Generation of Peptides for Highly Efficient Proximity Utilizing Site-Specific Biotinylation in Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Workplan A
2.2.1. Vector Preparation
2.2.2. Construction of BAP1070 DNA Fragment
2.2.3. Subcloning of Fragment, Containing BAP1070-H2Az into pcDNA3.1(+) Vector
2.3. Workplan B
2.4. Cell Culture, Transient Transfection, and Biotin Labeling In Vivo (Generalized Protocol)
2.5. Cell Lysis and Sample Preparation
3. Results
3.1. Assembling of BAP1070 Fragment (Workplan A)
3.2. Assembling of BAP1108 Fragment (Workplan B)
4. Discussion
4.1. Site-Specific Biotinylation in the Model System BirA + BAP-X
4.2. Site-Specific Biotinylation in the Model System BirA-X + BAP-Y
- Reduced substrate specificity of biotin acceptor domain in BAP;
- Addition of His-tag to the sequence of BAP;
- Addition of two flanking arginines for generation of tryptic peptides and monitoring of biotinylation level of BAP by LC-MS/MS;
- Using humanized wild-type biotin ligase BirA;
- Coexpression in mammalian cells from 2 plasmids instead of a single bicistronic vector;
- Availability of vectors with strong (CMV) and weak (MoMuLV) promoters;
- Plasmid constructs have elements for both transient and stable expression in mammalian cells.
4.3. Design, Generation, and Properties of BAP Peptides
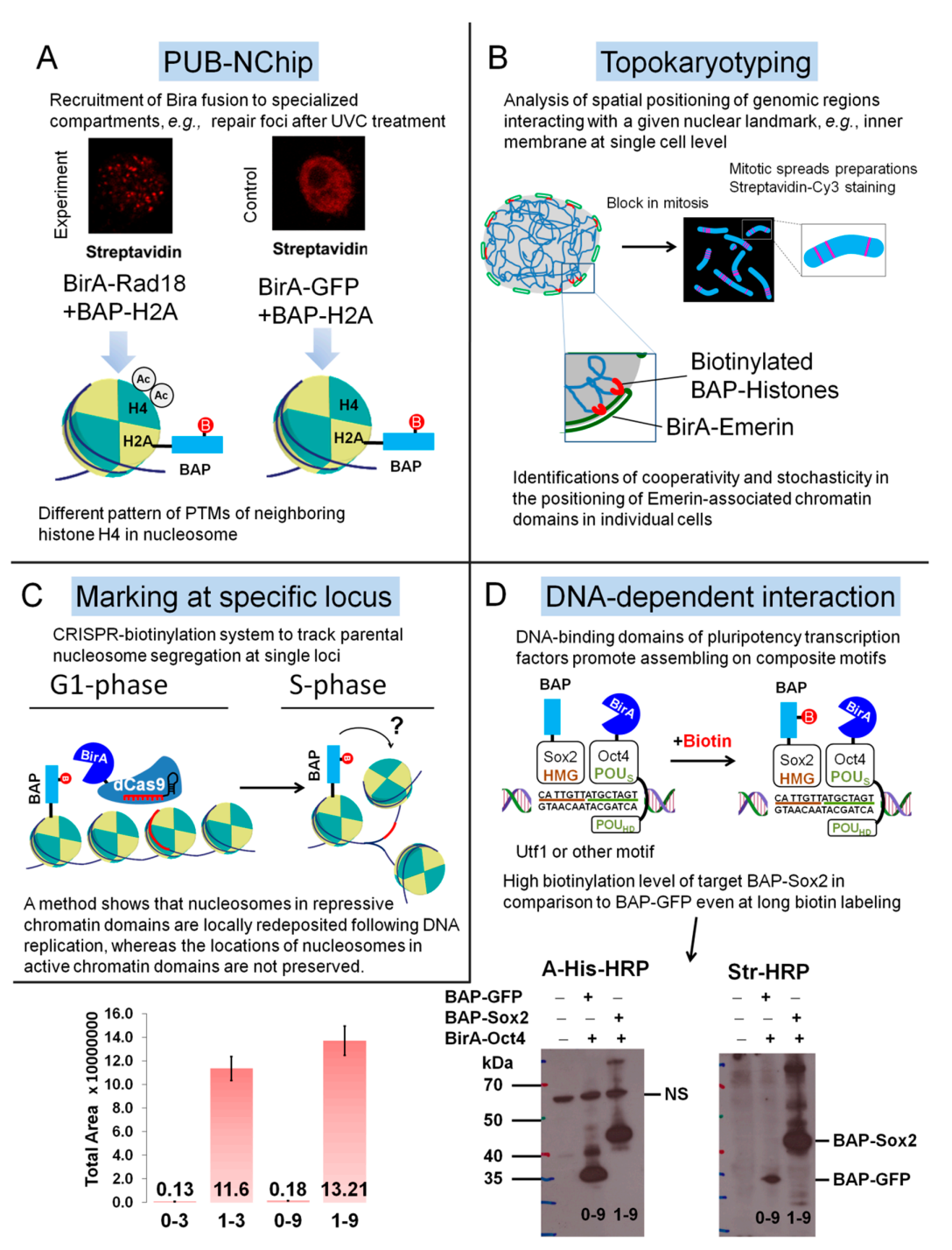
4.4. Examples of Applications of PUB Method
4.5. Differences in Two Approaches and Perspectives of BirA-Catalyzed In Vivo Labeling for Biological Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sunbul, M.; Yin, J. Site specific protein labeling by enzymatic posttranslational modification. Org. Biomol. Chem. 2009, 7, 3361–3371. [Google Scholar] [CrossRef] [PubMed]
- Ilangovan, U.; Ton-That, H.; Iwahara, J.; Schneewind, O.; Clubb, R.T. Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2001, 98, 6056–6061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wells, J.A.; Estell, D.A. Subtilisin—An Enzyme Designed to Be Engineered. Trends Biochem. Sci. 1988, 13, 291–297. [Google Scholar] [CrossRef]
- Milczek, E.M. Commercial Applications for Enzyme-Mediated Protein Conjugation: New Developments in Enzymatic Processes to Deliver Functionalized Proteins on the Commercial Scale. Chem. Rev. 2018, 118, 119–141. [Google Scholar] [CrossRef]
- Ersfeld, K.; Wehland, J.; Plessmann, U.; Dodemont, H.; Gerke, V.; Weber, K. Characterization of the Tubulin Tyrosine Ligase. J. Cell Biol. 1993, 120, 725–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, T.W.; Reed, K.E.; Cronan, J.E. Identification of the Gene Encoding Lipoate-Protein Ligase-a of Escherichia-Coli—Molecular-Cloning and Characterization of the Lpla Gene and Gene-Product. J. Biol. Chem. 1994, 269, 16091–16100. [Google Scholar] [CrossRef]
- Chen, I.; Howarth, M.; Lin, W.; Ting, A.Y. Site-Specific labeling of cell surface proteins with biophysical probes using biotin ligase. Nat. Methods 2005, 2, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Chapman-Smith, A.; Cronan, J.E. The enzymatic biotinylation of proteins: A post-translational modification of exceptional specificity. Trends Biochem. Sci. 1999, 24, 359–363. [Google Scholar] [CrossRef]
- Fairhead, M.; Howarth, M. Site-Specific biotinylation of purified proteins using BirA. Methods Mol. Biol. 2015, 1266, 171–184. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Park, K.Y.; Suazo, K.F.; Distefano, M.D. Recent progress in enzymatic protein labelling techniques and their applications. Chem. Soc. Rev. 2018, 47, 9106–9136. [Google Scholar] [CrossRef]
- Zhang, G.; Zheng, S.; Liu, H.; Chen, P.R. Illuminating biological processes through site-specific protein labeling. Chem Soc. Rev. 2015, 44, 3405–3417. [Google Scholar] [CrossRef]
- Lotze, J.; Reinhardt, U.; Seitz, O.; Beck-Sickinger, A.G. Peptide-Tags for site-specific protein labelling in vitro and in vivo. Mol. Biosyst. 2016, 12, 1731–1745. [Google Scholar] [CrossRef] [Green Version]
- Berki, T.; Bakunts, A.; Duret, D.; Fabre, L.; Ladaviere, C.; Orsi, A.; Charreyre, M.T.; Raimondi, A.; van Anken, E.; Favier, A. Advanced Fluorescent Polymer Probes for the Site-Specific Labeling of Proteins in Live Cells Using the HaloTag Technology. ACS Omega 2019, 4, 12841–12847. [Google Scholar] [CrossRef] [Green Version]
- Zubarev, R.A. The challenge of the proteome dynamic range and its implications for in-depth proteomics. Proteomics 2013, 13, 723–726. [Google Scholar] [CrossRef]
- Milo, R. What is the total number of protein molecules per cell volume? A call to rethink some published values. Bioessays 2013, 35, 1050–1055. [Google Scholar] [CrossRef] [Green Version]
- Geiger, T.; Wehner, A.; Schaab, C.; Cox, J.; Mann, M. Comparative proteomic analysis of eleven common cell lines reveals ubiquitous but varying expression of most proteins. Mol. Cell. Proteom. 2012, 11, 014050. [Google Scholar] [CrossRef] [Green Version]
- Rees, J.S.; Li, X.W.; Perrett, S.; Lilley, K.S.; Jackson, A.P. Protein Neighbors and Proximity Proteomics. Mol. Cell. Proteom. 2015, 14, 2848–2856. [Google Scholar] [CrossRef] [Green Version]
- Kulyyassov, A.; Shoaib, M.; Pichugin, A.; Kannouche, P.; Ramanculov, E.; Lipinski, M.; Ogryzko, V. PUB-MS: A Mass Spectrometry-based Method to Monitor Protein-Protein Proximity in vivo. J. Proteome Res. 2011, 10, 4416–4427. [Google Scholar] [CrossRef]
- Fernandez-Suarez, M.; Chen, T.S.; Ting, A.Y. Protein-Protein interaction detection in vitro and in cells by proximity biotinylation. J. Am. Chem. Soc. 2008, 130, 9251–9253. [Google Scholar] [CrossRef] [Green Version]
- Slavoff, S.A.; Liu, D.S.; Cohen, J.D.; Ting, A.Y. Imaging protein-protein interactions inside living cells via interaction-dependent fluorophore ligation. J. Am. Chem. Soc. 2011, 133, 19769–19776. [Google Scholar] [CrossRef] [Green Version]
- Green, M.R.; Sambrook, J. The Hanahan Method for Preparation and Transformation of Competent Escherichia coli: High-Efficiency Transformation. Cold Spring Harb. Protoc. 2018, 2018, 183–190. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Isolation of DNA Fragments from Polyacrylamide Gels by the Crush and Soak Method. Cold Spring Harb. Protoc. 2019, 2019, 143–146. [Google Scholar] [CrossRef] [PubMed]
- Green, M.R.; Sambrook, J. Screening Colonies by Polymerase Chain Reaction (PCR). Cold Spring Harb. Protoc. 2019, 2019, 516–518. [Google Scholar] [CrossRef] [PubMed]
- Mechold, U.; Gilbert, C.; Ogryzko, V. Codon optimization of the BirA enzyme gene leads to higher expression and an improved efficiency of biotinylation of target proteins in mammalian cells. J. Biotechnol. 2005, 116, 245–249. [Google Scholar] [CrossRef]
- Fang, M.; Ivanisevic, J.; Benton, H.P.; Johnson, C.H.; Patti, G.J.; Hoang, L.T.; Uritboonthai, W.; Kurczy, M.E.; Siuzdak, G. Thermal Degradation of Small Molecules: A Global Metabolomic Investigation. Anal. Chem. 2015, 87, 10935–10941. [Google Scholar] [CrossRef]
- Chapman-Smith, A.; Cronan, J.E., Jr. In vivo enzymatic protein biotinylation. Biomol. Eng. 1999, 16, 119–125. [Google Scholar] [CrossRef]
- Chen, I.; Ting, A.Y. Site-specific labeling of proteins with small molecules in live cells. Curr. Opin. Biotechnol. 2005, 16, 35–40. [Google Scholar] [CrossRef]
- de Boer, E.; Rodriguez, P.; Bonte, E.; Krijgsveld, J.; Katsantoni, E.; Heck, A.; Grosveld, F.; Strouboulis, J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. USA 2003, 100, 7480–7485. [Google Scholar] [CrossRef] [Green Version]
- Viens, A.; Mechold, U.; Lehrmann, H.; Harel-Bellan, A.; Ogryzko, V. Use of protein biotinylation in vivo for chromatin immunoprecipitation. Anal. Biochem. 2004, 325, 68–76. [Google Scholar] [CrossRef]
- Kim, J.; Cantor, A.B.; Orkin, S.H.; Wang, J. Use of in vivo biotinylation to study protein-protein and protein-DNA interactions in mouse embryonic stem cells. Nat. Protoc. 2009, 4, 506–517. [Google Scholar] [CrossRef]
- Wang, J.; Rao, S.; Chu, J.; Shen, X.; Levasseur, D.N.; Theunissen, T.W.; Orkin, S.H. A protein interaction network for pluripotency of embryonic stem cells. Nature 2006, 444, 364–368. [Google Scholar] [CrossRef]
- Tenzer, S.; Moro, A.; Kuharev, J.; Francis, A.C.; Vidalino, L.; Provenzani, A.; Macchi, P. Proteome-wide characterization of the RNA-binding protein RALY-interactome using the in vivo-biotinylation-pulldown-quant (iBioPQ) approach. J. Proteome Res. 2013, 12, 2869–2884. [Google Scholar] [CrossRef]
- Li, Y. The tandem affinity purification technology: An overview. Biotechnol. Lett. 2011, 33, 1487–1499. [Google Scholar] [CrossRef]
- Titeca, K.; Lemmens, I.; Tavernier, J.; Eyckerman, S. Discovering cellular protein-protein interactions: Technological strategies and opportunities. Mass Spectrom. Rev. 2019, 38, 79–111. [Google Scholar] [CrossRef] [Green Version]
- Schatz, P.J. Use of Peptide Libraries to Map the Substrate-Specificity of a Peptide-Modifying Enzyme—A 13 Residue Consensus Peptide Specifies Biotinylation in Escherichia-Coli. Bio/Technology 1993, 11, 1138–1143. [Google Scholar] [CrossRef]
- Beckett, D.; Kovaleva, E.; Schatz, P.J. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999, 8, 921–929. [Google Scholar] [CrossRef] [Green Version]
- Nakatani, Y.; Ogryzko, V. Immunoaffinity purification of mammalian protein complexes. Method Enzym. 2003, 370, 430–444. [Google Scholar]
- Kulyyassov, A.; Ogryzko, V. In Vivo Quantitative Estimation of DNA-Dependent Interaction of Sox2 and Oct4 Using BirA-Catalyzed Site-Specific Biotinylation. Biomolecules 2020, 10, 142. [Google Scholar] [CrossRef] [Green Version]
- Robin, P.; Fritsch, L.; Philipot, O.; Svinarchuk, F.; Ait-Si-Ali, S. Post-Translational modifications of histones H3 and H4 associated with the histone methyltransferases Suv39h1 and G9a. Genome Biol. 2007, 8, R270. [Google Scholar] [CrossRef] [Green Version]
- Shoaib, M.; Kulyyassov, A.; Robin, C.; Winczura, K.; Tarlykov, P.; Despas, E.; Kannouche, P.; Ramanculov, E.; Lipinski, M.; Ogryzko, V. PUB-NChIP-"in vivo biotinylation" approach to study chromatin in proximity to a protein of interest. Genome Res. 2013, 23, 331–340. [Google Scholar] [CrossRef] [Green Version]
- Jurisic, A.; Robin, C.; Tarlykov, P.; Siggens, L.; Schoell, B.; Jauch, A.; Ekwall, K.; Sorensen, C.S.; Lipinski, M.; Shoaib, M.; et al. Topokaryotyping demonstrates single cell variability and stress dependent variations in nuclear envelope associated domains. Nucleic Acids Res. 2018, 46, e135. [Google Scholar] [CrossRef] [PubMed]
- Escobar, T.M.; Oksuz, O.; Saldana-Meyer, R.; Descostes, N.; Bonasio, R.; Reinberg, D. Active and Repressed Chromatin Domains Exhibit Distinct Nucleosome Segregation during DNA Replication. Cell 2019, 179, 953–963. [Google Scholar] [CrossRef]
- Wuebben, E.L.; Rizzino, A. The dark side of SOX2: Cancer—A comprehensive overview. Oncotarget 2017, 8, 44917–44943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulyyassov, A.; Kalendar, R. In Silico Estimation of the Abundance and Phylogenetic Significance of the Composite Oct4-Sox2 Binding Motifs within a Wide Range of Species. Data 2020, 5, 111. [Google Scholar] [CrossRef]
- Kulyyassov, A.; Fresnais, M.; Longuespee, R. Targeted liquid chromatography-tandem mass spectrometry analysis of proteins: Basic principles, applications, and perspectives. Proteomics 2021, 21, e2100153. [Google Scholar] [CrossRef]
- Kulyyassov, A. Application of Skyline for Analysis of Protein-Protein Interactions In Vivo. Molecules 2021, 26, 7170. [Google Scholar] [CrossRef]
- Veyron-Churlet, R.; Locht, C. In Vivo Methods to Study Protein-Protein Interactions as Key Players in Mycobacterium Tuberculosis Virulence. Pathogens 2019, 8, 173. [Google Scholar] [CrossRef] [Green Version]
- Roux, K.J.; Kim, D.I.; Raida, M.; Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 2012, 196, 801–810. [Google Scholar] [CrossRef] [Green Version]
- Branon, T.C.; Bosch, J.A.; Sanchez, A.D.; Udeshi, N.D.; Svinkina, T.; Carr, S.A.; Feldman, J.L.; Perrimon, N.; Ting, A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018, 36, 880–887. [Google Scholar] [CrossRef]
- Kulyyassov, A.T.; Ramanculov, Y.M.; Ogryzko, V.V. Cloning and expression of recombinant protein of SUMO, fused with biotin acceptor peptide. Eurasian J. Appl. Biotechnol. 2018, 1, 37–41. [Google Scholar] [CrossRef]
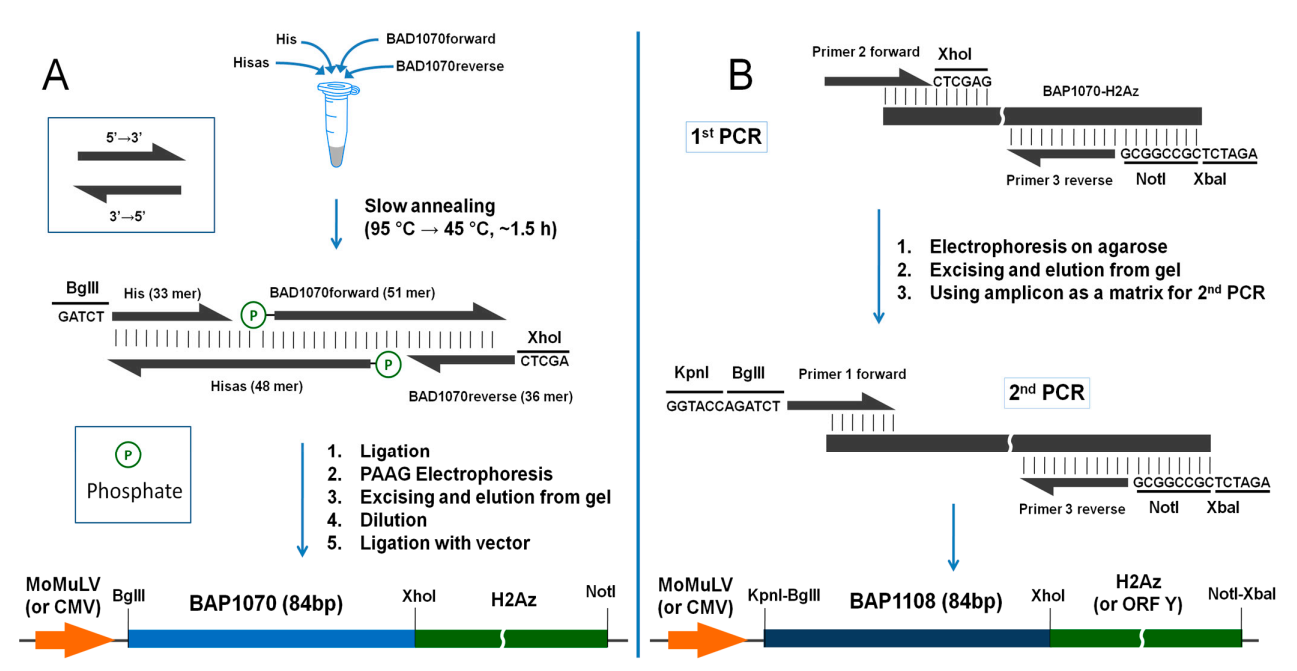
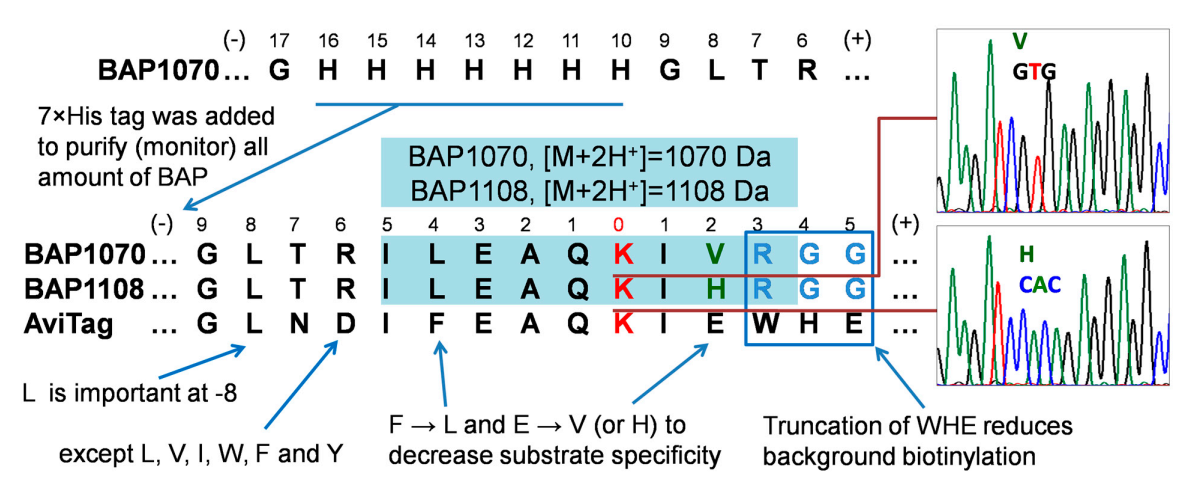
| Abbreviation | Primer Sequence (5′-3′ End) | Workplan |
|---|---|---|
| His | GATCTTGAACCATGGGACACCATCACCATCACC | A |
| Hisas | PhosATTCTTGTCAGGCCATGATGGTGATGGTGATGGTGTCCCATGGTTCAA | A |
| BAD1070 forward | PhosATCATGGCCTGACAAGAATCCTGGAAGCTCAGAAGATCGTGAGAGGAGGCC | A |
| BAD1070 reverse | TCGAGGCCTCCTCTCACGATCTTCTGAGCTTCCAGGA | A |
| Primer BADPCR | CATCATGGCCTGACAAGAATCCTG | A |
| Primer 1 NKpnIBAD | CACACACAGGTACCAGATCTTGAACCATGGGACACCATCACCATCACCATCATGGCCTGACA | B |
| Primer 2 NXhoIBAD | CATCACCATCATGGCCTGACAAGAATCCTGGAAGCTCAGAAGATCCACAGAGGAGGCCTCGAG | B |
| Primer 3CH2AZNotXbaIAS | TGTGTGTGTCTAGAGCGGCCGCTAGACAGTCTTCTGTTGTC | B |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kulyyassov, A.; Ramankulov, Y.; Ogryzko, V. Generation of Peptides for Highly Efficient Proximity Utilizing Site-Specific Biotinylation in Cells. Life 2022, 12, 300. https://doi.org/10.3390/life12020300
Kulyyassov A, Ramankulov Y, Ogryzko V. Generation of Peptides for Highly Efficient Proximity Utilizing Site-Specific Biotinylation in Cells. Life. 2022; 12(2):300. https://doi.org/10.3390/life12020300
Chicago/Turabian StyleKulyyassov, Arman, Yerlan Ramankulov, and Vasily Ogryzko. 2022. "Generation of Peptides for Highly Efficient Proximity Utilizing Site-Specific Biotinylation in Cells" Life 12, no. 2: 300. https://doi.org/10.3390/life12020300
APA StyleKulyyassov, A., Ramankulov, Y., & Ogryzko, V. (2022). Generation of Peptides for Highly Efficient Proximity Utilizing Site-Specific Biotinylation in Cells. Life, 12(2), 300. https://doi.org/10.3390/life12020300







