Staphylococcus epidermidis Controls Opportunistic Pathogens in the Nose, Could It Help to Regulate SARS-CoV-2 (COVID-19) Infection?
Abstract
:1. Introduction
2. Overview of the Nasal Cavity
3. Microbiota of the Nasal Cavity
4. Generalities and Genetic Characteristics of S. epidermidis
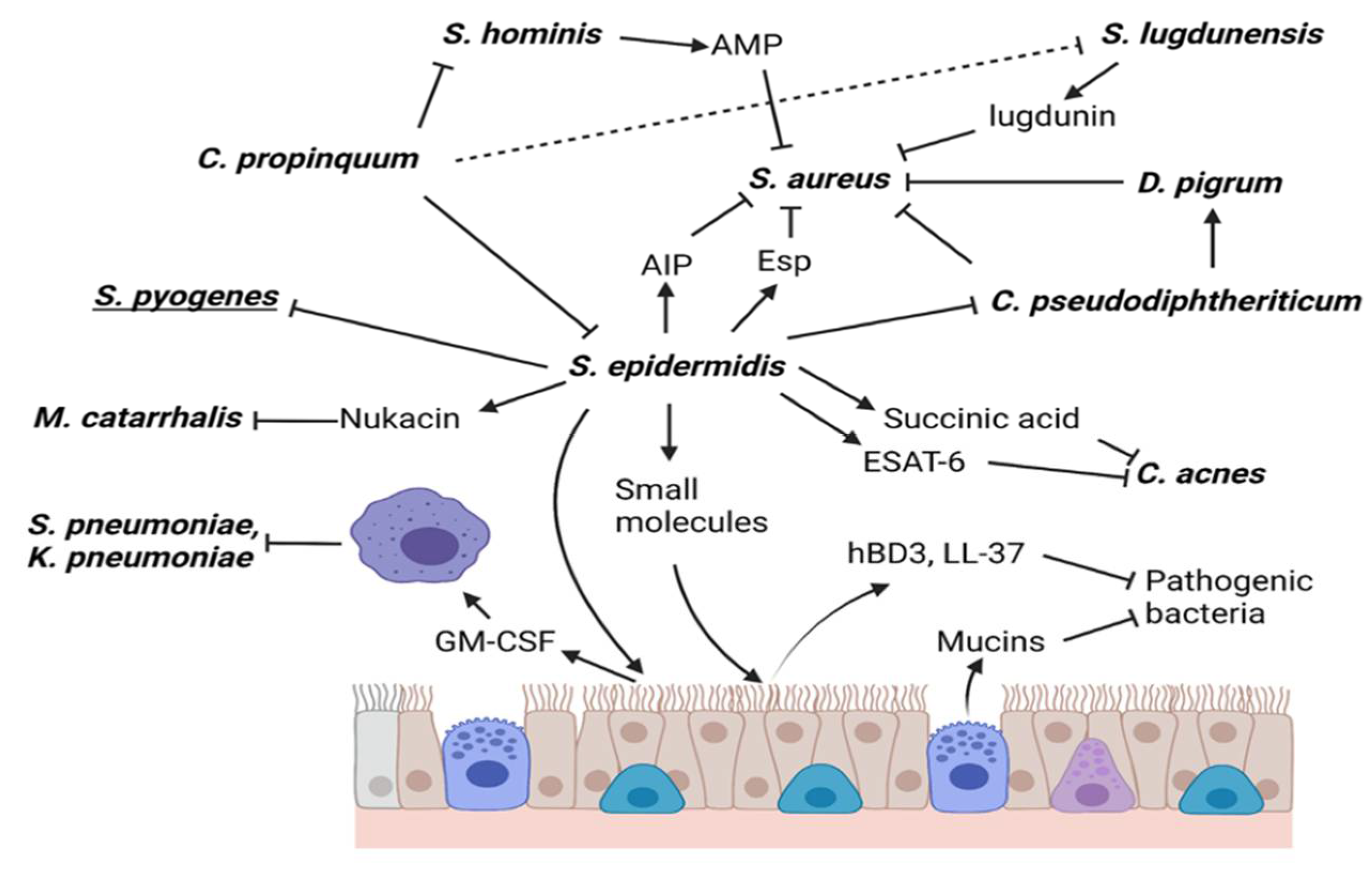
5. S. epidermidis Directly or Indirectly Kills Respiratory Bacteria
5.1. Staphylococcus aureus
5.2. Corynebacterium pseudodiphtheriticum
5.3. Cutibacterium
5.4. Moraxella catarrhalis
5.5. Streptococcus pyogenes
6. Other Mechanisms of Action of S. epidermidis
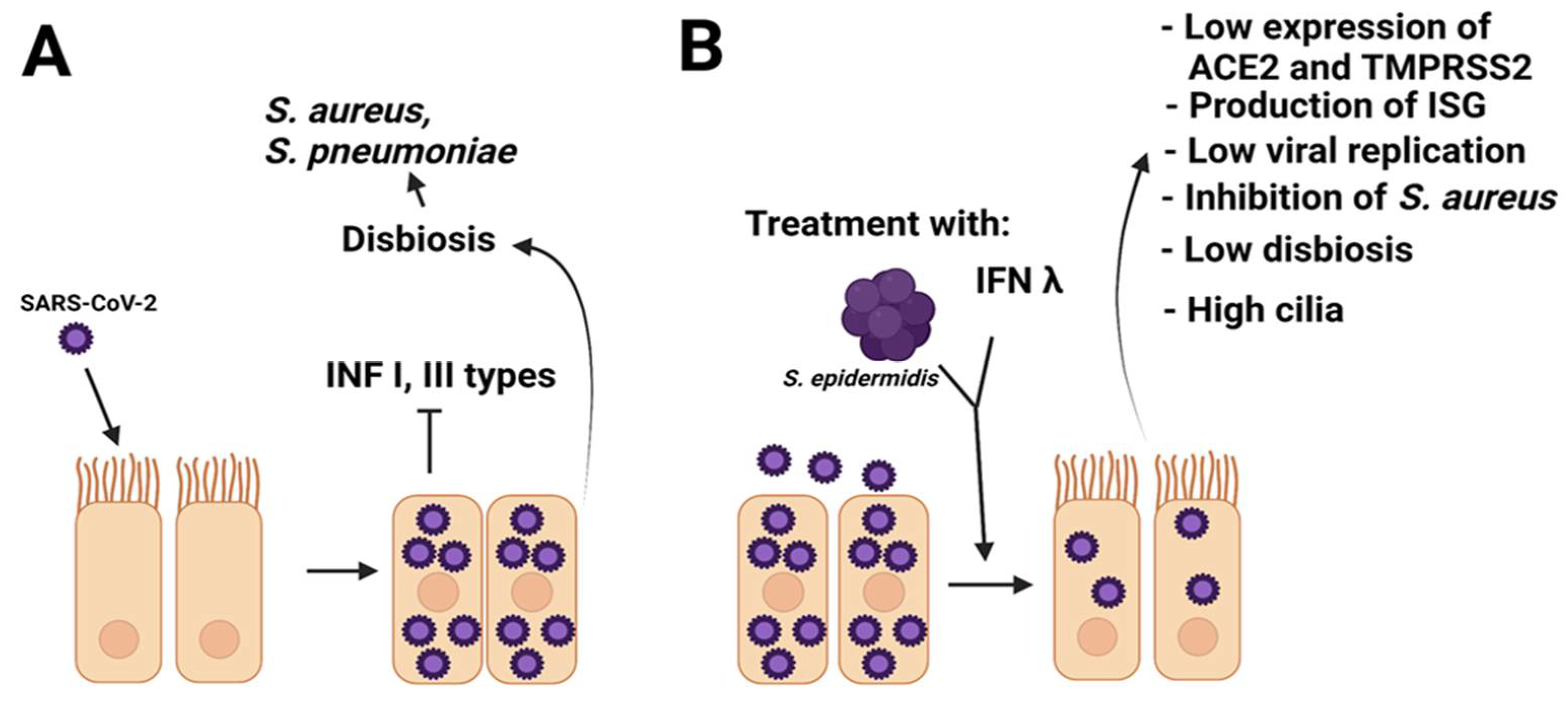
7. S. epidermidis as a Regulator of Respiratory Viral Infections
8. Possible Involvement of S. epidermidis in COVID-19 Disease
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shahani, L.; Ariza-Heredia, E.J.; Chemaly, R.F. Antiviral therapy for respiratory viral infections in immunocompromised patients. Expert Rev. Anti Infect. Ther. 2017, 15, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Tregoning, J.S.; Schwarze, J. Respiratory viral infections in infants: Causes, clinical symptoms, virology, and immunology. Clin. Microbiol. Rev. 2010, 23, 74–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriyama, M.; Hugentobler, W.J.; Iwasaki, A. Seasonality of Respiratory Viral Infections. Annu. Rev. Virol. 2020, 7, 83–101. [Google Scholar] [CrossRef] [PubMed]
- Leung, N.H.L. Transmissibility and transmission of respiratory viruses. Nat. Rev. Microbiol. 2021, 19, 528–545. [Google Scholar] [CrossRef]
- Martini, M.; Gazzaniga, V.; Bragazzi, N.L.; Barberis, I. The Spanish Influenza Pandemic: A lesson from history 100 years after 1918. J. Prev. Med. Hyg. 2019, 29, E64–E67. [Google Scholar] [CrossRef]
- Viboud, C.; Grais, R.F.; Lafont, B.A.; Miller, M.A.; Simonsen, L.; Multinational Influenza Seasonal Mortality Study Group. Multinational impact of the 1968 Hong Kong influenza pandemic: Evidence for a smoldering pandemic. J. Infect. Dis. 2005, 15, 233–248. [Google Scholar] [CrossRef]
- Peiris, J.S.; Yuen, K.Y.; Osterhaus, A.D.; Stöhr, K. The severe acute respiratory syndrome. N. Engl. J. Med. 2003, 18, 2431–2441. [Google Scholar] [CrossRef] [Green Version]
- Zumla, A.; Hui, D.S.; Perlman, S. Middle East respiratory syndrome. Lancet 2015, 5, 995–1007. [Google Scholar] [CrossRef] [Green Version]
- Fraser, C.; Donnelly, C.A.; Cauchemez, S.; Hanage, W.P.; Van Kerkhove, M.D.; Hollingsworth, T.D.; Griffin, J.; Baggaley, R.F.; Jenkins, H.E.; Lyons, E.J.; et al. Pandemic potential of a strain of influenza A (H1N1): Early findings. Science 2009, 19, 1557–1561. [Google Scholar] [CrossRef] [Green Version]
- Din, A.U.; Mazhar, M.; Waseem, M.; Ahmad, W.; Bibi, A.; Hassan, A.; Ali, N.; Gang, W.; Qian, G.; Ullah, R.; et al. SARS-CoV-2 microbiome dysbiosis linked disorders and possible probiotics role. Biomed. Pharmacother. 2021, 133, 110947. [Google Scholar] [CrossRef]
- Gralinski, L.E.; Baric, R.S. Molecular pathology of emerging coronavirus infections. J. Pathol. 2015, 235, 185–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, C.; Yao, X.; Zhao, Y.; Wu, J.; Huang, P.; Pan, C.; Liu, S.; Pan, C. Comparative review of respiratory diseases caused by coronaviruses and influenza A viruses during epidemic season. Microbes Infect. 2020, 22, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Heinonen, S.; Rodriguez-Fernandez, R.; Diaz, A.; Rodriguez-Pastor, S.O.; Ramilo, O.; Mejias., A. Infant Immune Response to Respiratory Viral Infections. Immunol. Allergy Clin. North Am. 2019, 39, 361–376. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Pérez, O.; Merino, E.; Leon-Ramirez, J.M.; Andres, M.; Ramos, J.M.; Arenas-Jiménez, J.; Asensio, S.; Sanchez, R.; Ruiz-Torregrosa, P.; Galan, I.; et al. Post-acute COVID-19 syndrome. Incidence and risk factors: A Mediterranean cohort study. J. Infect. 2021, 82, 378–383. [Google Scholar] [CrossRef] [PubMed]
- Abdelrahman, Z.; Li, M.; Wang, X. Comparative Review of SARS-CoV-2, SARS-CoV, MERS-CoV, and Influenza A Respiratory Viruses. Front. Immunol. 2020, 11, 552909. [Google Scholar] [CrossRef]
- Lee, K.H.; Gordon, A.; Shedden, K.; Kuan, G.; Ng, S.; Balmaseda, A.; Foxman, B. The respiratory microbiome and susceptibility to influenza virus infection. PLoS ONE 2019, 9, e0207898. [Google Scholar] [CrossRef] [Green Version]
- Ogunrinola, G.A.; Oyewale, J.O.; Oshamika, O.O.; Olasehinde, G.I. The Human Microbiome and Its Impacts on Health. Int. J. Microbiol. 2020, 12, 8045646. [Google Scholar] [CrossRef]
- Man, W.H.; de Steenhuijsen Piters, W.A.; Bogaert, D. The microbiota of the respiratory tract: Gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef]
- Kumpitsch, C.; Koskinen, K.; Schöpf, V.; Moissl-Eichinger, C. The microbiome of the upper respiratory tract in health and disease. BMC Biol. 2019, 7, 87. [Google Scholar] [CrossRef] [Green Version]
- Clark, S.E. Commensal bacteria in the upper respiratory tract regulate susceptibility to infection. Curr. Opin. Immunol. 2020, 66, 42–49. [Google Scholar] [CrossRef]
- Tsang, T.K.; Lee, K.H.; Foxman, B.; Balmaseda, A.; Gresh, L.; Sanchez, N.; Ojeda, S.; Lopez, R.; Yang, Y.; Kuan, G.; et al. Association Between the Respiratory Microbiome and Susceptibility to Influenza Virus Infection. Clin. Infect. Dis. 2020, 22, 1195–1203. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Liu, Q.; Meng, H.; Lv, H.; Liu, Y.; Liu, J.; Wang, H.; He, L.; Qin, J.; Wang, Y.; et al. Staphylococcus epidermidis Contributes to Healthy Maturation of the Nasal Microbiome by Stimulating Antimicrobial Peptide Production. Cell Host Microbe 2020, 8, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Raita, Y.; Toivonen, L.; Schuez-Havupalo, L.; Karppinen, S.; Waris, M.; Hoffman, K.L.; Camargo, C.A., Jr.; Peltola, V.; Hasegawa, K. Maturation of nasal microbiota and antibiotic exposures during early childhood: A population-based cohort study. Clin. Microbiol. Infect. 2021, 27, e1–e283. [Google Scholar] [CrossRef]
- Chu, D.M.; Ma, J.; Prince, A.L.; Antony, K.M.; Seferovic, M.D.; Aagaard, K.M. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 2017, 23, 314–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Durack, J.; Lynch, S.V.; Nariya, S.; Bhakta, N.R.; Beigelman, A.; Castro, M.; Dyer, A.M.; Israel, E.; Kraft, M.; Martin, R.J.; et al. Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J. Allergy Clin. Immunol. 2017, 140, 63–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sundararaman, A.; Ray, M.; Ravindra, P.V.; Halami, P.M. Role of probiotics to combat viral infections with emphasis on COVID-19. Appl. Microbiol. Biotechnol. 2020, 104, 8089–8104. [Google Scholar] [CrossRef] [PubMed]
- Al Kassaa, I.; Hober, D.; Hamze, M.; Chihib, N.E.; Drider, D. Antiviral potential of lactic acid bacteria and their bacteriocins. Probiotics Antimicrob. Proteins 2014, 6, 177–185. [Google Scholar] [CrossRef]
- Suez, J.; Zmora, N.; Segal, E.; Elinav, E. The pros, cons, and many unknowns of probiotics. Nat. Med. 2019, 25, 716–729. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Deming, C.; Conlan, S.; NISC Comparative Sequencing Program; Kong, H.H.; Segre, J.A. Biogeography and individuality shape function in the human skin metagenome. Nature 2014, 514, 59–64. [Google Scholar] [CrossRef] [Green Version]
- Ueta, M.; Iida, T.; Sakamoto, M.; Sotozono, C.; Takahashi, J.; Kojima, K.; Okada, K.; Chen, C.; Kinoshita, S.; Honda, T. Polyclonality of Staphylococcus epidermidis residing on the healthy ocular surface. J. Med. Microbiol. 2007, 56, 77–82. [Google Scholar] [CrossRef] [Green Version]
- Su, F.; Tian, R.; Yang, Y.; Li, H.; Sun, G.; Li, Y.; Han, B.; Xu, X.; Chen, X.; Zhao, G.; et al. Comparative Genome Analysis Reveals the Molecular Basis of Niche Adaptation of Staphylococcus epidermidis Strains. Front. Genet. 2020, 9, 566080. [Google Scholar] [CrossRef] [PubMed]
- Swaney, M.H.; Kalan, L.R. Living in Your Skin: Microbes, Molecules, and Mechanisms. Infect. Immun. 2021, 17, e00695-20. [Google Scholar] [CrossRef]
- Johansen, F.E.; Kaetzel, C.S. Regulation of the polymeric immunoglobulin receptor and IgA transport: New advances in environmental factors that stimulate pIgR expression and its role in mucosal immunity. Mucosal. Immunol. 2011, 4, 598–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanner, A.; Salathé, M.; O’Riordan, T.G. Mucociliary clearance in the airways. Am. J. Respir. Crit. Care Med. 1996, 154, 1868–1902. [Google Scholar] [CrossRef]
- Hariri, B.M.; Cohen, N.A. New insights into upper airway innate immunity. Am. J. Rhinol. Allergy 2016, 30, 319–323. [Google Scholar] [CrossRef] [Green Version]
- Basil, M.C.; Katzen, J.; Engler, A.E.; Guo, M.; Herriges, M.J.; Kathiriya, J.J.; Windmueller, R.; Ysasi, A.B.; Zacharias, W.J.; Chapman, A.H.; et al. The Cellular and Physiological Basis for Lung Repair and Regeneration: Past, Present, and Future. Cell Stem Cell 2020, 26, 482–502. [Google Scholar] [CrossRef] [PubMed]
- Vieira Braga, F.A.; Gozde, K.; Berg, M.; Carpaij, O.A.; Polanski, K.; Simon, L.M.; Brouwer, S.; Gomes, T.; Hesse, L.; Jiang, J.; et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med. 2019, 25, 1153–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deprez, M.; Zaragosi, L.E.; Truchi, M.; Becavin, C.; Ruiz García, S.; Arguel, M.J.; Plaisant, M.; Magnone, V.; Lebrigand, K.; Abelanet, S.; et al. A Single-Cell Atlas of the Human Healthy Airways. Am. J. Respir. Crit. Care Med. 2020, 202, 1636–1645. [Google Scholar] [CrossRef]
- Fahy, J.V.; Dickey, B.F. Airway mucus function and dysfunction. N. Engl. J. Med. 2010, 363, 2233–2247. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, R.; Müller, M.M.; Klassert, T.E.; Driesch, D.; Stock, M.; Heinrich, A.; Conrad, T.; Moore, C.; Schier, U.K.; Guthke, R.; et al. Differential regulation of the transcriptomic and secretomic landscape of sensor and effector functions of human airway epithelial cells. Mucosal. Immunol. 2018, 11, 627–642. [Google Scholar] [CrossRef] [Green Version]
- Morse, C.; Tabib, T.; Sembrat, J.; Buschur, K.L.; Bittar, T.H.; Valenzi, E.; Yale Jiang, Y.; Kass, D.J.; Gibson, K.; Chen, W.; et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur. Respir. J. 2019, 54, 1802441. [Google Scholar] [CrossRef]
- Puttur, F.; Gregory, L.G.; Lloyd, C.M. Airway macrophages as the guardians of tissue repair in the lung. Immunol. Cell Biol. 2019, 97, 246–257. [Google Scholar] [CrossRef]
- Masopust, D.; Soerens, A.G. Tissue-resident T cells and other resident leukocytes. Annu. Rev. Immunol. 2019, 37, 521–546. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.V.; Ma, W.; Miron, M.; Granot, T.; Guyer, R.S.; Carpenter, D.J.; Senda, T.; Sun, X.; Ho, S.H.; Lerner, H.; et al. Human tissue-resident memory T cells are defined by core transcriptional and functional signatures in lymphoid and mucosal sites. Cell Rep. 2017, 20, 2921–2934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, D.L.; Bickham, K.L.; Thome, J.J.; Kim, C.Y.; D’Ovidio, F.; Wherry, E.J.; Farber, D.L. Lung niches for the generation and maintenance of tissue-resident memory T cells. Mucosal. Immunol. 2014, 7, 501–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinks, T.S.; Wallington, J.C.; Williams, A.P.; Djukanović, R.; Staples, K.J.; Wilkinson, T.M. Steroid-induced deficiency of mucosal-associated invariant T cells in the chronic obstructive pulmonary disease lung. Implications for nontypeable haemophilus influenzae infection. Am. J. Respir. Crit. Care Med. 2016, 194, 1208–1218. [Google Scholar] [CrossRef] [Green Version]
- Toubal, A.; Nel, I.; Lotersztajn, S.; Lehuen, A. Mucosal-associated invariant T cells and disease. Nat. Rev. Immunol. 2019, 19, 643–657. [Google Scholar] [CrossRef]
- Wang, H.; D’Souza, C.; Lim, X.Y.; Kostenko, L.; Pediongco, T.J.; Eckle, S.B.G.; Meehan, B.S.; Mai Shi, M.; Wang, N.; Li, S.; et al. MAIT cells protect against pulmonary Legionella longbeachae infection. Nat. Commun. 2018, 9, 3350. [Google Scholar] [CrossRef] [Green Version]
- Hinks, T.S.C.; Marchi, E.; Jabeen, M.; Olshansky, M.; Kurioka, A.; Pediongco, T.J.; Meehan, B.S.; Kostenko, L.; Turner, S.j.; Corbett, A.J.; et al. Activation and in vivo evolution of the MAIT cell transcriptome in mice and humans reveals tissue repair functionality. Cell Rep. 2019, 28, 3249–3262. [Google Scholar] [CrossRef] [Green Version]
- Krismer, B.; Liebeke, M.; Janek, D.; Nega, M.; Rautenberg, M.; Hornig, G.; Unger, C.; Weidenmaier, C.; Lalk, M.; Peschel, A. Nutrient limitation governs Staphylococcus aureus metabolism and niche adaptation in the human nose. PLoS Pathog. 2014, 10, e1003862. [Google Scholar] [CrossRef]
- Geurkink, N. Nasal anatomy, physiology, and function. J. Allergy Clin. Immunol. 1983, 72, 123–128. [Google Scholar] [CrossRef]
- Biswas, K.; Hoggard, M.; Jain, R.; Taylor, M.W.; Douglas, R.G. The nasal microbiota in health and disease: Variation within and between subjects. Front. Microbiol. 2015, 9, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bassis, C.M.; Tang, A.L.; Young, V.B.; Pynnonen, M.A. The nasal cavity microbiota of healthy adults. Microbiome 2014, 11, 27. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Mihindukulasuriya, K.A.; Gao, H.; La Rosa, P.S.; Wylie, K.M.; Martin, J.C.; Kota, K.; Shannon, W.D.; Mitreva, M.; Sodergren, E.; et al. Exploration of bacterial community classes in major human habitats. Genome Biol. 2014, 7, R66. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, V.R.; Feazel, L.M.; Gitomer, S.A.; Ir, D.; Robertson, C.E.; Frank, D.N. The microbiome of the middle meatus in healthy adults. PLoS ONE 2013, 30, e85507. [Google Scholar] [CrossRef] [PubMed]
- Boutin, S.; Depner, M.; Stahl, M.; Graeber, S.Y.; Dittrich, S.A.; Legatzki, A.; von Mutius, E.; Mall, M.; Dalpke, A.H. Comparison of Oropharyngeal Microbiota from Children with Asthma and Cystic Fibrosis. Mediat. Inflamm. 2017, 2017, 5047403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camarinha-Silva, A.; Jáuregui, R.; Pieper, D.H.; Wos-Oxley, M.L. The temporal dynamics of bacterial communities across human anterior nares. Environ. Microbiol. Rep. 2012, 4, 126–132. [Google Scholar] [CrossRef]
- Biesbroek, G.; Tsivtsivadze, E.; Sanders, E.A.; Montijn, R.; Veenhoven, R.H.; Keijser, B.J.; Bogaert, D. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am. J. Respir. Crit. Care Med. 2014, 190, 1283–1292. [Google Scholar] [CrossRef]
- Yan, M.; Pamp, S.J.; Fukuyama, J.; Hwang, P.H.; Cho, D.Y.; Holmes, S.; Relman, D.A. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe 2013, 14, 631–640. [Google Scholar] [CrossRef] [Green Version]
- Johnson, R.C.; Ellis, M.W.; Lanier, J.B.; Schlett, C.D.; Cui, T.; Merrell, D.S. Correlation between nasal microbiome composition and remote purulent skin and soft tissue infections. Infect. Immun. 2015, 83, 802–811. [Google Scholar] [CrossRef] [Green Version]
- Ta, L.D.H.; Yap, G.C.; Tay, C.J.X.; Lim, A.S.M.; Huang, C.-H.; Chu, C.W.; De Sessions, P.F.; Shek, L.P.; Goh, A.; Van Bever, H.P.; et al. Establishment of the nasal microbiota in the first 18 months of life: Correlation with early-onset rhinitis and wheezing. J. Allergy Clin. Immunol. 2018, 142, 86–95. [Google Scholar] [CrossRef] [Green Version]
- Ahluwalia, J.; Borok, J.; Haddock, E.S.; Ahluwalia, R.S.; Schwartz, E.W.; Hosseini, D.; Amini, S.; Eichenfield, L.F. The microbiome in preadolescent acne: Assessment and prospective analysis of the influence of benzoyl peroxide. Pediatr. Dermatol. 2019, 36, 200–206. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.M.; Price, L.B.; Hungate, B.A.; Abraham, A.G.; Larsen, L.A.; Christensen, K.; Stegger, M.; Skov, R.; Andersen, P.S. Staphylococcus aureus and the ecology of the nasal microbiome. Sci. Adv. 2015, 1, e1400216. [Google Scholar] [CrossRef] [Green Version]
- Teo, S.M.; Mok, D.; Pham, K.; Kusel, M.; Serralha, M.; Troy, N.; Holt, B.J.; Hales, B.J.; Walker, M.L.; Hollams, E.; et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 2015, 17, 704–715. [Google Scholar] [CrossRef] [Green Version]
- Toivonen, L.; Hasegawa, K.; Waris, M.; Ajami, N.J.; Petrosino, J.F.; Camargo, C.A., Jr.; Peltola, V. Early nasal microbiota and acute respiratory infections during the first years of life. Thorax 2019, 74, 592–599. [Google Scholar] [CrossRef]
- Di Stadio, A.; Costantini, C.; Renga, G.; Pariano, M.; Ricci, G.; Romani, L. The Microbiota/Host Immune System Interaction in the Nose to Protect from COVID-19. Life 2020, 11, 345. [Google Scholar] [CrossRef]
- Tai, J.; Han, M.S.; Kwak, J.; Kim, T.H. Association Between Microbiota and Nasal Mucosal Diseases in terms of Immunity. Int. J. Mol. Sci. 2021, 29, 4744. [Google Scholar] [CrossRef]
- Baishya, J.; Bisht, K.; Rimbey, J.N.; Yihunie, K.D.; Islam, S.; Mahmud, H.A.; Waller, J.E.; Wakeman, C.A. The Impact of Intraspecies and Interspecies Bacterial Interactions on Disease Outcome. Pathogens 2021, 21, 96. [Google Scholar] [CrossRef]
- Conlan, S.; Mijares, L.A.; NISC Comparative Sequencing Program; Becker, J.; Blakesley, R.W.; Bouffard, G.; Brooks, S.; Coleman, H.; Gupta, J.; Natalie Gurson, N.; et al. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 2012, 25, R64. [Google Scholar] [CrossRef] [Green Version]
- Espadinha, D.; Sobral, R.G.; Mendes, C.I.; Méric, G.; Sheppard, S.K.; Carriço, J.A.; de Lencastre, H.; Miragaia, M. Distinct Phenotypic and Genomic Signatures Underlie Contrasting Pathogenic Potential of Staphylococcus epidermidis Clonal Lineages. Front. Microbiol. 2019, 27, 1971. [Google Scholar] [CrossRef]
- Khan, R.; Peterse, F.C.; Shekhar, S. Commensal Bacteria: An Emerging Player in Defense Against Respiratory Pathogens. Front. Immunol. 2019, 31, 1203. [Google Scholar] [CrossRef] [Green Version]
- Mergenhagen, K.A.; Starr, K.E.; Wattengel, B.A.; Lesse, A.J.; Sumon, Z.; Sellick, J.A. Determining the Utility of Methicillin-Resistant Staphylococcus aureus Nares Screening in Antimicrobial Stewardship. Clin. Infect. Dis. 2020, 22, 1142–1148. [Google Scholar] [CrossRef]
- Park, B.; Iwase, T.; Liu, G.Y. Intranasal application of S. epidermidis prevents colonization by methicillin-resistant Staphylococcus aureus in mice. PLoS ONE 2011, 6, e25880. [Google Scholar] [CrossRef] [Green Version]
- Foster, T.J. Surface Proteins of Staphylococcus aureus. Microbiol. Spectr. 2019, 7, 7–14. [Google Scholar] [CrossRef]
- Yin, W.; Xu, S.; Wang, Y.; Zhang, Y.; Chou, S.H.; Galperin, M.Y.; He, J. Ways to control harmful biofilms: Prevention, inhibition, and eradication. Crit. Rev. Microbiol. 2021, 47, 57–78. [Google Scholar] [CrossRef]
- Sugimoto, S.; Iwamoto, T.; Takada, K.; Okuda, K.; Tajima, A.; Iwase, T.; Mizunoe, Y. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host-pathogen interaction. J. Bacteriol. 2013, 195, 1645–1655. [Google Scholar] [CrossRef] [Green Version]
- Corrigan, R.M.; Miajlovic, H.; Foster, T.J. Surface proteins that promote adherence of Staphylococcus aureus to human desquamated nasal epithelial cells. BMC Microbio. 2009, 30, 22. [Google Scholar] [CrossRef] [Green Version]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010, 20, 346–349. [Google Scholar] [CrossRef]
- Glatthardt, T.; Campos, J.C.M.; Chamon, R.C.; de Sá Coimbra, T.F.; Rocha, G.A.; de Melo, M.A.F.; Parente, T.E.; Lobo, L.A.; Antunes, L.C.M.; Dos Santos, K.R.N.; et al. Small Molecules Produced by Commensal Staphylococcus epidermidis Disrupt Formation of Biofilms by Staphylococcus aureus. Appl. Environ. Microbiol. 2020, 18, e02539-19. [Google Scholar] [CrossRef]
- Otto, M. Quorum-sensing control in Staphylococci—A target for antimicrobial drug therapy? FEMS Microbiol. Lett. 2004, 15, 135–141. [Google Scholar] [CrossRef]
- Kong, K.F.; Vuong, C.; Otto, M. Staphylococcus quorum sensing in biofilm formation and infection. Int. J. Med. Microbiol. 2006, 296, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Otto, M.; Süssmuth, R.; Vuong, C.; Jung, G.; Götz, F. Inhibition of virulence factor expression in Staphylococcus aureus by the Staphylococcus epidermidis agr pheromone and derivatives. FEBS Lett. 1999, 7, 257–262. [Google Scholar] [CrossRef] [Green Version]
- Otto, M.; Echner, H.; Voelter, W.; Götz, F. Pheromone cross-inhibition between Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 2001, 69, 1957–1960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-García, S.; Ortiz-García, C.I.; Cruz-Aguilar, M.; Zenteno, J.C.; Murrieta-Coxca, J.M.; Pérez-Tapia, S.M.; Rodríguez-Martínez, S.; Cancino-Diaz, M.E.; Cancino-Diaz, J.C. Competition/antagonism associations of biofilm formation among Staphylococcus epidermidis Agr groups I, II, and III. J. Microbiol. 2019, 57, 143–153. [Google Scholar] [CrossRef]
- Nakatsuji, T.; Chen, T.H.; Narala, S.; Chun, K.A.; Two, A.M.; Yun, T.; Shafiq, F.; Kotol, P.F.; Bouslimani, A.; Melnik, A.V.; et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl. Med. 2017, 9, eaah4680. [Google Scholar] [CrossRef] [Green Version]
- Zipperer, A.; Konnerth, M.C.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M.; et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535, 511–516. [Google Scholar] [CrossRef]
- Stubbendieck, R.M.; May, D.S.; Chevrette, M.G.; Temkin, M.I.; Wendt-Pienkowski, E.; Cagnazzo, J.; Carlson, C.M.; Gern, J.E.; Currie, C.R. Competition among nasal bacteria suggests a role for siderophore-mediated interactions in shaping the human nasal microbiota. Appl. Environ. Microbiol. 2019, 85, e02406–e02418. [Google Scholar] [CrossRef] [Green Version]
- Ramsey, M.M.; Freire, M.O.; Gabrilska, R.A.; Rumbaugh, K.P.; Lemon, K.P. Staphylococcus aureus shifts toward commensalism in response to Corynebacterium species. Front. Microbiol. 2016, 7, 1230. [Google Scholar] [CrossRef] [Green Version]
- Brugger, S.D.; Eslami, S.M.; Pettigrew, M.M.; Escapa, I.F.; Henke, M.T.; Kong, Y.; Lemon, K.P. Dolosigranulum pigrum cooperation and competition in human nasal microbiota. mSphere 2020, 5, e00852-20. [Google Scholar] [CrossRef]
- Pericone, C.D.; Overweg, K.; Hermans, P.W.; Weiser, J.N. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect. Immun. 2000, 68, 3990–3997. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Gordon, O.; Jiang, W.; Antezana, B.S.; Angulo-Zamudio, U.A.; del Rio, C.; Moller, A.; Brissac, T.; Tierney, A.R.P.; Warncke, K.; et al. Interaction between Streptococcus pneumoniae and Staphylococcus aureus generates OH radicals that rapidly kill Staphylococcus aureus strains. J. Bacteriol. 2019, 201, e00474-19. [Google Scholar] [CrossRef] [PubMed]
- Bomar, L.; Brugger, S.D.; Yost, B.H.; Davies, S.S.; Lemon, K.P. Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio 2016, 7, e01725-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo, C.W.; Lai, Y.K.; Liu, Y.T.; Gallo, R.L.; Huang, C.M. Staphylococcus aureus hijacks a skin commensal to intensify its virulence: Immunization targeting-hemolysin and CAMP factor. J. Investig. Dermatol. 2011, 131, 401–409. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, I.M.; Hartford, O.; Foster, T.; Tarkowski, A. Alpha-toxin and gamma-toxin jointly promote Staphylococcus aureus virulence in murine septic arthritis. Infect. Immun. 1999, 67, 1045–1049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakhnovich, E.A.; King, S.J.; Weiser, J.N. Neuraminidase expressed by Streptococcus pneumoniae desialylates the lipopolysaccharide of Neisseria meningitidis and Haemophilus influenzae: A paradigm for interbacterial competition among pathogens of the human respiratory tract. Infect. Immun. 2002, 70, 7161–7164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiryukhina, N.V.; Melnikov, V.G.; Suvorov, A.V.; Morozova, Y.A.; Ilyin, V.K. Use of Corynebacterium pseudodiphtheriticum for elimination of Staphylococcus aureus from the nasal cavity in volunteers exposed to abnormal microclimate and altered gaseous environment. Probiotics Antimicrob. Proteins 2013, 5, 233–238. [Google Scholar] [CrossRef]
- Hardy, B.L.; Dickey, S.W.; Plaut, R.D.; Riggins, D.P.; Stibitz, S.; Otto, M.; Merrell, D.S. Corynebacterium pseudodiphtheriticum exploits Staphylococcus aureus virulence components in a novel polymicrobial defense strategy. mBio 2019, 10, e02491-18. [Google Scholar] [CrossRef] [Green Version]
- Janek, D.; Zipperer, A.; Kulik, A.; Krismer, B.; Peschel, A. High frequency and diversity of antimicrobial activities produced by nasal Staphylococcus strains against bacterial competitors. PLoS Pathog. 2016, 12, e1005812. [Google Scholar] [CrossRef] [Green Version]
- Wollenberg, M.S.; Claesen, J.; Escapa, I.F.; Aldridge, K.L.; Fischbach, M.A.; Lemon, K.P. Propionibacterium-produced coproporphyrin III induces Staphylococcus aureus aggregation and biofilm formation. mBio 2014, 5, e01286-14. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Kuo, S.; Shu, M.; Yu, J.; Huang, S.; Dai, A.; Two, A.; Gallo, R.L.; Huang, C.M. Staphylococcus epidermidis in the human skin microbiome mediates fermentation to inhibit the growth of Propionibacterium acnes: Implications of probiotics in acne vulgaris. Appl. Microbiol. Biotechnol. 2014, 98, 411–424. [Google Scholar] [CrossRef] [Green Version]
- Christensen, G.J.M.; Scholz, C.F.P.; Enghild, J.; Rohde, H.; Kilian, M.; Thürmer, A.; Brzuszkiewicz, E.; Lomholt, H.B.; Brüggemann, H. Antagonism between Staphylococcus epidermidis and Propionibacterium acnes and its genomic basis. BMC Genom. 2016, 17, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geitani, R.; Moubareck, C.A.; Xu, Z.; Sarkis, D.K.; Touqui, L. Expression and Roles of Antimicrobial Peptides in Innate Defense of Airway Mucosa: Potential Implication in Cystic Fibrosis. Front. Immunol. 2020, 30, 1198. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Cogen, A.L.; Radek, K.A.; Park, H.J.; Macleod, D.T.; Leichtle, A.; Ryan, A.F.; Di Nardo, A.; Gallo, R.L. Activation of TLR2 by a small molecule produced by Staphylococcus epidermidis increases antimicrobial defense against bacterial skin infections. J. Investig. Dermatol. 2010, 130, 2211–2221. [Google Scholar] [CrossRef] [Green Version]
- Brown, R.L.; Sequeira, R.P.; Clarke, T.B. The microbiota protects against respiratory infection via GM-CSF signaling. Nat. Commun. 2017, 15, 1512. [Google Scholar] [CrossRef] [PubMed]
- Kanmani, P.; Clua, P.; Vizoso-Pinto, M.G.; Rodriguez, C.; Alvarez, S.; Melnikov, V.; Takahashi, H.; Kitazawa, H.; Villena, J. Respiratory commensal bacteria Corynebacterium pseudodiphtheriticum improves resistance of infant mice to respiratory syncytial virus and Streptococcus pneumoniae superinfection. Front. Microbiol. 2017, 8, 1613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Rosa-Ramos, M.A.; Salcedo-Hernández, R.; Sarmiento-Silva, R.E.; Aguilera-Arreola, M.G.; Alcántar-Curiel, M.D.; Betanzos-Cabrera, G.; Rodríguez-Mártinez, S.; Cancino-Diaz, M.E.; Cancino-Díaz, J.C. Non-epidermidis coagulase-negative Staphylococcus isolated from farm animals can inhibit the hemagglutinating activity of Newcastle disease virus and bovine parainfluenza virus type 3. Comp. Immunol. Microbiol. Infect. Dis. 2021, 76, 101649. [Google Scholar] [CrossRef]
- Chen, H.W.; Liu, P.F.; Liu, Y.T.; Kuo, S.; Zhang, X.Q.; Schooley, R.T.; Rohde, H.; Gallo, R.L.; Huang, C.M. Nasal commensal Staphylococcus epidermidis counteracts influenza virus. Sci. Rep. 2016, 16, 27870. [Google Scholar] [CrossRef] [Green Version]
- Stanifer, M.L.; Guo, C.; Doldan, P.; Boulant, S. Importance of Type I and III Interferons at Respiratory and Intestinal Barrier Surfaces. Front. Immunol. 2020, 11, 608645. [Google Scholar] [CrossRef]
- Kim, H.J.; Jo, A.; Jeon, Y.J.; An, S.; Lee, K.M.; Yoon, S.S.; Choi, J.Y. Nasal commensal Staphylococcus epidermidis enhances interferon-λ-dependent immunity against influenza virus. Microbiome 2019, 30, 80. [Google Scholar] [CrossRef] [Green Version]
- Jeon, Y.J.; Gil, C.H.; Jo, A.; Won, J.; Ki, S.; Kim, H.J. The influence of interferon-lambda on restricting Middle East Respiratory Syndrome Coronavirus replication in the respiratory epithelium. Antivir. Res. 2020, 180, 104860. [Google Scholar] [CrossRef]
- Hartenian, E.; Nandakumar, D.; Lari, A.; Ly, M.; Tucker, J.M.; Glaunsinger, B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020, 11, 12910–12934. [Google Scholar] [CrossRef] [PubMed]
- Bridges, J.P.; Vladar, E.K.; Huang, H.; Mason, R.J. Respiratory epithelial cell responses to SARS-CoV-2 in COVID-19. Thorax 2021, 17, 2021–217561. [Google Scholar] [CrossRef] [PubMed]
- Hurst, J.H.; McCumber, A.W.; Aquino, J.N.; Rodriguez, J.; Heston, S.M.; Lugo, D.J.; Rotta, A.T.; Turner, N.A.; Pfeiffer, T.S.; Gurley, T.C.; et al. Age-related changes in the upper respiratory microbiome are associated with SARS-CoV-2 susceptibility and illness severity. medRxiv 2021, in press. [Google Scholar] [CrossRef]
- Mostafa, H.H.; Fissel, J.A.; Fanelli, B.; Bergman, Y.; Gniazdowski, V.; Dadlani, M.; Carroll, K.C.; Colwell, R.R.; Simner, P.J. Metagenomic Next-Generation Sequencing of Nasopharyngeal Specimens Collected from Confirmed and Suspect COVID-19 Patients. mBio 2020, 20, e01969-20. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Karyakarte, R.; Joshi, S.; Das, R.; Jani, K.; Shouche, Y.; Sharma, A. Nasopharyngeal microbiome reveals the prevalence of opportunistic pathogens in SARS-CoV-2 infected individuals and their association with host types. Microbes Infect. 2021, 20, 104880. [Google Scholar] [CrossRef]
- Zhong, H.; Wang, Y.; Shi, Z.; Zhang, L.; Ren, H.; He, W.; Zhang, Z.; Zhu, A.; Zhao, J.; Xiao, F.; et al. Characterization of respiratory microbial dysbiosis in hospitalized COVID-19 patients. Cell Discov. 2021, 13, 23. [Google Scholar] [CrossRef]
- Ogawa, Y.; Ote, H.; Arai, T.; Kazama, R.; Kimura, K.; Nagata, T.; Kumasawa, J.; Kohno, M.; Kohata, H.; Nishida, K.; et al. Corynebacterium pseudodiphtheriticum as a pathogen in bacterial co-infection in COVID-19 patients with mechanical ventilation. Jpn. J. Infec. Dis. 2021, JJID-2021. [Google Scholar] [CrossRef]
- Sungnak, W.; Huang, N.; Bécavin, C.; Berg, M.; Queen, R.; Litvinukova, M.; Talavera-López, C.; Maatz, H.; Reichart, D.; Sampaziotis, F.; et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020, 26, 681–687. [Google Scholar] [CrossRef] [Green Version]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 2020, 181, 1036–1045. [Google Scholar] [CrossRef]
- Onabajo, O.O.; Banday, A.R.; Stanifer, M.L.; Yan, W.; Obajemu, A.; Santer, D.M.; Florez-Vargas, O.; Piontkivska, H.; Vargas, J.M.; Ring, T.J.; et al. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat. Genet. 2020, 52, 1283–1293. [Google Scholar] [CrossRef]
- Stanifer, M.L.; Kee, C.; Cortese, M.; Zumaran, C.M.; Triana, S.; Mukenhirn, M.; Kraeusslich, H.G.; Alexandrov, T.; Bartenschlager, R.; Boulant, S. Critical Role of Type III Interferon in Controlling SARS-CoV-2 Infection in Human Intestinal Epithelial Cells. Cell Rep. 2020, 32, 107863. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.Y.; Jo, A.; Won, J.; Gil, C.H.; Shin, H.; Kim, S.; Jeon, Y.J.; Kim, H.J. The nasal symbiont Staphylococcus species restricts the transcription of SARS-CoV-2 entry factors in human nasal epithelium. iScience 2021, 22, 103172. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, T.R.; Thomas, D.L.; Jackson, S.S.; Prokunina-Olsson, L.; Donnelly, R.P.; Rune Hartmann, R. Weak Induction of Interferon Expression by Severe Acute Respiratory Syndrome Coronavirus 2 Supports Clinical Trials of Interferon-λ to Treat Early Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 12, 1410–1412. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.; Mommert, M.; Mouton, W.; Pizzorno, A.; Brengel-Pesce, K.; Mezidi, M.; Villard, M.; Lina, B.; Richard, J.C.; Fassier, J.B.; et al. Early nasal type I IFN immunity against SARS-CoV-2 is compromised in patients with autoantibodies against type I IFNs. J. Exp. Med. 2021, 218, e20211211. [Google Scholar] [CrossRef]
- Winkley, K.; Banerjee, D.; Bradley, T.; Koseva, B.; Cheung, W.A.; Selvarangan, R.; Pastinen, T.; Grundberg, E. Immune cell residency in the nasal mucosa may partially explain respiratory disease severity across the age range. Sci. Rep. 2021, 5, 15927. [Google Scholar] [CrossRef]
- Zhang, Q.; Bastard, P.; Liu, Z.; Le Pen, J.; Moncada-Velez, M.; Chen, J.; Ogishi, M.; Sabli, I.K.D.; Hodeib, S.; Cecilia Korol, C.; et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 2020, 370, eabd4570. [Google Scholar] [CrossRef]
- Channappanavar, R.; Fehr, A.R.; Vijay, R.; Mack, M.; Zhao, J.; Meyerholz, D.K.; Perlman, S. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoVinfected mice. Cell Host Microbe 2016, 19, 181–193. [Google Scholar] [CrossRef] [Green Version]
- Negari, I.P.; Keshari, S.; Huang, C.M. Probiotic Activity of Staphylococcus epidermidis Induces Collagen Type I Production through FFaR2/p-ERK Signaling. Int. J. Mol. Sci. 2021, 22, 1414. [Google Scholar] [CrossRef]
| Strains or Products/Reference | Pathogen | Pre-Clinical Studies on Human and Animal Models | Main Findings |
|---|---|---|---|
| S. epidermidis (ST9-N442); commensal, [22]. | S. epidermidis (ST2); infection. S. aureus; infection. | Murine model of Respiratory Tract Infection (RTI), first pre-colonization intranasally with S. epidermidis (ST9-N442) or pathogens. | S. epidermidis (ST9-N44-2) increase more expresión of CRAMP. S. epidermidis (ST9-N44-2) efficiently outcompeted the two pathogenic bacteria S. aureus and M. catarrhalis in vivo and led to decreased signs of infection caused by these pathogens. |
| M. catarrhalis; infection. | Murine model RTI co-colonized intranasally with S. epidermidis (ST9-N442) and pathogens. | ||
| S. epidermidis NRS122, [73]. | S. aureus BD02-31 | Murine model RTI pre-colonized intranasally with S. epidermidis NRS122 followed by intranasally challenge with S. aureus BD02-31. | Pre-colonization of mouse nares with S. epidermidis NRS122 reduces colonization of S. aureus BD02-31. |
| S. epidermidis (wild type, JK16); Esp-positive. | S. aureus | In a pilot sudy, S. epidermidis cells or purified Esp were introduced into anterior nares of volunters who were S. aureus carriers. | S. epidermidis (JK16) and purified Esp eliminated S.aureus colonization. S. epidermidis Esp-deficient and S. epidermidis (JK11) did not reduce colonization of S. aureus. |
| S. epidermidis Esp-deficient. | |||
| S. epidermidis (JK11); Esp-negative. | |||
| Purified Esp, [78]. | |||
| S. epidermidis AMT-A9, [85]. | S. aureus | In a pilot study, S. epidermidis AMT-A9 was inoculated into wound atopic dermatitis (AD) of volunteers who were S. aureus carriers. | S. epidermidis AMT-A9 eliminated S. aureus from the wound (AD), and clinical manifestation of AD improved. |
| S. epidermidis 1457, [103] | Group A Streptococcus (GAS) | Murine skin infection model with GAS treated with S. epidermidis 1457. | S. epidermidis 1457 protects mice against GAS by the activation of TLR2 and induction of hBDs 2 y 3. |
| S. epidermidis (human), [104]. | S. pneumoniae | Murine model RTI pre-colonized intranasally with S. epidermidis followed by intranasal challenge with S. pneumoniae or K. pneumoniae. | Pre-colonization of mouse nares with S. epidermidis limited the spread of S. pneumoniae and K. pneumoniae by the activation of Nod2 receptor, production of IL-17A, release of GM-CSF, and activation of alveolar macrophages. |
| K. pneumoniae | |||
| rEMbp6599 of S. epidermidis, [107]. | IVA | Chicken model of RTI pre-colonized intranasally with rEMbp6599 followed by intranasal challenge with IVA. | rEMbp6599 protects against RTI (IVA) by reducing the tissue viral load and inducing robust expression of antiviral cytokines (IFN-α, IL-6, and Mx) |
| S. epidermidis (human), [109]. | IVA | Murine model of RTI pre-colonized with S. epidermidis followed by intransal challenge with IVA | Pre-colonization of mouse nares with S. epidermidis limited the spread of IVA to the lungs by stimulating innate immunity in which IFN-λ suppresses the replication of IVA in the nasal mucosa. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortega-Peña, S.; Rodríguez-Martínez, S.; Cancino-Diaz, M.E.; Cancino-Diaz, J.C. Staphylococcus epidermidis Controls Opportunistic Pathogens in the Nose, Could It Help to Regulate SARS-CoV-2 (COVID-19) Infection? Life 2022, 12, 341. https://doi.org/10.3390/life12030341
Ortega-Peña S, Rodríguez-Martínez S, Cancino-Diaz ME, Cancino-Diaz JC. Staphylococcus epidermidis Controls Opportunistic Pathogens in the Nose, Could It Help to Regulate SARS-CoV-2 (COVID-19) Infection? Life. 2022; 12(3):341. https://doi.org/10.3390/life12030341
Chicago/Turabian StyleOrtega-Peña, Silvestre, Sandra Rodríguez-Martínez, Mario E. Cancino-Diaz, and Juan C. Cancino-Diaz. 2022. "Staphylococcus epidermidis Controls Opportunistic Pathogens in the Nose, Could It Help to Regulate SARS-CoV-2 (COVID-19) Infection?" Life 12, no. 3: 341. https://doi.org/10.3390/life12030341
APA StyleOrtega-Peña, S., Rodríguez-Martínez, S., Cancino-Diaz, M. E., & Cancino-Diaz, J. C. (2022). Staphylococcus epidermidis Controls Opportunistic Pathogens in the Nose, Could It Help to Regulate SARS-CoV-2 (COVID-19) Infection? Life, 12(3), 341. https://doi.org/10.3390/life12030341







