Characterization of Serum Metabolome and Proteome Profiles Identifies SNX5 Specific for Pregnancy Failure in Holstein Heifers
Abstract
:1. Introduction
2. Results
2.1. Metabolic Analysis of Bovine Peripheral Blood during the Peri-Implantation Period
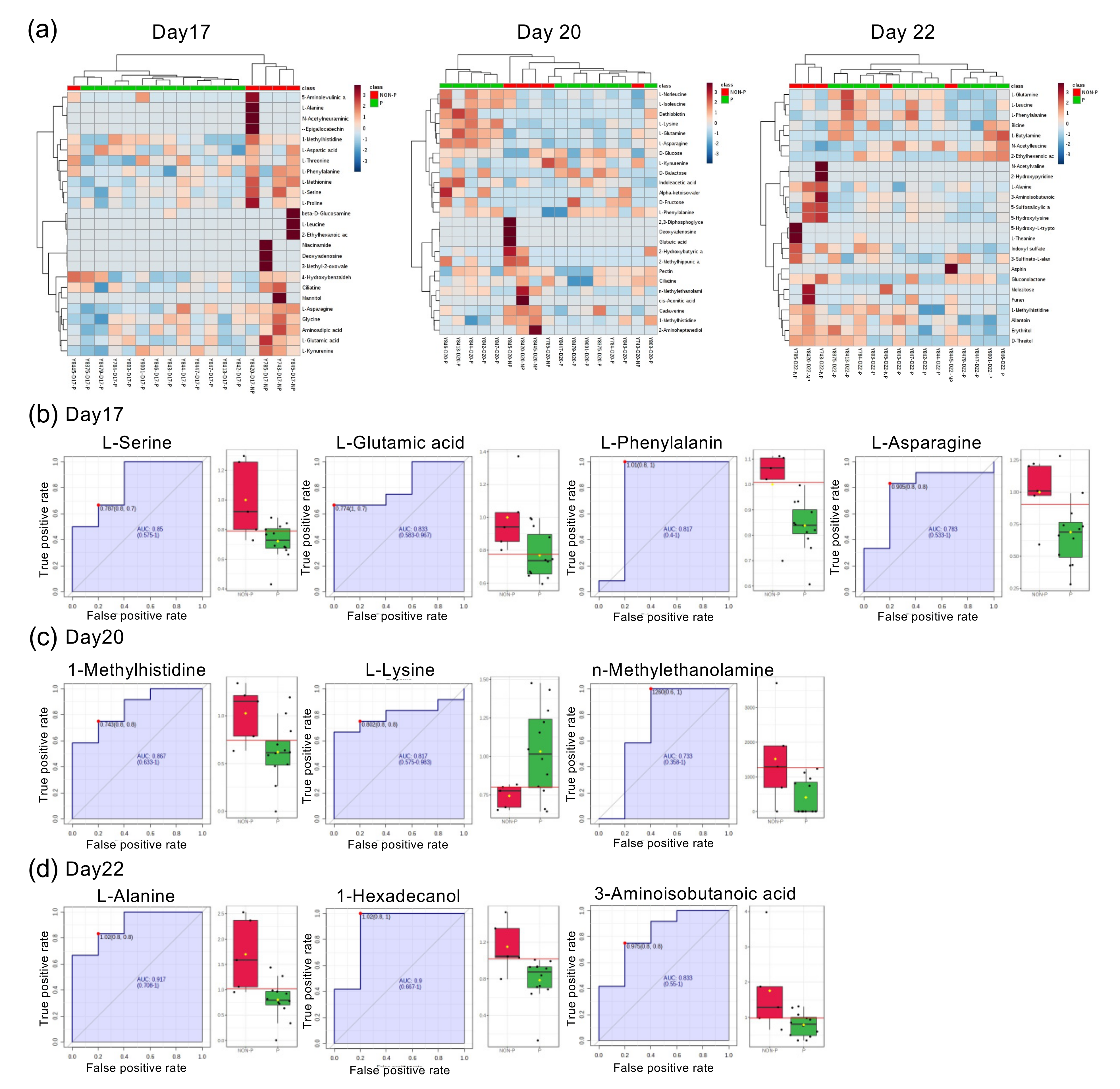
2.2. Comparison of Serum Composition between Pregnant, NP Heifers, and Those with the Estrous Cycle
2.3. Global Proteome Analysis of Serum from Day 20 Pregnant and NP Heifers
2.4. Identification of Specific Proteins in Day-22 Blood Serum Detecting NP Heifers
3. Discussion
4. Materials and Methods
4.1. Collection of Bovine Blood Samples
4.2. Metabolome Analysis
4.3. iTRAQ Analysis
4.4. Western Blot Analysis
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roberts, R.M.; Cross, J.C.; Leaman, D.W. Interferons as hormones of pregnancy. Endocr. Rev. 1992, 13, 432–452. [Google Scholar] [CrossRef] [PubMed]
- Spencer, T.E.; Becker, W.C.; George, P.; Mirando, M.A.; Ogle, T.F.; Bazer, F.W. Ovine interferon-tau regulates expression of endometrial receptors for estrogen and oxytocin but not progesterone. Biol. Reprod. 1995, 53, 732–745. [Google Scholar] [CrossRef] [PubMed]
- Spencer, T.E.; Bazer, F.W. Ovine interferon tau suppresses transcription of the estrogen receptor and oxytocin receptor genes in the ovine endometrium. Endocrinology 1996, 137, 1144–1147. [Google Scholar] [CrossRef] [PubMed]
- Forde, N.; Lonergan, P. Interferon-tau and fertility in ruminants. Reproduction 2017, 154, F33–F43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sponchiado, M.; Gomes, N.S.; Fontes, P.K.; Martins, T.; Del Collado, M.; Pastore, A.A.; Pugliesi, G.; Nogueira, M.F.G.; Binelli, M. Pre-hatching embryo-dependent and -independent programming of endometrial function in cattle. PLoS ONE 2017, 12, e0175954. [Google Scholar] [CrossRef] [PubMed]
- Bridi, A.; Bertolin, K.; Rissi, V.B.; Mujica, L.K.S.; Glanzner, W.G.; de Macedo, M.P.; Comim, F.V.; Gonçalves, P.B.D.; Antoniazzi, A.Q. Parthenogenetic bovine embryos secrete type I interferon capable of stimulating ISG15 in luteal cell culture. Anim. Reprod. 2018, 15, 1268–1277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruhmann, B.; Giller, K.; Hankele, A.K.; Ulbrich, S.E.; Schmicke, M. Interferon-τ induced gene expression in bovine hepatocytes during early pregnancy. Theriogenology 2017, 104, 198–204. [Google Scholar] [CrossRef]
- Kizaki, K.; Shichijo-Kizaki, A.; Furusawa, T.; Takahashi, T.; Hosoe, M.; Hashizume, K. Differential neutrophil gene expression in early bovine pregnancy. Reprod. Biol. Endocrinol. 2013, 11, 6. [Google Scholar] [CrossRef] [Green Version]
- Soumya, N.P.; Das, D.N.; Jeyakumar, S.; Mondal, S.; Mor, A.; Mundhe, U.T. Differential expression of ISG 15 mRNA in peripheral blood mononuclear cells of nulliparous and multiparous pregnant versus non-pregnant Bos indicus cattle. Reprod. Domest. Anim. Zuchthyg. 2017, 52, 97–106. [Google Scholar] [CrossRef]
- Pugliesi, G.; Miagawa, B.T.; Paiva, Y.N.; França, M.R.; Silva, L.A.; Binelli, M. Conceptus-induced changes in the gene expression of blood immune cells and the ultrasound-accessed luteal function in beef cattle: How early can we detect pregnancy? Biol. Reprod. 2014, 91, 95. [Google Scholar] [CrossRef]
- Hansen, T.R.; Sinedino, L.D.P.; Spencer, T.E. Paracrine and endocrine actions of interferon tau (IFNT). Reproduction 2017, 154, F45–F59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha, C.C.; da Silva Andrade, S.C.; de Melo, G.D.; Motta, I.G.; Coutinho, L.L.; Gonella-Diaza, A.M.; Binelli, M.; Pugliesi, G. Early pregnancy-induced transcripts in peripheral blood immune cells in Bos indicus heifers. Sci. Rep. 2020, 10, 13733. [Google Scholar] [CrossRef] [PubMed]
- Bazer, F.W.; Spencer, T.E.; Johnson, G.A.; Burghardt, R.C.; Wu, G. Comparative aspects of implantation. Reproduction 2009, 138, 195–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trousdale, C.; Kim, K. Retromer: Structure, function, and roles in mammalian disease. Eur. J. Cell Biol. 2015, 94, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Bonifacino, J.S.; Hurley, J.H. Retromer. Curr. Opin. Cell Biol. 2008, 20, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Itai, N.; Shimazu, T.; Kimura, T.; Ibe, I.; Yamashita, R.; Kaburagi, Y.; Dohi, T.; Tonozuka, T.; Takao, T.; Nishikawa, A. The phosphorylation of sorting nexin 5 at serine 226 regulates retrograde transport and macropinocytosis. PLoS ONE 2018, 13, e0207205. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Cao, G.; Chen, D.; Liu, J.; Yu, B.; Liu, M.; Li, Y.X.; Cao, B.; Sadovsky, Y.; Wang, Y.L. Placental trophoblast syncytialization potentiates macropinocytosis via mTOR signaling to adapt to reduced amino acid supply. Proc. Natl. Acad. Sci. USA 2021, 118, e2017092118. [Google Scholar] [CrossRef]
- Palm, W.; Park, Y.; Wright, K.; Pavlova, N.N.; Tuveson, D.A.; Thompson, C.B. The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell 2015, 162, 259–270. [Google Scholar] [CrossRef] [Green Version]
- Dong, X.; Yang, Y.; Zou, Z.; Zhao, Y.; Ci, B.; Zhong, L.; Bhave, M.; Wang, L.; Kuo, Y.C.; Zang, X.; et al. Sorting nexin 5 mediates virus-induced autophagy and immunity. Nature 2021, 589, 456–461. [Google Scholar] [CrossRef]
- Forde, N.; Simintiras, C.A.; Sturmey, R.; Mamo, S.; Kelly, A.K.; Spencer, T.E.; Bazer, F.W.; Lonergan, P. Amino acids in the uterine luminal fluid reflects the temporal changes in transporter expression in the endometrium and conceptus during early pregnancy in cattle. PLoS ONE 2014, 9, e100010. [Google Scholar] [CrossRef]
- Gilbreath, K.R.; Bazer, F.W.; Satterfield, M.C.; Wu, G. Amino Acid Nutrition and Reproductive Performance in Ruminants. Adv Exp. Med. Biol. 2021, 1285, 43–61. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Mitchell, M.D.; Walker, C.G.; Roche, J.R.; Verkerk, G.A. Amino acid concentrations in uterine fluid during early pregnancy differ in fertile and subfertile dairy cow strains. J. Dairy Sci. 2014, 97, 1364–1376. [Google Scholar] [CrossRef] [Green Version]
- Pohler, K.G.; Reese, S.T.; Franco, G.A.; Oliveira, R.V.; Paiva, R.; Fernandez, L.; de Melo, G.; Vasconcelos, J.L.M.; Cooke, R.; Poole, R.K. New approaches to diagnose and target reproductive failure in cattle. Anim. Reprod. 2020, 17, e20200057. [Google Scholar] [CrossRef] [PubMed]
- Ott, T.L. Immunological detection of pregnancy: Evidence for systemic immune modulation during early pregnancy in ruminants. Theriogenology 2020, 150, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Ott, T.L. Symposium review: Immunological detection of the bovine conceptus during early pregnancy. J. Dairy Sci. 2019, 102, 3766–3777. [Google Scholar] [CrossRef] [Green Version]
- Kunii, H.; Kubo, T.; Asaoka, N.; Balboula, A.Z.; Hamaguchi, Y.; Shimasaki, T.; Bai, H.; Kawahara, M.; Kobayashi, H.; Ogawa, H.; et al. Loop-mediated isothermal amplification (LAMP) and machine learning application for early pregnancy detection using bovine vaginal mucosal membrane. Biochem. Biophys. Res. Commun. 2021, 569, 179–186. [Google Scholar] [CrossRef]
- Kunii, H.; Koyama, K.; Ito, T.; Suzuki, T.; Balboula, A.Z.; Shirozu, T.; Bai, H.; Nagano, M.; Kawahara, M.; Takahashi, M. Hot topic: Pregnancy-induced expression of interferon-stimulated genes in the cervical and vaginal mucosal membranes. J. Dairy Sci. 2018, 101, 8396–8400. [Google Scholar] [CrossRef]
- Green, J.C.; Okamura, C.S.; Poock, S.E.; Lucy, M.C. Measurement of interferon-tau (IFN-tau) stimulated gene expression in blood leukocytes for pregnancy diagnosis within 18-20d after insemination in dairy cattle. Anim. Reprod. Sci. 2010, 121, 24–33. [Google Scholar] [CrossRef]
- Green, J.A.; Parks, T.E.; Avalle, M.P.; Telugu, B.P.; McLain, A.L.; Peterson, A.J.; McMillan, W.; Mathialagan, N.; Hook, R.R.; Xie, S.; et al. The establishment of an ELISA for the detection of pregnancy-associated glycoproteins (PAGs) in the serum of pregnant cows and heifers. Theriogenology 2005, 63, 1481–1503. [Google Scholar] [CrossRef]
- Wooding, F.; Roberts, R.; Green, J. Light and electron microscope immunocytochemical studies of the distribution of pregnancy associated glycoproteins (PAGs) throughout pregnancy in the cow: Possible functional implications. Placenta 2005, 26, 807–827. [Google Scholar] [CrossRef]
- Wallace, R.M.; Pohler, K.G.; Smith, M.F.; Green, J.A. Placental PAGs: Gene origins, expression patterns, and use as markers of pregnancy. Reproduction 2015, 149, R115–R126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Green, J.A.; Xie, S.; Roberts, R.M. Pepsin-related molecules secreted by trophoblast. Rev. Reprod. 1998, 3, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Ideta, A.; Urakawa, M.; Aoyagi, Y.; Saeki, K. Early development in utero of bovine nuclear transfer embryos using early G1 and G0 phase cells. Cloning Stem Cells 2007, 9, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Pu, S.; Usuda, K.; Nagaoka, K.; Watanabe, G. Heat challenge influences serum metabolites concentrations and liver lipid metabolism in Japanese quail (Coturnix japonica). J. Vet. Med. Sci. 2019, 81, 77–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kusama, K.I.; Kazuhiko, I. Supplementary Tables.xlsx. Figshare. Dataset. 2021. Available online: https://doi.org/10.6084/m9.figshare.16778674.v1 (accessed on 9 October 2021).
- Xia, J.; Wishart, D.S. Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat. Protoc. 2011, 6, 743–760. [Google Scholar] [CrossRef]
- Kusama, K.; Bai, R.; Ideta, A.; Aoyagi, Y.; Okuda, K.; Imakawa, K. Regulation of epithelial to mesenchymal transition in bovine conceptuses through the interaction between follistatin and activin A. Mol. Cell. Endocrinol. 2016, 434, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Kusama, K.; Bai, R.; Imakawa, K. Regulation of human trophoblast cell syncytialization by transcription factors STAT5B and NR4A3. J. Cell. Biochem. 2018, 119, 4918–4927. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kusama, K.; Bai, R.; Matsuno, Y.; Ideta, A.; Sakurai, T.; Nagaoka, K.; Hori, M.; Imakawa, K. Characterization of Serum Metabolome and Proteome Profiles Identifies SNX5 Specific for Pregnancy Failure in Holstein Heifers. Life 2022, 12, 309. https://doi.org/10.3390/life12020309
Kusama K, Bai R, Matsuno Y, Ideta A, Sakurai T, Nagaoka K, Hori M, Imakawa K. Characterization of Serum Metabolome and Proteome Profiles Identifies SNX5 Specific for Pregnancy Failure in Holstein Heifers. Life. 2022; 12(2):309. https://doi.org/10.3390/life12020309
Chicago/Turabian StyleKusama, Kazuya, Rulan Bai, Yuta Matsuno, Atsushi Ideta, Toshihiro Sakurai, Kentaro Nagaoka, Masatoshi Hori, and Kazuhiko Imakawa. 2022. "Characterization of Serum Metabolome and Proteome Profiles Identifies SNX5 Specific for Pregnancy Failure in Holstein Heifers" Life 12, no. 2: 309. https://doi.org/10.3390/life12020309
APA StyleKusama, K., Bai, R., Matsuno, Y., Ideta, A., Sakurai, T., Nagaoka, K., Hori, M., & Imakawa, K. (2022). Characterization of Serum Metabolome and Proteome Profiles Identifies SNX5 Specific for Pregnancy Failure in Holstein Heifers. Life, 12(2), 309. https://doi.org/10.3390/life12020309





