui Feng, Daoyu Hu, Zhen Li, and Yonghua Niu. 2022. "Whole-Lesion CT Texture Analysis as a Quantitative Biomarker for the Identification of Homogeneous Renal Tumors" Life 12, no. 12: 2148. https://doi.org/10.3390/life12122148
APA StyleMeng, X., Li, S., Feng, C., Hu, D., Li, Z., & Niu, Y. (2022). Whole-Lesion CT Texture Analysis as a Quantitative Biomarker for the Identification of Homogeneous Renal Tumors. Life, 12(12), 2148. https://doi.org/10.3390/life12122148







