The R Language: An Engine for Bioinformatics and Data Science
Abstract
1. Introduction
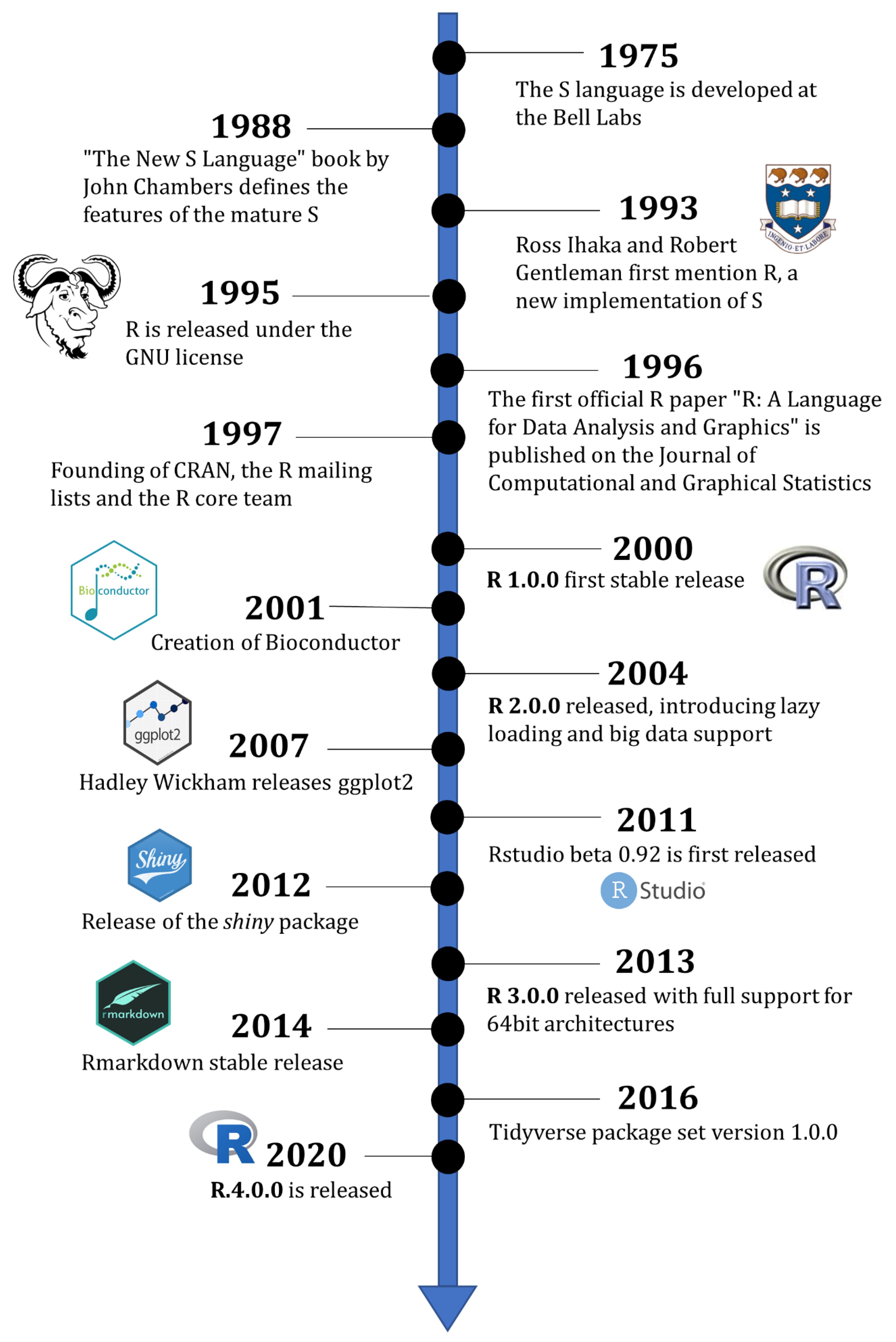
2. History of the R Programming Language
2.1. The S Language
2.2. The Birth of R
2.3. The R Community
2.4. The Official R Project after the Year 2000
2.5. The Founding of Bioconductor
2.6. Expanding R: ggplot2 and the Tidyverse
2.7. Beyond Statistics: RStudio, Shiny and Rmarkdown
3. The R Repositories
3.1. CRAN
3.2. Bioconductor
3.3. R-Forge
3.4. Github
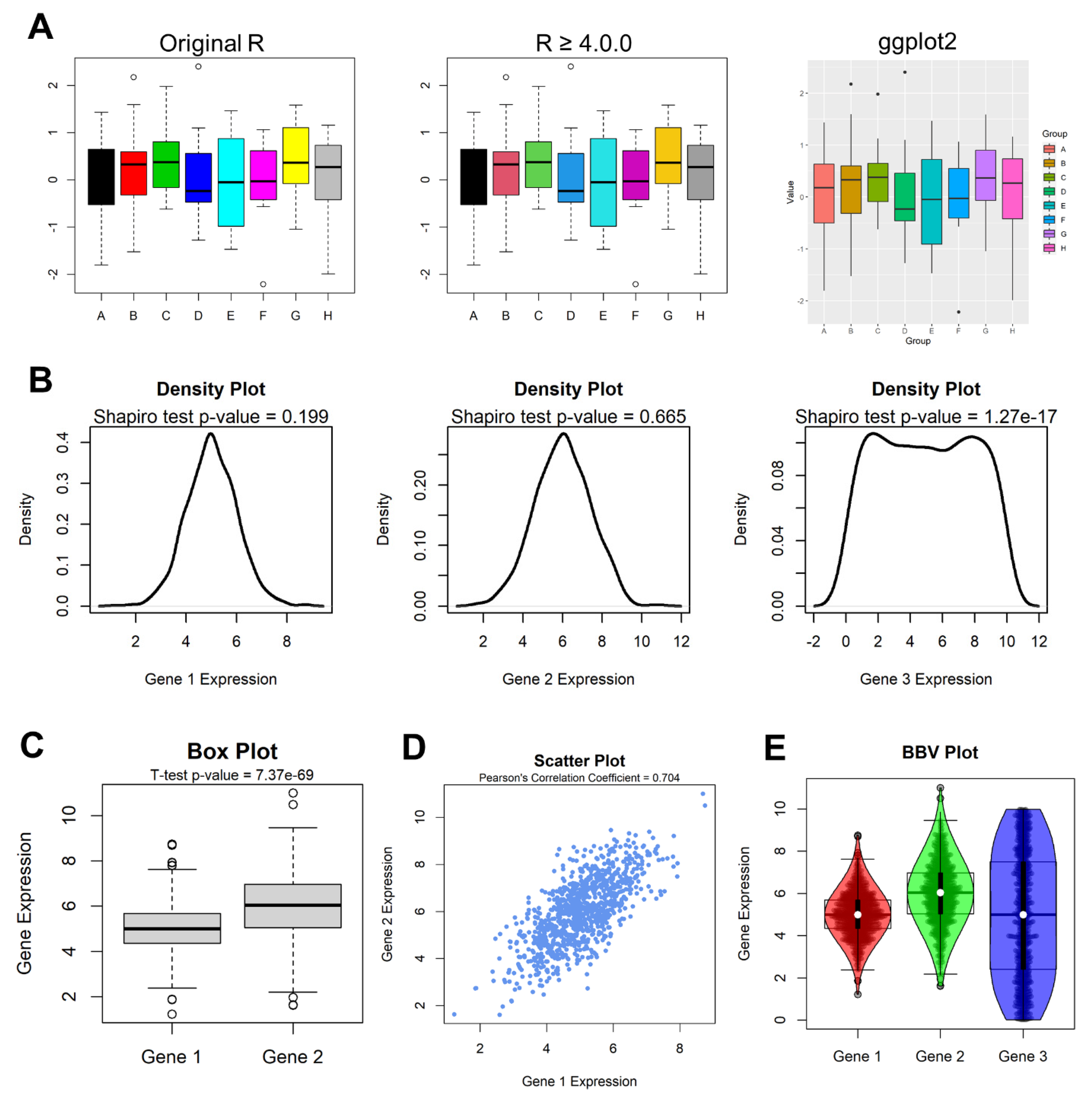
4. Practical R
4.1. Data Interaction
4.2. Analysis
4.3. Visualization
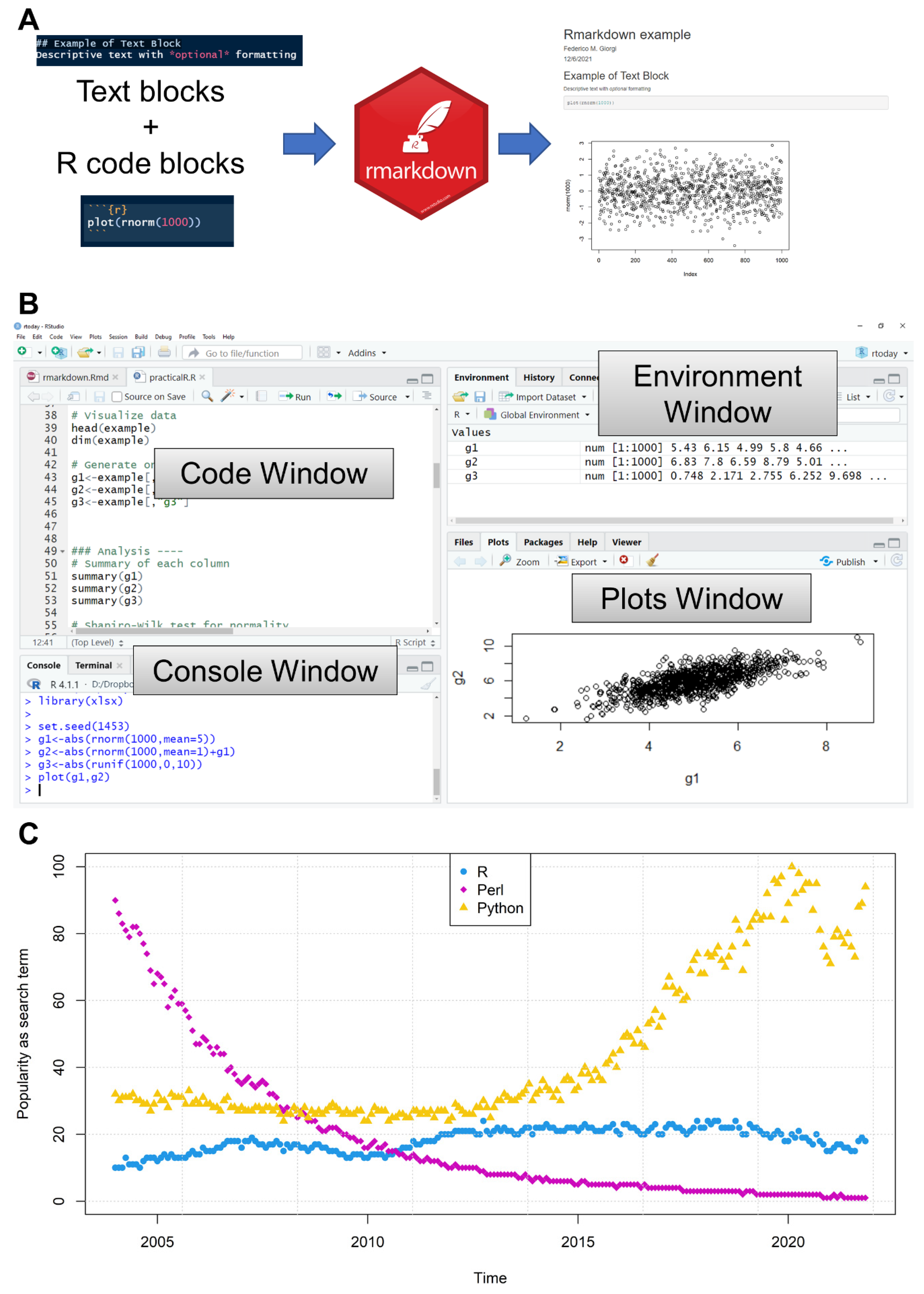
5. Rmarkdown and the Role of R in Scientific Reproducibility
6. Writing R: Editors and Environments
6.1. IDEs/GUIs
6.1.1. RStudio
6.1.2. Jupyter Notebook
6.1.3. RKward
6.1.4. StatET (Eclipse Plugin)
6.1.5. Google Colab
6.1.6. Visual Studio Code
6.2. Text Editors
6.2.1. Vi/Vim
6.2.2. Emacs ESS
6.2.3. Sublime Text
6.2.4. Notepad++
7. R and Other Programming Languages
8. Machine Learning and Artificial Intelligence through R
9. R on the World Wide Web: The Shiny Framework
10. User-Friendly Resources for Learning R
10.1. Books
10.2. Online Resources
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ihaka, R.; Gentleman, R. R: A Language for Data Analysis and Graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar] [CrossRef]
- Becker, R.A. A Brief History of S. In Computational Statistics; Dirschedl, P., Ostermann, R., Eds.; Contributions to Statistics; Physica-Verlag HD: Heidelberg, Germany, 1994; pp. 81–110. ISBN 978-3-7908-0813-1. [Google Scholar]
- Chambers, J.M. Programming with Data: A Guide to the S Language; Springer Science & Business Media: Berlin/Heidelberg, Germany, 1998; ISBN 978-0-387-98503-9. [Google Scholar]
- Becker, R.A. The New S Language; CRC Press: Boca Raton, FL, USA, 2018; ISBN 978-1-351-09188-6. [Google Scholar]
- Ihaka, R. The R Project: A Brief History and Thoughts about the Future. Univ. Auckl. 2017, 4, 22. [Google Scholar]
- Morandat, F.; Hill, B.; Osvald, L.; Vitek, J. Evaluating the Design of the R Language. In Proceedings of the ECOOP 2012—Object-Oriented Programming; Noble, J., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 104–131. [Google Scholar]
- Ihaka, R. R: Past and Future History. Comput. Sci. Stat. 1998, 392396. Available online: https://cran.r-project.org/doc/html/interface98-paper/paper.html (accessed on 21 April 2022).
- Hornik, K. R Frequently Asked Questions. Available online: https://cran.r-project.org/doc/FAQ/R-FAQ.html#What-are-the-differences-between-R-and-S_003f (accessed on 8 December 2021).
- Carbonnelle, P. PYPL PopularitY of Programming Language Index. Available online: https://pypl.github.io/PYPL.html (accessed on 9 December 2021).
- Maechler, M. “R-Announce”, “R-Help”, “R-Devel”: 3 Mailing Lists for R. Available online: https://stat.ethz.ch/pipermail/r-announce/1997/000000.html (accessed on 8 December 2021).
- Hornik, K. Post from the R-Announce Mailing List: “ANNOUNCE: CRAN”. Available online: https://stat.ethz.ch/pipermail/r-announce/1997/000001.html (accessed on 9 December 2021).
- R: Contributors. Available online: https://www.r-project.org/contributors.html (accessed on 9 December 2021).
- Bates, D. Post from the R-Announce Mailing List: “New Domain—r-Project.Org”. Available online: https://stat.ethz.ch/pipermail/r-announce/1999/000103.html (accessed on 9 December 2021).
- Dalgaard, P. Post from the R-Announce Mailing List: “R-1.0.0 Is Released”. Available online: https://stat.ethz.ch/pipermail/r-announce/2000/000127.html (accessed on 9 December 2021).
- Leisch, F. Post from the R-Announce Mailing List: “R Foundation for Statistical Computing”. Available online: https://stat.ethz.ch/pipermail/r-announce/2003/000385.html (accessed on 9 December 2021).
- The R Foundation Statute. Available online: https://www.r-project.org/foundation/Rfoundation-statutes.pdf (accessed on 9 December 2021).
- Roh, S.W.; Abell, G.C.; Kim, K.-H.; Nam, Y.-D.; Bae, J.-W. Comparing Microarrays and Next-Generation Sequencing Technologies for Microbial Ecology Research. Trends Biotechnol. 2010, 28, 291–299. [Google Scholar] [CrossRef]
- Galili, T. R 3.0.0 Is Released! (What’s New, and How to Upgrade)|R-Statistics Blog. 2013. Available online: https://www.r-statistics.com/2013/04/r-3-0-0-is-released-whats-new-and-how-to-upgrade/ (accessed on 21 April 2022).
- Smith, D. R 4.0.0 Now Available, and a Look Back at R’s History. Available online: https://blog.revolutionanalytics.com/2020/04/r-400-is-released.html (accessed on 9 December 2021).
- Lockstone, H.E. Exon Array Data Analysis Using Affymetrix Power Tools and R Statistical Software. Brief. Bioinform. 2011, 12, 634–644. [Google Scholar] [CrossRef]
- Heather, J.M.; Chain, B. The Sequence of Sequencers: The History of Sequencing DNA. Genomics 2016, 107, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, Robert 2002 Annual Report for the Bioconductor Project. Available online: https://www.bioconductor.org/about/annual-reports/AnnRep2002.pdf (accessed on 9 December 2021).
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open Software Development for Computational Biology and Bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef]
- Kopf, D. Ggplot2 Is 10 Years Old: The Program That Brought Data Visualization to the Masses. Available online: https://qz.com/1007328/all-hail-ggplot2-the-code-powering-all-those-excellent-charts-is-10-years-old/ (accessed on 9 December 2021).
- Villanueva, R.A.M.; Chen, Z.J. Ggplot2: Elegant Graphics for Data Analysis (2nd Ed.). Meas. Interdiscip. Res. Perspect. 2019, 17, 160–167. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- RStudio GitHub Repository. Available online: https://github.com/rstudio (accessed on 9 December 2021).
- RStudio Team RStudio, New Open-Source IDE for R. Available online: https://rstudio.comhttps://www.rstudio.com/blog/rstudio-new-open-source-ide-for-r/ (accessed on 9 December 2021).
- Smith, D. RStudio Releases Shiny|R-Bloggers. 2012. Available online: https://www.r-bloggers.com/2012/11/rstudio-releases-shiny/ (accessed on 21 April 2022).
- Mercatelli, D.; Holding, A.N.; Giorgi, F.M. Web Tools to Fight Pandemics: The COVID-19 Experience. Brief. Bioinform. 2021, 22, 690–700. [Google Scholar] [CrossRef]
- Xie, Y.; Allaire, J.J.; Grolemund, G. R Markdown: The Definitive Guide, 1st ed.; Chapman and Hall/CRC: London, UK, 2018; ISBN 978-1-138-35944-4. [Google Scholar]
- Baumer, B.; Udwin, D. R Markdown. WIREs Comput. Stat. 2015, 7, 167–177. [Google Scholar] [CrossRef]
- Xu, W.; Huang, R.; Zhang, H.; El-Khamra, Y.; Walling, D. Empowering R with High Performance Computing Resources for Big Data Analytics. In Conquering Big Data with High Performance Computing; Arora, R., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 191–217. ISBN 978-3-319-33742-5. [Google Scholar]
- Schäfer, J.; Opgen-Rhein, R.; Strimmer, K. Reverse Engineering Genetic Networks Using the GeneNet Package. Newsl. R Proj. 2006, 6, 50. [Google Scholar]
- Hornik, K. Are There Too Many R Packages? Austrian J. Stat. 2012, 41, 59–66. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Smyth, G.K. Limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Springer: Berlin/Heidelberg, Germany, 2005; pp. 397–420. [Google Scholar]
- Lawrence, M.; Huber, W.; Pages, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.T.; Carey, V.J. Software for Computing and Annotating Genomic Ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Mercatelli, D.; Lopez-Garcia, G.; Giorgi, F.M. Corto: A Lightweight R Package for Gene Network Inference and Master Regulator Analysis. Bioinformatics 2020, 36, 3916–3917. [Google Scholar] [CrossRef]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial Reconstruction of Single-Cell Gene Expression Data. Nat. Biotechnol. 2015, 33, 495–502. [Google Scholar] [CrossRef]
- R-Forge Home Page. Available online: https://r-forge.r-project.org/ (accessed on 9 December 2021).
- Zapponi, C. GitHut—Programming Languages and GitHub. Available online: https://githut.info/ (accessed on 9 December 2021).
- Lopez, G.; Egolf, L.E.; Giorgi, F.M.; Diskin, S.J.; Margolin, A.A. Svpluscnv: Analysis and Visualization of Complex Structural Variation Data. Bioinformatics 2021, 37, 1912–1914. [Google Scholar] [CrossRef]
- Su, K.; Wu, Z.; Wu, H. Simulation, Power Evaluation and Sample Size Recommendation for Single-Cell RNA-Seq. Bioinformatics 2020, 36, 4860–4868. [Google Scholar] [CrossRef]
- Gillespie, C. Understanding the Parquet File Format. Available online: https://www.jumpingrivers.com/blog/parquet-file-format-big-data-r/ (accessed on 9 December 2021).
- Royston, P. Approximating the Shapiro-Wilk W-Test for Non-Normality. Stat. Comput. 1992, 2, 117–119. [Google Scholar] [CrossRef]
- Gosset, W.S. The Probable Error of a Mean. Biometrika 1908, 6, 1–25. [Google Scholar] [CrossRef]
- Bonett, D.G.; Wright, T.A. Sample Size Requirements for Estimating Pearson, Kendall and Spearman Correlations. Psychometrika 2000, 65, 23–28. [Google Scholar] [CrossRef]
- Wilcoxon, F. Individual Comparisons by Ranking Methods. Biom. Bull. 1945, 1, 80–83. [Google Scholar] [CrossRef]
- Mercatelli, D.; Balboni, N.; Palma, A.; Aleo, E.; Sanna, P.P.; Perini, G.; Giorgi, F.M. Single-Cell Gene Network Analysis and Transcriptional Landscape of MYCN-Amplified Neuroblastoma Cell Lines. Biomolecules 2021, 11, 177. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, M.; Wildenhain, J.; Rappsilber, J.; Tyers, M. BoxPlotR: A Web Tool for Generation of Box Plots. Nat. Methods 2014, 11, 121–122. [Google Scholar] [CrossRef]
- Kenny, M.; Schoen, I. Violin SuperPlots: Visualizing Replicate Heterogeneity in Large Data Sets. MBoC 2021, 32, 1333–1334. [Google Scholar] [CrossRef]
- Hintze, J.L.; Nelson, R.D. Violin Plots: A Box Plot-Density Trace Synergism. Am. Stat. 1998, 52, 181–184. [Google Scholar] [CrossRef]
- Leisch, F. Sweave: Dynamic Generation of Statistical Reports Using Literate Data Analysis. In Proceedings of the Compstat; Härdle, W., Rönz, B., Eds.; Physica-Verlag HD: Heidelberg, Germany, 2002; pp. 575–580. [Google Scholar]
- Xie, Y. Dynamic Documents with R and Knitr; Chapman and Hall/CRC: London, UK, 2016; ISBN 978-0-429-17103-1. [Google Scholar]
- Markowetz, F. Five Selfish Reasons to Work Reproducibly. Genome Biol. 2015, 16, 274. [Google Scholar] [CrossRef]
- Murrell, P. R Graphics; Chapman and Hall/CRC: New York, NY, USA, 2005; ISBN 978-0-429-19610-2. [Google Scholar]
- Stander, J.; Dalla Valle, L. On Enthusing Students About Big Data and Social Media Visualization and Analysis Using R, RStudio, and RMarkdown. J. Stat. Educ. 2017, 25, 60–67. [Google Scholar] [CrossRef]
- Rödiger, S.; Friedrichsmeier, T.; Kapat, P.; Michalke, M. RKWard: A Comprehensive Graphical User Interface and Integrated Development Environment for Statistical Analysis with R. J. Stat. Softw. 2012, 49, 1–34. [Google Scholar] [CrossRef][Green Version]
- Lam, L. A Guide to Eclipse and the R Plug-in StatET. Available online: https://usermanual.wiki/Document/A20guide20to20Eclipse20and20the20R20plugin20StatET.1831954166 (accessed on 21 April 2022).
- Wahlbrink, S.; Verbeke, T. An Open Source Visual R Debugger in StatET. In Proceedings of the R User Conference, Coventry, UK, 16–18 August 2011; University of Warwick: Coventry, UK; p. 71. [Google Scholar]
- Nelson, M.J.; Hoover, A.K. Notes on Using Google Colaboratory in AI Education. In Proceedings of the 2020 ACM Conference on Innovation and Technology in Computer Science Education, Trondheim, Norway, 15–19 June 2020; pp. 533–534. [Google Scholar]
- Beard, B. Setup and Installation of R Tools for Visual Studio. In Beginning SQL Server R Services; Springer: Berlin/Heidelberg, Germany, 2016; pp. 33–71. [Google Scholar]
- Ueda, Y. R Extension for Visual Studio Code. Available online: https://marketplace.visualstudio.com/items?itemName=Ikuyadeu.r (accessed on 9 December 2021).
- Stack Overflow Developer Survey 2021—Most Popular Integrated Development Environments. Available online: https://insights.stackoverflow.com/survey/2021#section-most-popular-technologies-integrated-development-environment (accessed on 9 December 2021).
- de Aquino, J.A. Jalvesaq/Nvim-R. Available online: https://github.com/jalvesaq/Nvim-R (accessed on 21 April 2022).
- Bell, C.G.; Mudge, J.C.; McNamara, J.E. Digital Equipment Corporation. In Computer Engineering: A DEC View of Hardware Systems Design; Digital Press: Bedford, MA, USA, 1978; ISBN 978-0-932376-00-8. [Google Scholar]
- Kirkbride, P. Emacs and Vim. In Basic Linux Terminal Tips and Tricks; Springer: Berlin/Heidelberg, Germany, 2020; pp. 247–274. [Google Scholar]
- Hallen, J. Text Editor Performance Comparison. Available online: https://github.com/jhallen/joes-sandbox/tree/master/editor-perf (accessed on 9 December 2021).
- Sparapani, R. Revolutions Blog—Emacs, ESS and R for Zombies. Available online: https://blog.revolutionanalytics.com/2014/03/emacs-ess-and-r-for-zombies.html (accessed on 9 December 2021).
- Fourment, M.; Gillings, M.R. A Comparison of Common Programming Languages Used in Bioinformatics. BMC Bioinform. 2008, 9, 82. [Google Scholar] [CrossRef] [PubMed]
- Eddelbuettel, D.; Francois, R. Rcpp: Seamless R and C++ Integration. J. Stat. Softw. 2011, 40, 1–18. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Wu, Z.; Jaffee, H.A. Comparison of Affymetrix GeneChip Expression Measures. Bioinformatics 2006, 22, 789–794. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential Expression of RNA-Seq Data at the Gene Level–the DESeq Package. Heidelb. Ger. Eur. Mol. Biol. Lab. (EMBL) 2012, 10, f1000research. [Google Scholar]
- Eastwood, B. The 10 Most Popular Programming Languages to Learn in 2021. Available online: https://www.northeastern.edu/graduate/blog/most-popular-programming-languages/ (accessed on 9 December 2021).
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R Package for Comparing Biological Themes among Gene Clusters. Omics J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Durinck, S.; Spellman, P.T.; Birney, E.; Huber, W. Mapping Identifiers for the Integration of Genomic Datasets with the R/Bioconductor Package BiomaRt. Nat. Protoc. 2009, 4, 1184–1191. [Google Scholar] [CrossRef]
- Dowle, M. Benchmarks: Grouping · Rdatatable/Data.Table Wiki · GitHub. Available online: https://github.com/Rdatatable/data.table/wiki/Benchmarks-%3A-Grouping (accessed on 9 December 2021).
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning; Springer Texts in Statistics; Springer: New York, NY, USA, 2013; Volume 103, ISBN 978-1-4614-7137-0. [Google Scholar]
- Tibshirani, R. The Lasso Method for Variable Selection in the Cox Model. Stat. Med. 1997, 16, 385–395. [Google Scholar] [CrossRef]
- Vasilevski, A.; Giorgi, F.M.; Bertinetti, L.; Usadel, B. LASSO Modeling of the Arabidopsis Thaliana Seed/Seedling Transcriptome: A Model Case for Detection of Novel Mucilage and Pectin Metabolism Genes. Mol. BioSyst. 2012, 8, 2566. [Google Scholar] [CrossRef]
- Rawi, R.; Mall, R.; Kunji, K.; Shen, C.-H.; Kwong, P.D.; Chuang, G.-Y. PaRSnIP: Sequence-Based Protein Solubility Prediction Using Gradient Boosting Machine. Bioinformatics 2018, 34, 1092–1098. [Google Scholar] [CrossRef]
- Mercatelli, D.; Ray, F.; Giorgi, F.M. Pan-Cancer and Single-Cell Modeling of Genomic Alterations Through Gene Expression. Front. Genet. 2019, 10, 671. [Google Scholar] [CrossRef]
- Barter, R. Tidymodels: Tidy Machine Learning in R. Available online: https://www.rebeccabarter.com/blog/2020-03-25_machine_learning/ (accessed on 8 December 2021).
- LeDell, E.; Gill, N.; Aiello, S.; Fu, A.; Candel, A.; Click, C.; Kraljevic, T.; Nykodym, T.; Aboyoun, P.; Kurka, M.; et al. H2O: R Interface for the “H2O” Scalable Machine Learning Platform. Available online: https://docs.h2o.ai/h2o/latest-stable/h2o-r/docs/index.html (accessed on 21 April 2022).
- Lang, M.; Binder, M.; Richter, J.; Schratz, P.; Pfisterer, F.; Coors, S.; Au, Q.; Casalicchio, G.; Kotthoff, L.; Bischl, B. Mlr3: A Modern Object-Oriented Machine Learning Framework in R. J. Open Source Softw. 2019, 4, 1903. [Google Scholar] [CrossRef]
- Venables, W.N.; Ripley, B.D. Modern Applied Statistics with S, 4th ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Taylor, S.; Letham, B. Prophet: Automatic Forecasting Procedure. Available online: https://cran.r-project.org/web/packages/prophet/index.html (accessed on 21 April 2022).
- Papacharalampous, G.A.; Tyralis, H. Evaluation of Random Forests and Prophet for Daily Streamflow Forecasting. Adv. Geosci. 2018, 45, 201–208. [Google Scholar] [CrossRef]
- Rahimi, I.; Chen, F.; Gandomi, A.H. A Review on COVID-19 Forecasting Models. Neural Comput. Appl. 2021, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Berners-Lee, T.; Cailliau, R.; Groff, J.; Pollermann, B. World-Wide Web: The Information Universe. Internet Res. 1992, 2, 52–58. [Google Scholar] [CrossRef]
- Hendler, J. Web 3.0 Emerging. Computer 2009, 42, 111–113. [Google Scholar] [CrossRef]
- Becoming A Data-Driven CEO|Domo. Available online: https://www.domo.com/solution/data-never-sleeps-6 (accessed on 7 November 2021).
- Internet Users in the World. 2021. Available online: https://www.statista.com/statistics/617136/digital-population-worldwide/ (accessed on 7 November 2021).
- Brusic, V. The Growth of Bioinformatics. Brief. Bioinform. 2006, 8, 69–70. [Google Scholar] [CrossRef][Green Version]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. In Statistical Genomics: Methods and Protocols; Mathé, E., Davis, S., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2016; pp. 93–110. ISBN 978-1-4939-3578-9. [Google Scholar]
- Parkinson, H.; Kapushesky, M.; Shojatalab, M.; Abeygunawardena, N.; Coulson, R.; Farne, A.; Holloway, E.; Kolesnykov, N.; Lilja, P.; Lukk, M.; et al. ArrayExpress—A Public Database of Microarray Experiments and Gene Expression Profiles. Nucleic Acids Res. 2007, 35, D747–D750. [Google Scholar] [CrossRef]
- Hubbard, S.J.; Jones, A.R. (Eds.) Proteome Bioinformatics; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2010; Volume 604, ISBN 978-1-60761-443-2. [Google Scholar]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING Database in 2021: Customizable Protein–Protein Networks, and Functional Characterization of User-Uploaded Gene/Measurement Sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Stark, C.; Breitkreutz, B.-J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A General Repository for Interaction Datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef]
- Pal, S.; Mondal, S.; Das, G.; Khatua, S.; Ghosh, Z. Big Data in Biology: The Hope and Present-Day Challenges in It. Gene Rep. 2020, 21, 100869. [Google Scholar] [CrossRef]
- Jia, L.; Yao, W.; Jiang, Y.; Li, Y.; Wang, Z.; Li, H.; Huang, F.; Li, J.; Chen, T.; Zhang, H. Development of Interactive Biological Web Applications with R/Shiny. Brief. Bioinform. 2021, 23, bbab415. [Google Scholar] [CrossRef]
- Greene, C.S.; Tan, J.; Ung, M.; Moore, J.H.; Cheng, C. Big Data Bioinformatics. J. Cell. Physiol. 2014, 229, 1896–1900. [Google Scholar] [CrossRef]
- Mercatelli, D.; Triboli, L.; Fornasari, E.; Ray, F.; Giorgi, F.M. Coronapp: A Web Application to Annotate and Monitor SARS-CoV-2 Mutations. J. Med. Virol. 2021, 93, 3238–3245. [Google Scholar] [CrossRef]
- Menestrina, L.; Cabrelle, C.; Recanatini, M. COVIDrugNet: A Network-Based Web Tool to Investigate the Drugs Currently in Clinical Trial to Contrast COVID-19. Sci. Rep. 2021, 11, 19426. [Google Scholar] [CrossRef]
- Kasprzak, P.; Mitchell, L.; Kravchuk, O.; Timmins, A. Six Years of Shiny in Research—Collaborative Development of Web Tools in R. arXiv 2021, arXiv:2101.10948. [Google Scholar] [CrossRef]
- Salvaneschi, G.; Margara, A.; Tamburrelli, G. Reactive Programming: A Walkthrough. In Proceedings of the 2015 IEEE/ACM 37th IEEE International Conference on Software Engineering, Florence, Italy, 16–24 May 2015; Volume 2, pp. 953–954. [Google Scholar]
| Text Editor/IDE | Release Year | Web Link |
|---|---|---|
| RStudio | 2011 | https://www.rstudio.com |
| Jupyter Notebook | 2014 | https://jupyter.org |
| RKWard | 2002 | https://rkward.kde.org |
| Eclipse StatET | 2010 | https://projects.eclipse.org/projects/science.statet |
| Google Colab | 2017 | https://colab.research.google.com |
| Visual Studio Code | 2015 | https://code.visualstudio.com |
| vi/Vim | 1976 | https://www.vim.org/download.php |
| Emacs ESS | 1997 | https://ess.r-project.org/ |
| Sublime Text | 2008 | https://www.sublimetext.com/ |
| Notepad++ | 2003 | https://notepad-plus-plus.org/downloads/ |
| Rank | Language | Share | Trend |
|---|---|---|---|
| 1 | Python | 30.21% | −0.50% |
| 2 | Java | 17.82% | 1.30% |
| 3 | JavaScript | 9.16% | 0.60% |
| 4 | C# | 7.53% | 1.00% |
| 5 | C/C++ | 6.82% | 0.60% |
| 6 | PHP | 5.84% | −0.20% |
| 7 | R | 3.81% | 0.00% |
| 8 | Swift | 2.03% | −0.20% |
| 9 | Objective-C | 2.02% | −1.60% |
| 10 | MATLAB | 1.73% | −0.10% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giorgi, F.M.; Ceraolo, C.; Mercatelli, D. The R Language: An Engine for Bioinformatics and Data Science. Life 2022, 12, 648. https://doi.org/10.3390/life12050648
Giorgi FM, Ceraolo C, Mercatelli D. The R Language: An Engine for Bioinformatics and Data Science. Life. 2022; 12(5):648. https://doi.org/10.3390/life12050648
Chicago/Turabian StyleGiorgi, Federico M., Carmine Ceraolo, and Daniele Mercatelli. 2022. "The R Language: An Engine for Bioinformatics and Data Science" Life 12, no. 5: 648. https://doi.org/10.3390/life12050648
APA StyleGiorgi, F. M., Ceraolo, C., & Mercatelli, D. (2022). The R Language: An Engine for Bioinformatics and Data Science. Life, 12(5), 648. https://doi.org/10.3390/life12050648







