Karyotypical Confirmation of Natural Hybridization between Two Manatee Species, Trichechus manatus and Trichechus inunguis
Abstract
:1. Introduction
2. Materials and Methods
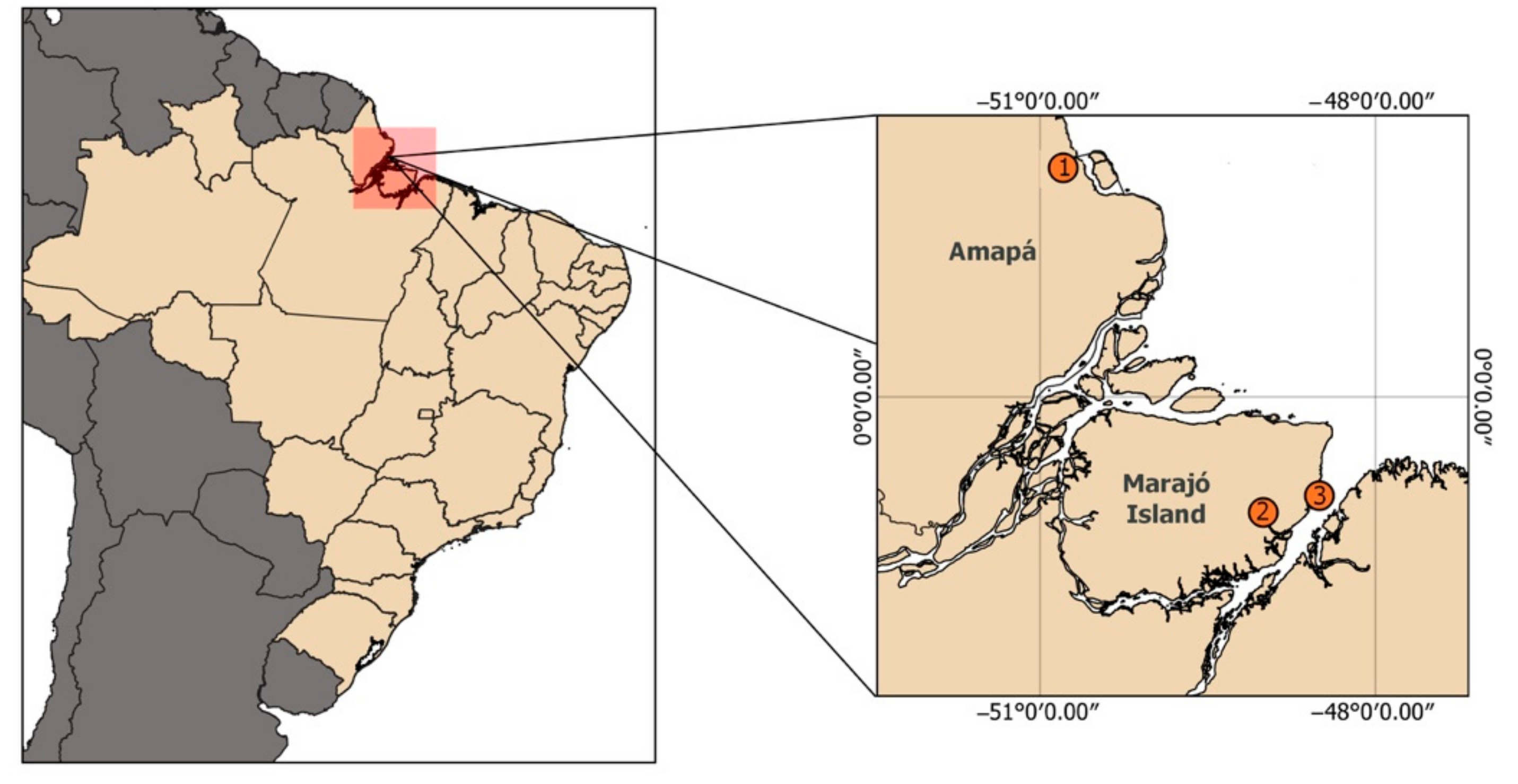
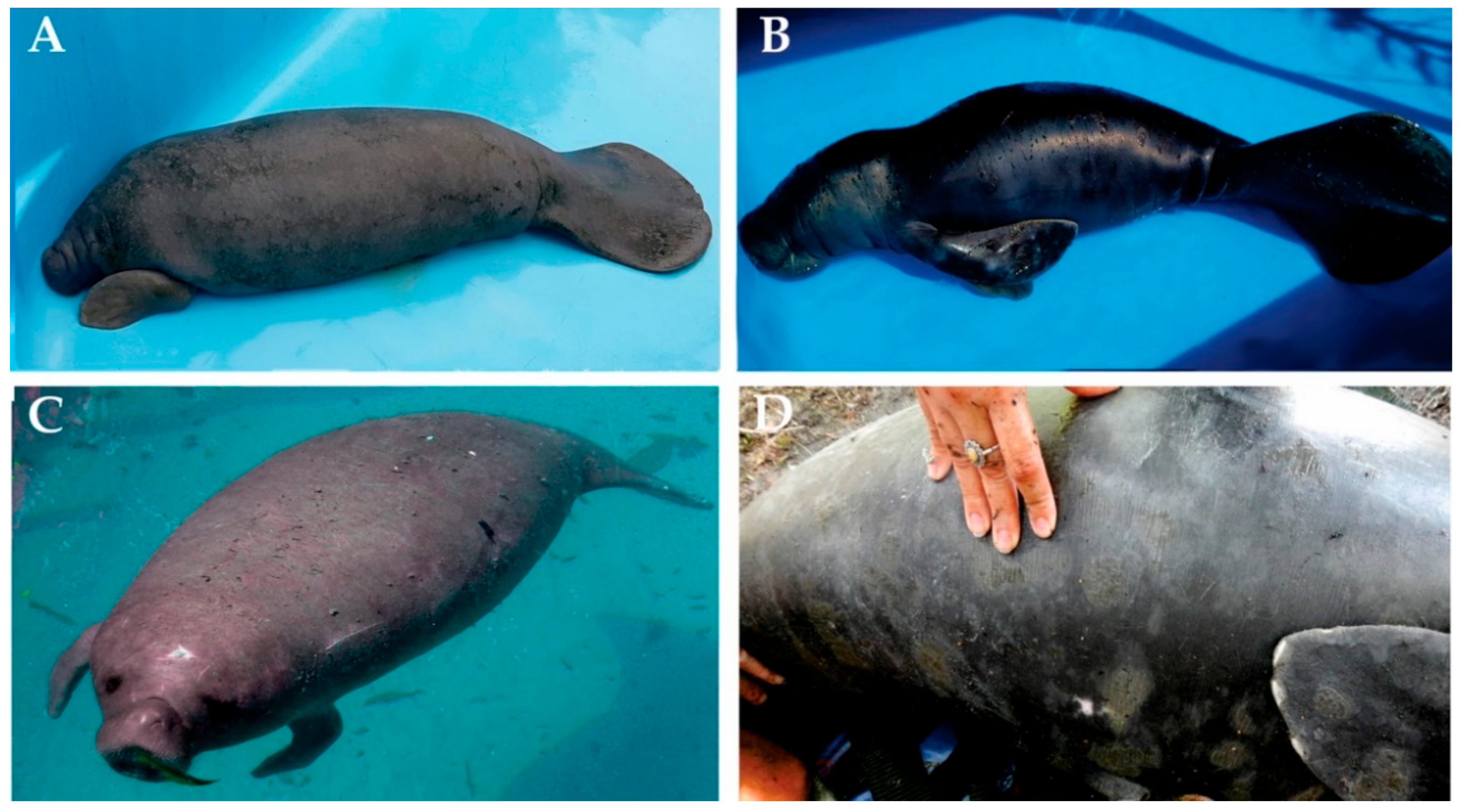
2.1. Samples and Chromosome Preparation
2.2. Classical Cytogenetic Techniques
2.3. Fluorescent In Situ Hybridization (FISH)
2.4. Microscopic Analyses and Image Capture and Processing
3. Results
3.1. Karyotype Description: Diploid Number, G-Banding Patterns and Distribution of Constitutive Heterochromatin Blocks
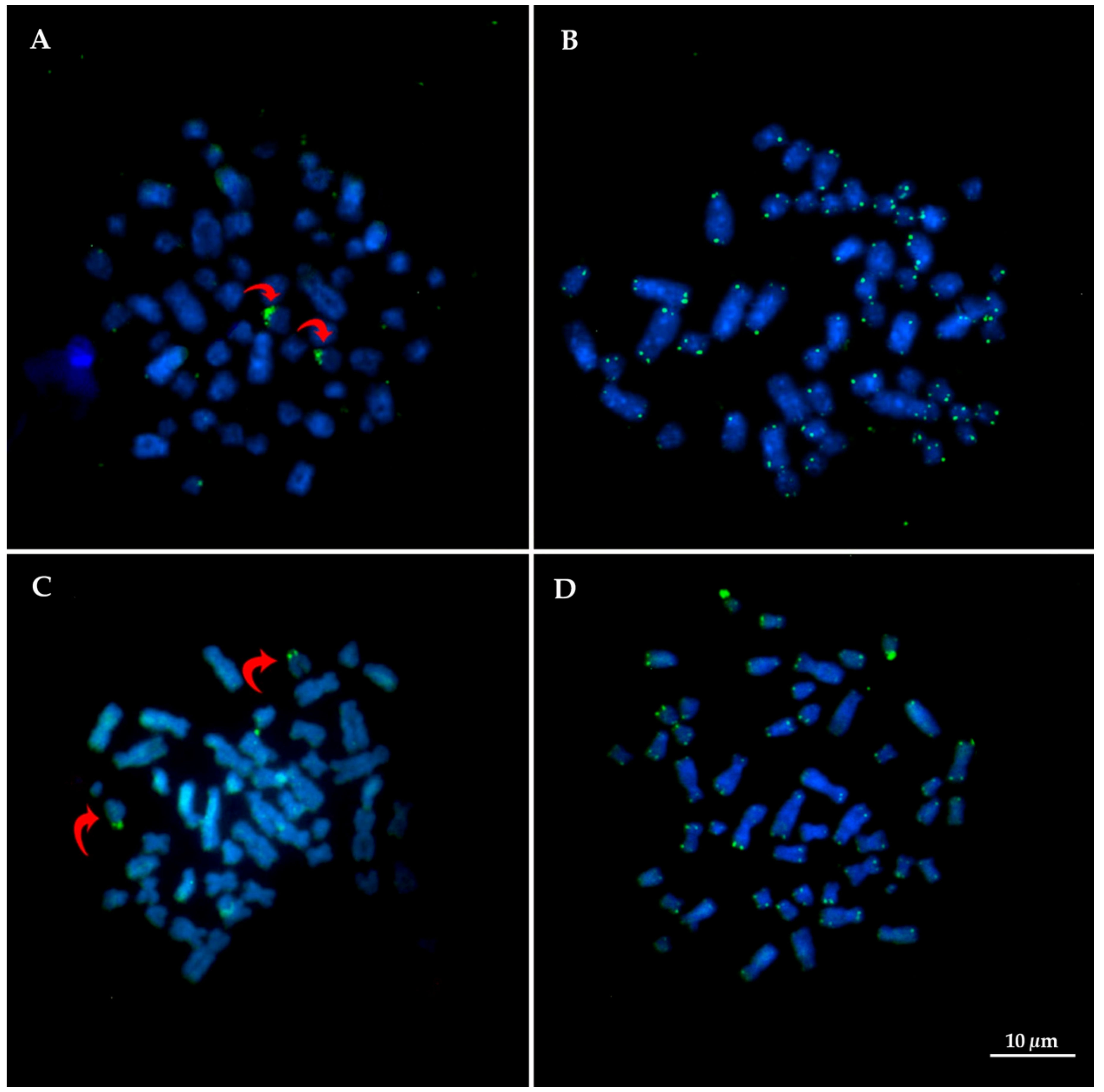
3.2. Distribution of Repetitive Sequences: 18/28rDNA, Telomeric Sequences and Microsatellites
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vianna, J.A.; Bonde, R.K.; Caballero, S.; Giraldo, J.P.; Lima, R.P.; Clark, A.; Marmontel, M.; Morales-Vela, B.; De Souza, M.J.; Parr, L.; et al. Phylogeography, phylogeny and hybridization in Trichechid sirenians: Implications for manatee conservation. Mol. Ecol. 2006, 15, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Marsh, H.; O’Shea, T.J.; Reynolds III, J.E. Ecology and Conservation of Sirenia: Dugongs and Manatees; Cambridge University Press: New York, NY, USA, 2011. [Google Scholar]
- Domning, D.P. Distribution and status of manatees Trichechus spp. near the mouth of the Amazon River, Brazil. Biol. Conserv. 1981, 19, 85–97. [Google Scholar] [CrossRef]
- Luna, F.O.; Lima, R.P.; Castro, D.F.; Vianna, J.A. Capture and utilization of the Amazonian manatee Trichechus inunguis in the state of Amazonas, Brasil. In Proceedings of the 14th Biennal Conference of the Biology of Marine Mammals, Vancouver, BC, Canada, 28 November–3 December 2001. [Google Scholar]
- Luna, F.O.; Araújo, J.P.; Oliveira, E.M.; Hage, L.M.; Passavante, J.Z.O. Distribuição do peixe-boi marinho, Trichechus manatus manatus, no litoral norte do Brasil. Arq. De Ciências Do Mar 2010, 43, 79–86. [Google Scholar]
- Sousa, M.E.M.; Martins, B.M.L.; Fernandes, M.E.B. Meeting the giants: The need for local ecological knowledge (LEK) as a tool for the participative management of manatees on Marajó Island, Brazilian Amazonian coast. Ocean Coast. Manag. 2013, 86, 53–60. [Google Scholar] [CrossRef]
- Lima, C.S.; Magalhaes, R.F.; Marmontel, M.; Meirelles, A.C.; Carvalho, V.L.; Lavergne, A.; Thoisy, B.D.; Santos, F.R. A hybrid swarm of manatees along the Guianas coastline, a peculiar environment under the influence of the Amazon River plume. An. Da Acad. Bras. De Ciências 2019, 91, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Luna, F.D.O.; Beaver, C.E.; Nourisson, C.; Bonde, R.K.; Attademo, F.L.; Miranda, A.V.; Torres-Florez, J.P.; de Sousa, G.P.; Passavante, J.Z.; Hunter, M.E. Genetic connectivity of the West Indian manatee in the southern range and limited evidence of hybridization with Amazonian manatees. Front. Mar Sci. 2021, 7, 574455. [Google Scholar] [CrossRef]
- De Sá, A.L.A.; Baker, P.K.B.; Breaux, B.; Oliveira, J.M.; de Macedo Klautau, A.G.C.; Legatzki, K.; de Oliveira Luna, F.; Attademo, F.L.N.; Hunter, M.E.; Criscitiello, M.F.; et al. Novel insights on aquatic mammal MHC evolution: Evidence from manatee DQB diversity. Dev. Comp. Immunol. 2022, 132, 104398. Available online: https://linkinghub.elsevier.com/retrieve/pii/S0145305X2200060X (accessed on 30 March 2022).
- Bulatova, N.; Shchipanov, N.; Searle, J. The Seliger Moscow hybrid zone between chromosome races of common shrews an initial description. Russ. J. Theriol. 2007, 6, 111–116. [Google Scholar] [CrossRef]
- Loughman, W.; Frye, F.; Herald, E. The chromosomes of a male manatee—Trichechus inunguis. Husb. Res. 1970, 10, 151–152. [Google Scholar]
- White, J.R.; Harknesst, D.R.; Isaackst, R.E.; Duffield, D.A. Some studies on blood of the Florida manatee, Trichechus manatus latirostris. Comp. Biochem. Physiol. 1976, 55A, 413–417. [Google Scholar] [CrossRef]
- Assis, M.; Best, R.; Barros, R.; Yassuda, Y. Cytogenetic study of Trichechus inunguis (Amazonian manatee). Rev. Bras. De Genética 1988, 11, 41–50. [Google Scholar]
- Gray, B.A.; Zori, R.T.; Mcguire, P.M.; Bonde, R.K. A first generation cytogenetic ideogram for the Florida manatee (Trichechus manatus latirostris) based on multiple chromosome banding techniques. Hereditas 2002, 137, 215–223. [Google Scholar] [CrossRef]
- Hunter, M.E.; Mignucci-Giannoni, A.A.; Tucker, K.P.; King, T.L.; Bonde, R.K.; Gray, B.A.; McGuire, P.M. Puerto Rico and Florida manatees represent genetically distinct groups. Conserv. Genet. 2012, 13, 1623–1635. [Google Scholar] [CrossRef]
- Barros, H.; Meirelles, A.; Luna, F.; Marmontel, M.; Cordeiro-Estrela, P.; Santos, N.; Astúa, D. Cranial and chromosomal geographic variation in manatees (Mammalia: Sirenia: Trichechidae) with the description of the Antillean manatee karyotype in Brazil. J. Zool. Syst. Evol. Res. 2017, 55, 73–87. [Google Scholar] [CrossRef]
- Kellogg, M.E.; Burkett, S.; Dennis, T.R.; Stone, G.; Gray, B.A.; McGuire, P.M.; Zori, R.T.; Stanyon, R. Chromosome painting in the manatee supports Afrotheria and Paenungulata. BMC Evol. Biol. 2007, 7, 6. [Google Scholar] [CrossRef] [Green Version]
- Pardini, A.T.; O’Brien, P.C.M.; Fu, B.; Bonde, R.K.; Elder, F.F.B.; Ferguson-Smith, M.A.; Yang, F.; Robinson, T.J. Chromosome painting among Proboscidea, Hyracoidea and Sirenia: Support for Paenungulata (Afrotheria, Mammalia) but not Tethytheria. Proc. R. Soc. B Biol. Sci. 2007, 274, 1333–1340. [Google Scholar] [CrossRef] [Green Version]
- Moorhead, P.; Nowell, P.; Mellman, W.; Battips, D.; Hungerford, D. Chromosome preparations of leukocites cultured from human peripheral blood. Exp. Cell Res. 1960, 20, 613–616. [Google Scholar] [CrossRef]
- Sumner, A. A simple technique for demonstrating centromeric heterochromatin. Exptl. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Seabright, M. A rapid banding technique for human chromosomes. Lancet 1971, 2, 971–972. [Google Scholar] [CrossRef]
- Cioffi, M.; Martins, C.; Centofante, L.; Jacobina, U.; Bertollo, L. Chromosomal variability among allopatric populations of erythrinidae fish Hoplias malabaricus: Mapping of three classes of repetitive DNAs. Cytogenet. Genome Res. 2009, 125, 132–141. [Google Scholar] [CrossRef]
- Ljdo, J.; Wells, R.; Baldini, A.; Reeders, S. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991, 19, 4780. [Google Scholar]
- Yang, E.; Carter, N.P.; Shiu, L.; Ferguson-Smith, M.A. A comparative study of karyotypes of muntjacs by chromosome painting. Chromosoma 1995, 103, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Kubat, Z.; Hobza, R.; Vyskot, B.; Kejnovsky, E. Microsatellite accumulation on the Y chromosome in Silene latifolia. Genome 2008, 51, 350–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graphodatsky, A.; Trifonov, V.; Stanyon, R. The genome diversity and karyotype evolution of mammals. Mol. Cytogenet. 2011, 4. Available online: http://www.molecularcytogenetics.org/content/4/1/22 (accessed on 30 March 2022). [CrossRef] [PubMed] [Green Version]
- Bakloushinskaya, I.Y.; Matveevsky, S.N.; Romanenko, S.A.; Serdukova, N.A.; Kolomiets, O.L.; Spangenberg, V.E.; Lyapunova, E.A.; Graphodatsky, A.S. A comparative analysis of the mole vole sibling species Ellobius tancrei and E. talpinus (Cricetidae, Rodentia) through chromosome painting and examination of synaptonemal complex structures in hybrids. Cytogenet. Genome Res. 2012, 136, 199–207. [Google Scholar] [CrossRef]
- Peres, W.A.M.; Bertollo, L.A.C.; Buckup, P.A.; Blanco, D.R.; Kantek, D.L.Z.; Moreira-Filho, O. Invasion, dispersion and hybridization of fish associated to river transposition: Karyotypic evidence in Astyanax “bimaculatus group” (Characiformes: Characidae). Rev. Fish Biol. Fish. 2012, 22, 519–526. [Google Scholar] [CrossRef]
- Garcia-Rodriguez, A.; Bowen, B.; Domning, D.; Mignucci-Giannoni, A.; Marmontel, M.; Montoyoa-Ospina, R.; Morales-Vela, B.; Rudin, M.; Bonde, R.K.; McGuire, P.M. Introduction Phylogeography of the West Indian manatee (Trichechus manatus): How many populations and how many taxa? Mol. Ecol. 1998, 7, 1137–1149. [Google Scholar] [CrossRef] [Green Version]
- Shurtliff, Q.R. Mammalian hybrid zones: A review. Mammal Rev. 2013, 43, 1–21. [Google Scholar] [CrossRef]
- Grize, S.A.; Wilwert, E.; Searle, J.B.; Lindholm, A.K. Measurements of hybrid fertility and a test of mate preference for two house mouse races with massive chromosomal divergence. BMC Evol. Biol. 2019, 19, 25. [Google Scholar] [CrossRef]
- Valeri, M.; Dias, G.; do Espírito Santo, A.; Moreira, C.; Yonenaga-Yassuda, Y.; Sommer, I.; Kuhn, G.; Svartman, M. First description of a satellite DNA in Manatees’ centromeric regions. Front. Genet. 2021, 12, 1537. [Google Scholar] [CrossRef]
- Horn, A.; Basset, P.; Yannic, G.; Banaszek, A.; Borodin, P.M.; Bulatova, N.S.; Jadwiszczak, K.; Jones, R.M.; Polyakov, A.V.; Ratkiewicz, M.; et al. Chromosomal rearrangements do not seem to affect the gene flow in hybrid zones between karyotypic races of the common shrew (Sorex araneus). Evolution 2012, 66, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Vrana, P.B. Genomic imprinting as a mechanism of reproductive isolation in mammals. J. Mammal. 2007, 88, 5–23. [Google Scholar] [CrossRef] [Green Version]
- Rieseberg, L. Chromosomal rearrangements andspeciation. TRENDS Ecol. Evol. 2001, 16, 351–358. [Google Scholar] [CrossRef]
- Sequeira, F.; Sodré, D.; Ferrand, N.; Bernardi, J.A.; Sampaio, I.; Schneider, H.; Vallinoto, M. Hybridization and Massive mtDNA Unidirectional Introgression between the Closely Related Neotropical Toads Rhinella marina and R. schneideri Inferred from mtDNA and Nuclear Markers. 2011. Available online: http://www.biomedcentral.com/1471-2148/11/264 (accessed on 5 March 2022).
- Mallet, J. Hybridization as an invasion of the genome. Trends Ecol. Evol. 2005, 20, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Toews, D.P.L.; Brelsford, A. The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 2012, 21, 3907–3930. [Google Scholar] [CrossRef]
- Vargas-Ramírez, M.; Carr, J.L.; Fritz, U. Complex phylogeography in Rhinoclemmys melanosterna: Conflicting mitochondrial and nuclear evidence suggests past hybridization (Testudines: Geoemydidae). Zootaxa 2013, 3670, 238. [Google Scholar] [CrossRef] [Green Version]
- Crossman, C.A.; Taylor, E.B.; Barrett-Lennard, L.G. Hybridization in the Cetacea: Widespread occurrence and associated morphological, behavioral, and ecological factors. Ecol. Evol. 2016, 6, 1293–1303. [Google Scholar] [CrossRef] [Green Version]
- Verkaar, E.L.C.; Nijman, I.J.; Beeke, M.; Hanekamp, E.; Lenstra, J.A. Maternal and paternal lineages in cross-breeding bovine species. Has wisent a hybrid origin? Mol. Biol. Evol. 2004, 21, 1165–1170. [Google Scholar] [CrossRef] [Green Version]
- Ropiquet, A.; Hassanin, A. Molecular phylogeny of caprines (Bovidae, Antilopinae): The question of their origin and diversification during the Miocene. J. Zool. Syst. Evol. Res. 2005, 43, 49–60. [Google Scholar] [CrossRef]
- Willows-Munro, S.; Robinson, T.J.; Matthee, C.A. Utility of nuclear DNA intron markers at lower taxonomic levels: Phylogenetic resolution among nine Tragelaphus spp. Mol. Phylogenetics Evol. 2005, 35, 624–636. [Google Scholar] [CrossRef]
- Hassanin, A.; Ropiquet, A.; Couloux, A.; Cruaud, C. Evolution of the mitochondrial genome in mammals living at high altitude: New insights from a study of the tribe Caprini (Bovidae, Antilopinae). J. Mol. Evol. 2009, 68, 293–310. [Google Scholar] [CrossRef] [PubMed]
- Hassanin, A.; An, J.; Ropiquet, A.; Nguyen, T.T.; Couloux, A. Combining multiple autosomal introns for studying shallow phylogeny and taxonomy of Laurasiatherian mammals: Application to the tribe Bovini (Cetartiodactyla, Bovidae). Mol. Phylogenetics Evol. 2013, 66, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Hassanin, A.; Houck, M.L.; Tshikung, D.; Kadjo, B.; Davis, H.; Ropiquet, A. Multi-locus phylogeny of the tribe Tragelaphini (Mammalia, Bovidae) and species delimitation in bushbuck: Evidence for chromosomal speciation mediated by interspecific hybridization. Mol. Phylogenetics Evol. 2018, 129, 96–105. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Oliveira, E.H.C.; Gomes, A.J.B.; Costa, A.F.; Emin-Lima, R.; Bonvicino, C.R.; Viana, M.C.; Reis, L.M.A.; Vidal, M.D.; Cavalcanti, M.V.G.; Attademo, F.L.N.; et al. Karyotypical Confirmation of Natural Hybridization between Two Manatee Species, Trichechus manatus and Trichechus inunguis. Life 2022, 12, 616. https://doi.org/10.3390/life12050616
de Oliveira EHC, Gomes AJB, Costa AF, Emin-Lima R, Bonvicino CR, Viana MC, Reis LMA, Vidal MD, Cavalcanti MVG, Attademo FLN, et al. Karyotypical Confirmation of Natural Hybridization between Two Manatee Species, Trichechus manatus and Trichechus inunguis. Life. 2022; 12(5):616. https://doi.org/10.3390/life12050616
Chicago/Turabian Stylede Oliveira, Edivaldo H. C., Anderson J. B. Gomes, Alexandra F. Costa, Renata Emin-Lima, Cibele R. Bonvicino, Maria C. Viana, Laura M. A. Reis, Marcelo D. Vidal, Mirella V. G. Cavalcanti, Fernanda L. N. Attademo, and et al. 2022. "Karyotypical Confirmation of Natural Hybridization between Two Manatee Species, Trichechus manatus and Trichechus inunguis" Life 12, no. 5: 616. https://doi.org/10.3390/life12050616
APA Stylede Oliveira, E. H. C., Gomes, A. J. B., Costa, A. F., Emin-Lima, R., Bonvicino, C. R., Viana, M. C., Reis, L. M. A., Vidal, M. D., Cavalcanti, M. V. G., Attademo, F. L. N., Luna, F. O., & Siciliano, S. (2022). Karyotypical Confirmation of Natural Hybridization between Two Manatee Species, Trichechus manatus and Trichechus inunguis. Life, 12(5), 616. https://doi.org/10.3390/life12050616






