Mineralogical and Genomic Constraints on the Origin of Microbial Mn Oxide Formation in Complexed Microbial Community at the Terrestrial Hot Spring
Abstract
:1. Introduction
1.1. General Background
1.2. Mn Oxidizing Enzyme
1.3. Purpose of the Present Study
2. Materials and Methods
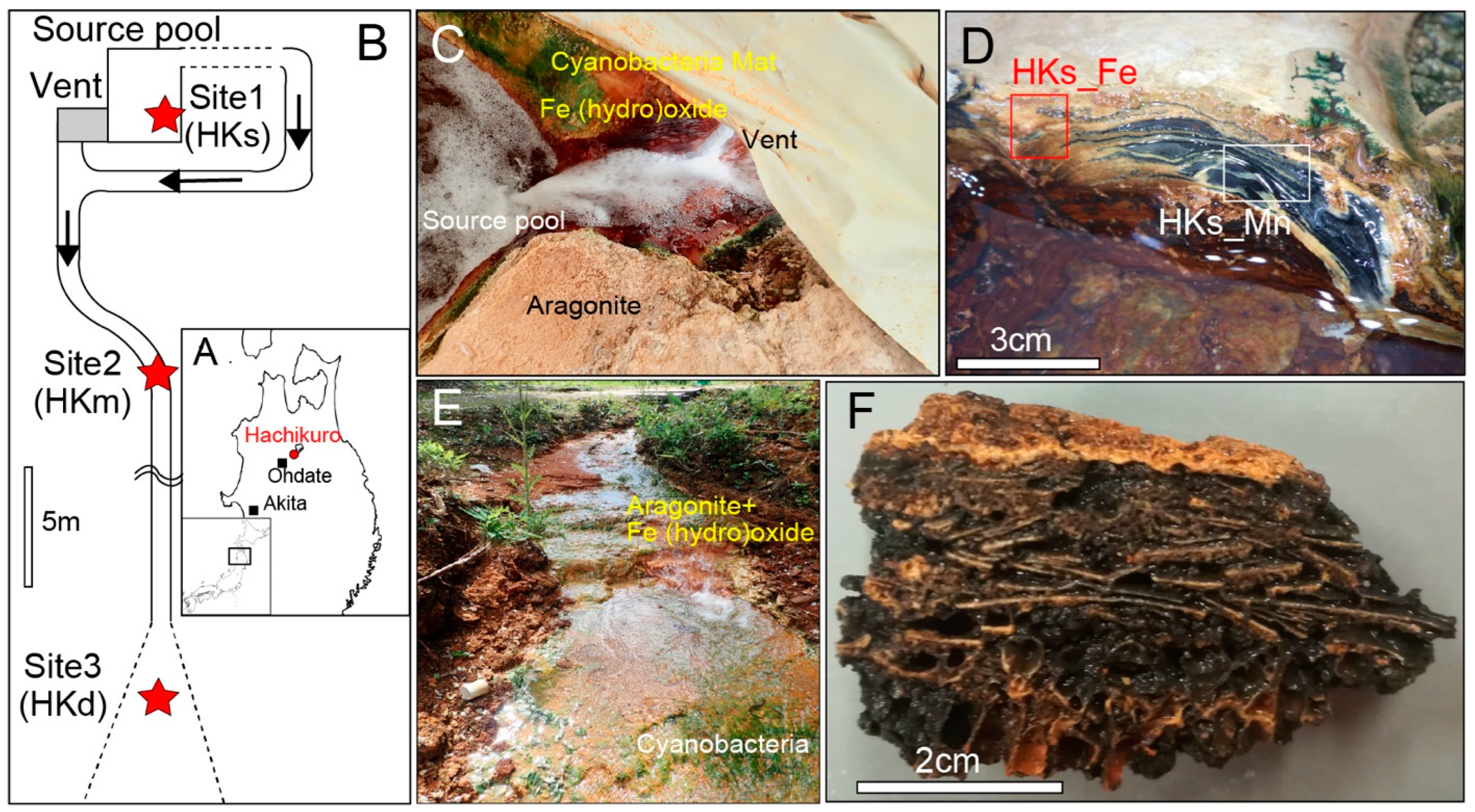
2.1. Site of Study
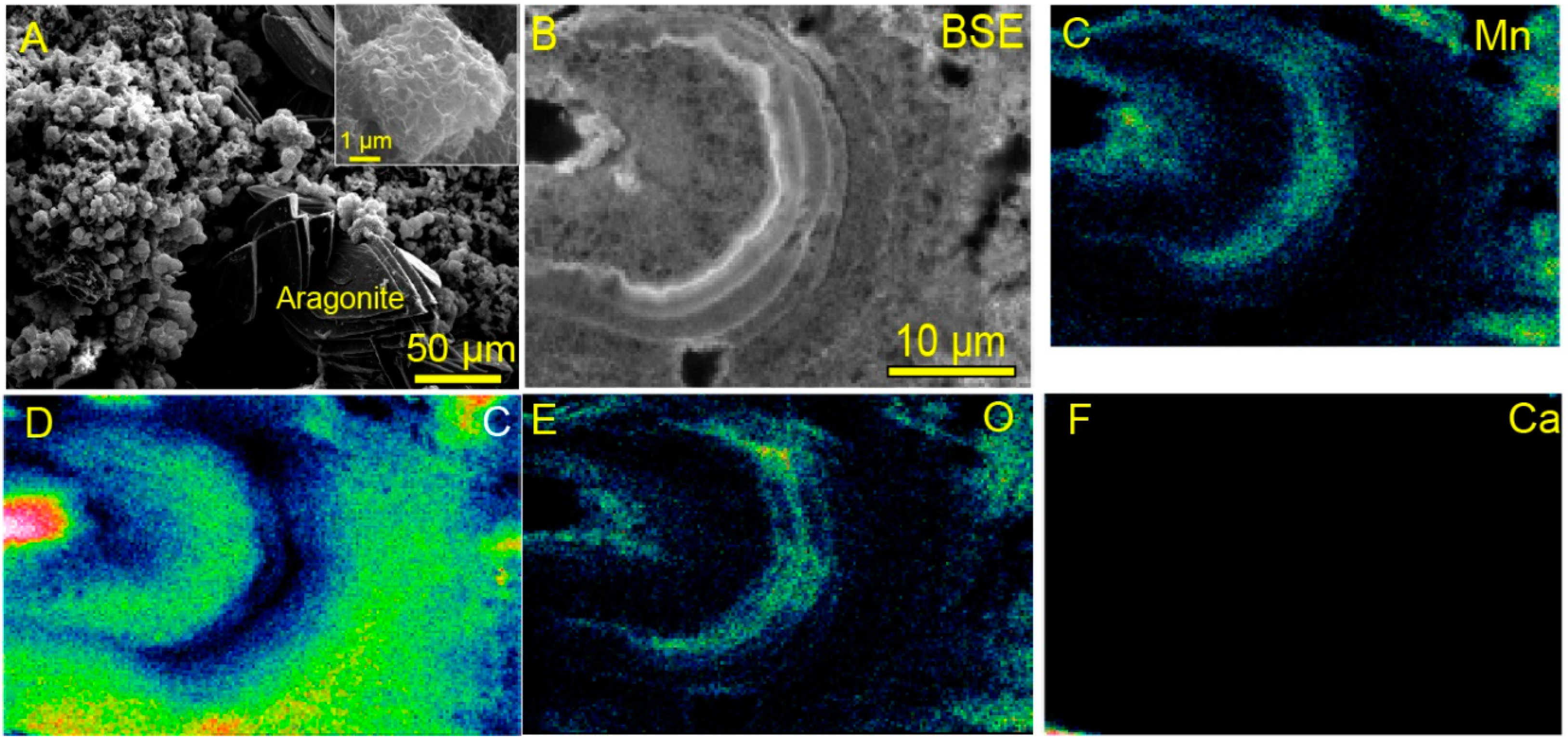
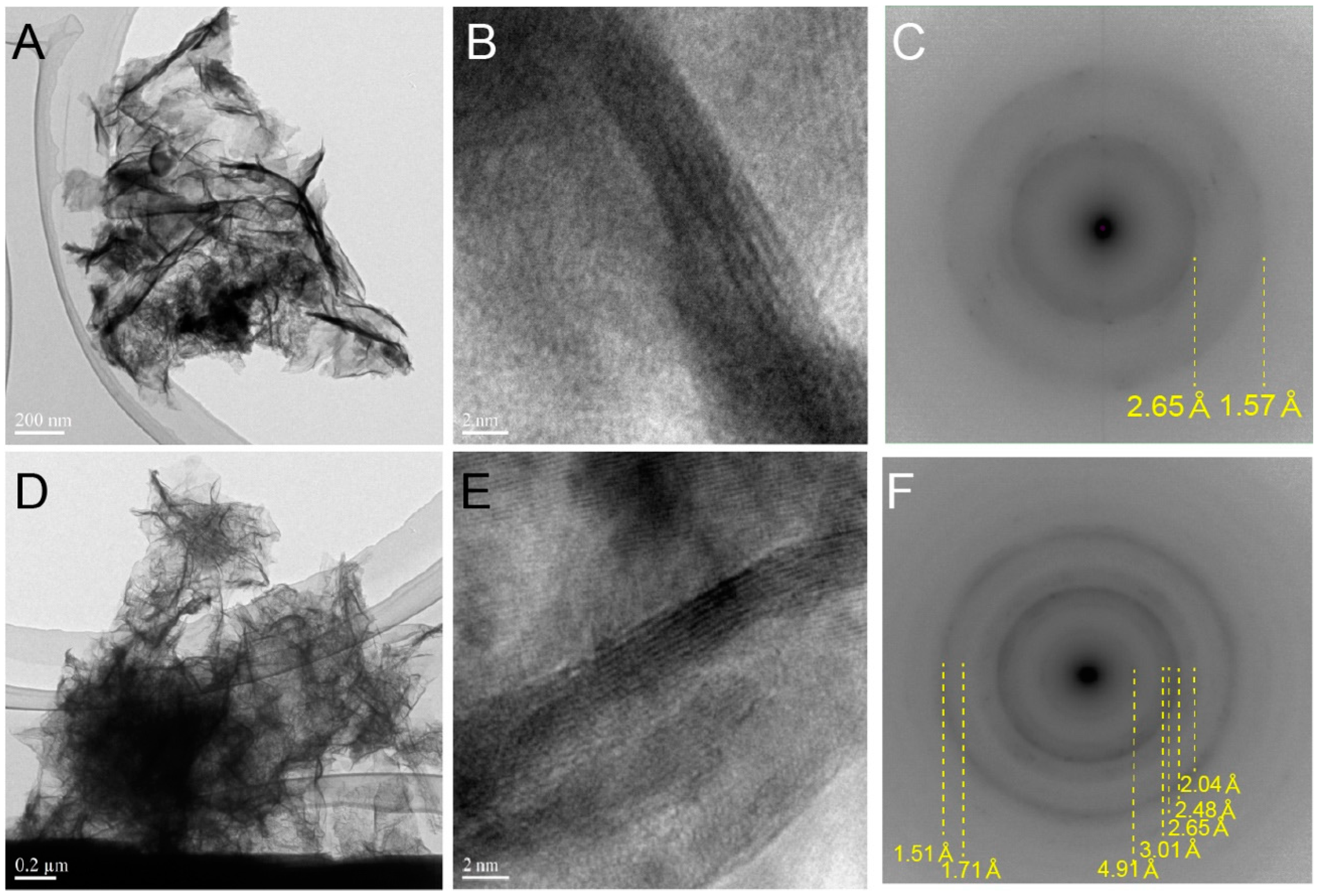
2.2. Electron Microscope Observation
2.3. Elemental and Stable Isotope Analyses
2.4. DNA Extraction, 16S rRNA Gene Amplicon Sequencing, and Shotgun Metagenomic Analyses
2.4.1. DNA Extraction and 16S rRNA Gene Amplicon Sequencing
2.4.2. Shotgun Metagenomic Analyses
3. Results and Discussion
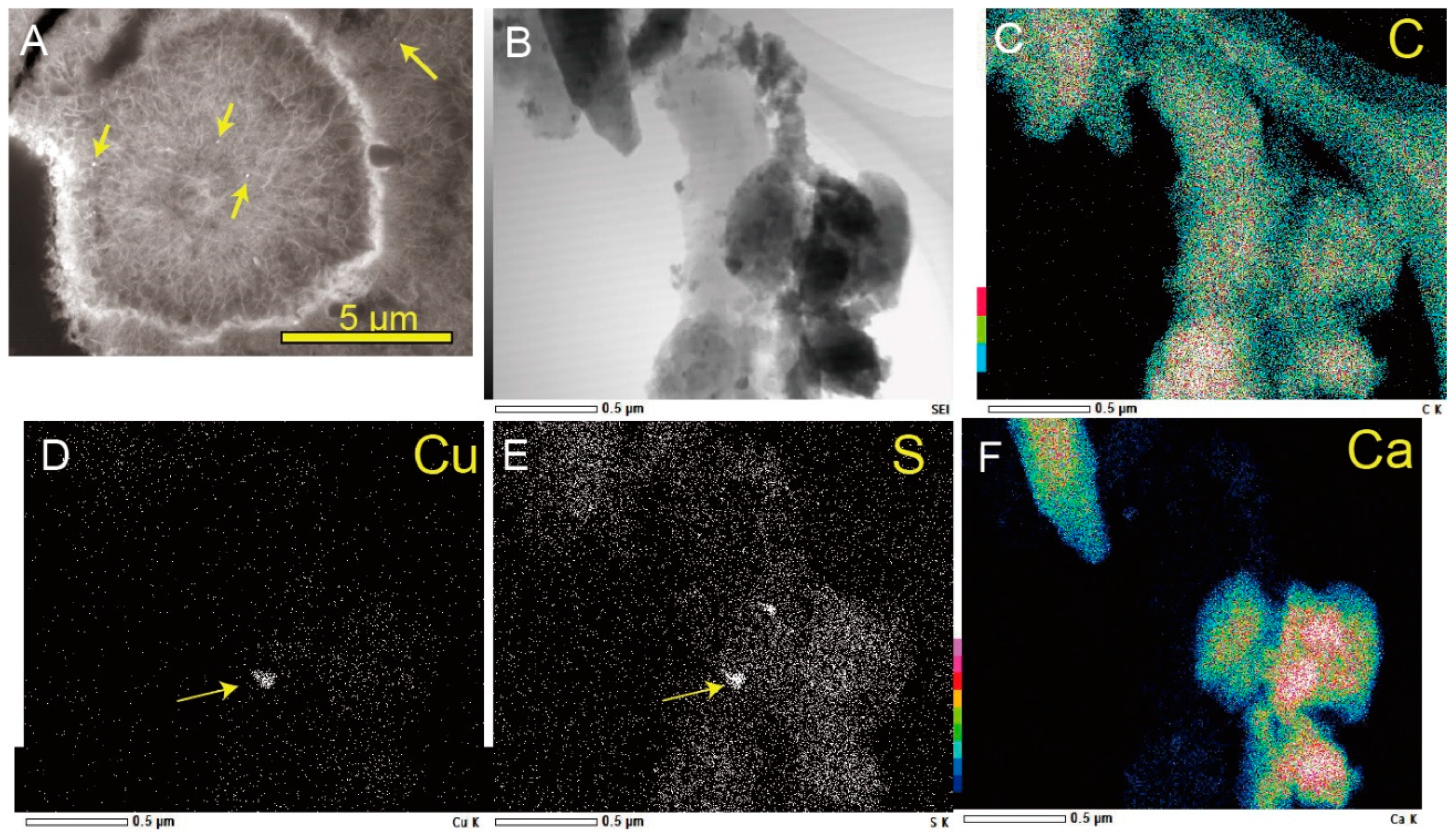
3.1. Biogenic Mn Oxides in Fe- and CO2-Rich Hot Spring
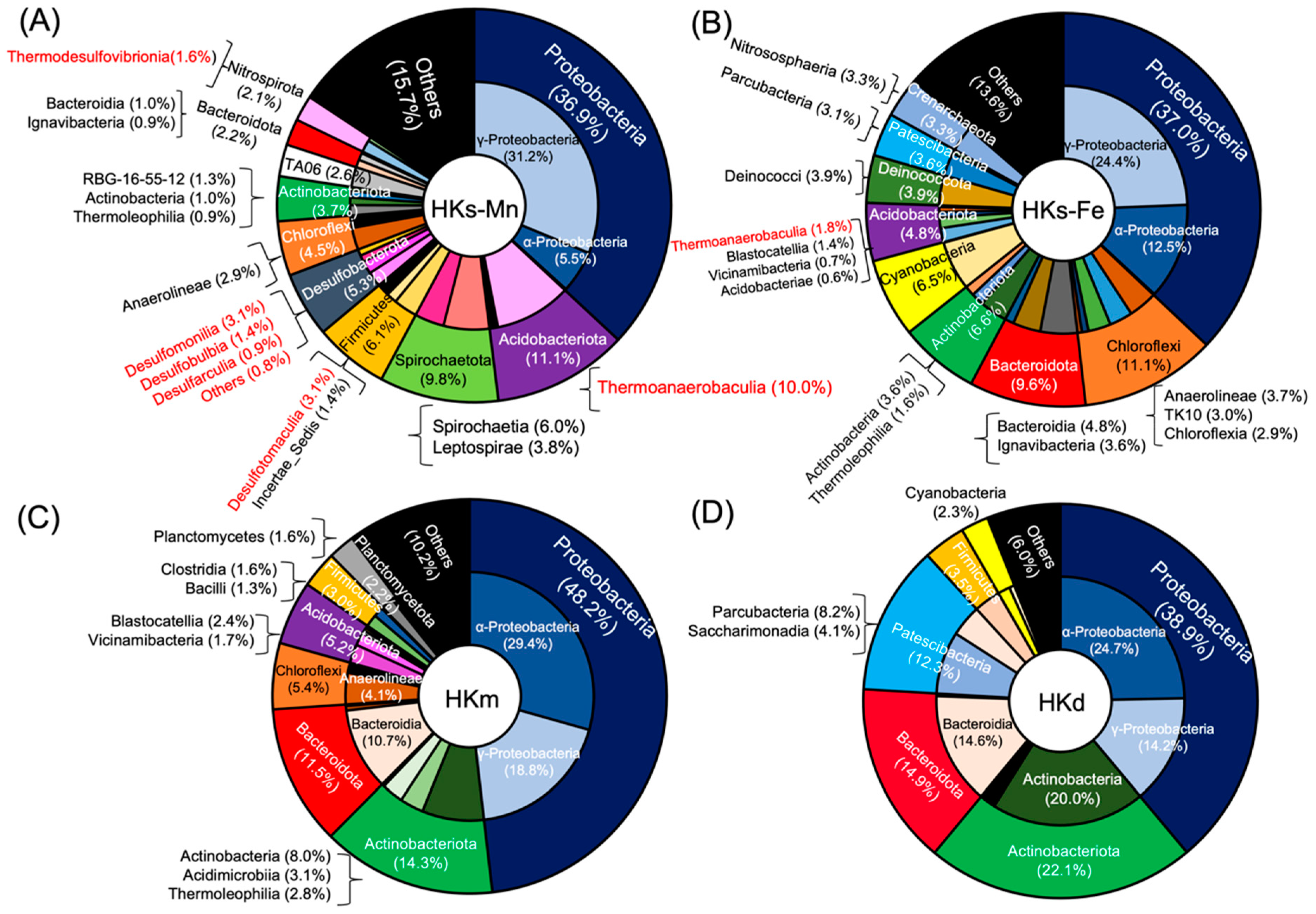
3.2. Complex Microbial Community at HK
3.3. Bacteria Associated with Mn Oxidation
3.4. MCOs Utilization for Biological Mn Oxidation in Nature
3.5. Role of Mn Oxidation in the Sinter Ecosystem
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tebo, B.M.; Bargar, J.R.; Clement, B.G.; Dick, G.J.; Murray, K.J.; Parker, D.; Verity, R.; Webb, S.M. Biogenic manganese oxides: Properties and mechanisms of formation. Annu. Rev. Earth Planet. Sci. 2004, 32, 287–328. [Google Scholar] [CrossRef] [Green Version]
- Akob, D.M.; Bohu, T.; Beyer, A.; Schäffner, F.; Händel, M.; Johnson, C.A.; Merten, D.; Büchel, G.; Totsche, K.U.; Küsel, K. Characterization of pH dependent Mn(II) oxidation strategies and formation of a bixbyite-like phase by Mesorhizobium australicum T-G1. Appl. Environ. Microbiol. 2014, 80, 5086–5097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bohu, T.; Santelli, C.M.; Akob, D.M.; Neu, T.R.; Ciobota, V.; Rösch, P.; Popp, J.; Nietzsche, S.; Küsel, K. Characterization of pH dependent Mn(II) oxidation strategies and formation of a bixbyite-like Phase by Mesorhizobium Australicum T-G1. Front. Microbiol. 2015, 6, 734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dick, G.J.; Lee, Y.E.; Tebo, B.M. Manganese(II)-oxidizing Bacillus spores in Guaymas basin hydrothermal sediments and plumes. Appl. Environ. Microbiol. 2006, 72, 3184–3190. [Google Scholar] [CrossRef] [Green Version]
- Dick, G.J.; Clement, B.G.; Webb, S.M.; Fodrie, F.J.; Bargar, J.R.; Tebo, B.M. Enzymatic microbial Mn(II) oxidation and Mn biooxide production in the Guaymas Basin deep-sea hydrothermal plume. Geochim. Cosmochim. Acta 2009, 73, 6517–6530. [Google Scholar] [CrossRef]
- Blöthe, M.; Wegorzewski, A.; Müller, C.; Simon, F.; Kuhn, T.; Schippers, A. Manganese-Cycling Microbial communities inside deep-sea manganese nodules. Environ. Sci. Technol. 2015, 49, 7692–7700. [Google Scholar] [CrossRef]
- Kato, S.; Hirai, M.; Ohkuma, M.; Suzuki, K. Microbial metabolisms in an abyssal ferromanganese crust from the Takuyo-Daigo Seamount as revealed by metagenomics. PLoS ONE 2019, 14, e0224888. [Google Scholar] [CrossRef]
- Wang, X.; Yu, M.; Wang, L.; Lin, H.; Li, B.; Xue, C.X.; Sun, H.; Zhang, X.H. Comparative genomic and metabolic analysis of manganese-oxidizing mechanisms in Celeribacter manganoxidans DY25T: Its adaptation to the environment of polymetallic nodules. Genomics 2020, 112, 2080–2091. [Google Scholar] [CrossRef]
- Takeda, M.; Kamagata, Y.; Ghiorse, W.C.; Hanada, S.; Koizumi, J.I. Caldimonas manganoxidans gen. nov., sp. nov., a poly(3-hydroxybutyrate)-degrading, manganese-oxidizing thermophile. Int. J. Syst. Evol. Microbiol. 2002, 52, 895–900. [Google Scholar]
- Okibe, N.; Maki, M.; Sasaki, K.; Hirajima, T. Mn(II)-oxidizing activity of pseudomonas sp. strain MM1 is involved in the formation of massive Mn sediments around sambe hot springs in Japan. Mater. Trans. 2013, 54, 2027–2031. [Google Scholar] [CrossRef] [Green Version]
- Shiraishi, F.; Matsumura, Y.; Chihara, R.; Okumura, T.; Itai, T.; Kashiwabara, T.; Kano, A.; Takahashi, Y. Depositional processes of microbially colonized manganese crusts, Sambe hot spring, Japan. Geochim. Cosmochim. Acta 2019, 258, 105624. [Google Scholar] [CrossRef]
- Peng, X.; Ta, K.; Chen, S.; Zhang, L.; Xu, H. Coexistence of Fe(II)- and Mn(II)-oxidizing bacteria govern the formation of deep sea umber deposits. Geochim. Cosmochim. Acta 2015, 169, 200–216. [Google Scholar] [CrossRef]
- Ossa Ossa, F.; Hofmann, A.; Wille, M.; Spangenberg, J.E.; Bekker, A.; Poulton, S.W.; Eickmann, B.; Schoenberg, R. Aerobic iron and manganese cycling in a redox-stratified Mesoarchean epicontinental sea. Earth Planet. Sci. Lett. 2018, 500, 28–40. [Google Scholar] [CrossRef] [Green Version]
- Biondi, J.C.; Polgári, M.; Gyollai, I.; Fintor, K.; Kovács, I.; Fekete, J.; Mojzsis, S.J. Biogenesis of the Neoproterozoic kremydilite manganese ores from Urucum (Brazil)–A new manganese ore type. Precambrian Res. 2020, 340, 105624. [Google Scholar] [CrossRef] [Green Version]
- Webb, S.M.; Dick, G.J.; Bargar, J.R.; Tebo, B.M. Evidence for the presence of Mn(III) intermediates in the bacterial oxidation of Mn(II). Proc. Natl. Acad. Sci. USA 2005, 102, 5558–5563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dick, G.J.; Torpey, J.W.; Beveridge, T.J.; Tebo, B.M. Direct identification of a bacterial manganese(II) oxidase, the multicopper oxidase MnxG, from spores of several different marine Bacillus species. Appl. Environ. Microbiol. 2008, 74, 1527–1534. [Google Scholar] [CrossRef] [Green Version]
- Soldatova, A.V.; Butterfield, C.; Oyerinde, O.F.; Tebo, B.M.; Spiro, T.G. Multicopper oxidase involvement in both Mn(II) and Mn(III) oxidation during bacterial formation of MnO2. J. Biol. Inorg. Chem. 2012, 17, 1151–1158. [Google Scholar] [CrossRef]
- Tebo, B.M.; Johnson, H.A.; McCarthy, J.K.; Templeton, A.S. Geomicrobiology of manganese(II) oxidation. Trends Microbiol. 2005, 13, 421–428. [Google Scholar] [CrossRef]
- Hullo, M.F.; Moszer, I.; Danchin, A.; Martin-Verstraete, I. CotA of Bacillus subtilis is a copper-dependent laccase. J. Bacteriol. 2001, 183, 5426–5430. [Google Scholar] [CrossRef] [Green Version]
- Banh, A.; Chavez, V.; Doi, J.; Nguyen, A.; Hernandez, S.; Ha, V.; Jimenez, P.; Espinoza, F.; Johnson, H.A. Manganese (Mn) Oxidation Increases Intracellular Mn in Pseudomonas putida GB-1. PLoS ONE 2013, 8, e77835. [Google Scholar] [CrossRef] [Green Version]
- Ghiorse, W.C. Biology of iron- and manganese-depositing bacteria. Annu. Rev. Microbiol. 1984, 38, 515–550. [Google Scholar] [CrossRef] [PubMed]
- Emerson, D. Ultrastructural Organization, Chemical Composition, and Manganese-Oxidizing Properties of the Sheath of Leptothrix Discophora SP-6. Ph.D. Thesis, Cornell University, Ithaca, NY, USA, 1989. [Google Scholar]
- Chernev, P.; Fischer, S.; Hoffmann, J.; Oliver, N.; Assunção, R.; Yu, B.; Burnap, R.L.; Zaharieva, I.; Nürnberg, D.J.; Haumann, M.; et al. Light-driven formation of manganese oxide by today’s photosystem II supports evolutionarily ancient manganese-oxidizing photosynthesis. Nat. Commun. 2020, 11, 6110. [Google Scholar] [CrossRef] [PubMed]
- Fischer, W.W.; Hemp, J.; Johnson, J.E. Evolution of oxygenic photosynthesis. Annu. Rev. Earth Planet. Sci. 2016, 44, 647–683. [Google Scholar] [CrossRef]
- Ridge, J.P.; Lin, M.; Larsen, E.I.; Fegan, M.; McEwan, A.G.; Sly, L.I. A Multicopper oxidase is essential for manganese oxidation and laccase-like activity in Pedomicrobium sp. ACM 3067. Environ. Microbiol. 2007, 9, 944–953. [Google Scholar] [CrossRef]
- Anderson, C.R.; Johnson, H.A.; Caputo, N.; Davis, R.E.; Torpey, J.W.; Tebo, B.M. Mn(II) Oxidation Is Catalyzed by Heme Peroxidases in “Aurantimonas Manganoxydans” Strain SI85-9A1 and Erythrobacter Sp. Strain SD-21. Appl. Environ. Microbiol. 2009, 75, 4130–4138. [Google Scholar] [CrossRef] [Green Version]
- Butterfield, C.N.; Soldatova, A.V.; Lee, S.W.; Spiro, T.G.; Tebo, B.M. Mn(II,III) oxidation and MnO2 mineralization by an expressed bacterial multicopper oxidase. Proc. Natl. Acad. Sci. USA 2013, 110, 11731–11735. [Google Scholar] [CrossRef] [Green Version]
- Geszvain, K.; McCarthy, J.K.; Tebo, B.M. Elimination of Manganese(II, III) Oxidation in Pseudomonas Putida GB-1 by a Double Knockout of Two Putative Multicopper Oxidase Genes. Appl. Environ. Microbiol. 2013, 79, 357–366. [Google Scholar] [CrossRef] [Green Version]
- Andeer, P.F.; Learman, D.R.; McIlvin, M.; Dunn, J.A.; Hansel, C.M. Extracellular haem peroxidases mediate Mn(II) oxidation in a marine Roseobacter bacterium via superoxide production. Environ. Microbiol. 2015, 17, 3925–3936. [Google Scholar] [CrossRef]
- Chaput, D.L.; Fowler, A.J.; Seo, O.; Duhn, K.; Hansel, C.M.; Santelli, C.M. Mn oxide formation by phototrophs: Spatial and temporal patterns, with evidence of an enzymatic superoxide-mediated pathway. Sci. Rep. 2019, 9, 18244. [Google Scholar]
- Granja-Travez, R.S.; Wilkinson, R.C.; Persinoti, G.F.; Squina, F.M.; Fülöp, V.; Bugg, T.D.H. Structural and functional characterisation of multi-copper oxidase CueO from lignin-degrading bacterium Ochrobactrum sp. reveal its activity towards lignin model compounds and lignosulfonate. FEBS J. 2018, 285, 1684–1700. [Google Scholar] [CrossRef] [Green Version]
- Hakulinen, N.; Kiiskinen, L.L.; Kruus, K.; Saloheimo, M.; Paanen, A.; Koivula, A.; Rouvinen, J. Crystal structure of a laccase from melanocarpus albomyces with an intact trinuclear copper site. Nat. Struct. Biol. 2002, 9, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Romano, C.A.; Zhou, M.; Song, Y.; Wysocki, V.H.; Dohnalkova, A.C.; Kovarik, L.; Paša-Tolić, L.; Tebo, B.M. Biogenic manganese oxide nanoparticle formation by a multimeric multicopper oxidase Mnx. Nat. Commun. 2017, 8, 746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crichton, R.R.; Pierre, J.L. Old Iron, Young Copper: From Mars to Venus. BioMetals 2001, 14, 99–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Requena, J.R.; Groth, D.; Legname, G.; Stadtman, E.R.; Prusiner, S.B.; Levine, R.L. Copper-catalyzed oxidation of the recombinant SHa(29-231) prion protein. Proc. Natl Acad. Sci. USA 2001, 98, 7170–7175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Outten, F.W.; Huffman, D.L.; Hale, J.A.; O’Halloran, T.V. The Independent cue and cus Systems Confer Copper Tolerance during Aerobic and Anaerobic Growth in Escherichia Coli. J. Biol. Chem. 2001, 276, 30670–30677. [Google Scholar] [CrossRef] [Green Version]
- Grass, G.; Rensing, C. CueO Is a Multi-Copper Oxidase That Confers Copper Tolerance in Escherichia Coli. Biochem. Biophys. Res. Commun. 2001, 286, 902–908. [Google Scholar] [CrossRef]
- Mita, N.; Miura, H. Evidence of microbial activity in the formation of manganese wads at the Asahidake hot spring in Hokkaido, Japan. Resour. Geol. 2003, 53, 233–238. [Google Scholar] [CrossRef]
- Yu, H.; Leadbetter, J.R. Bacterial chemolithoautotrophy via manganese oxidation. Nature 2020, 583, 453–458. [Google Scholar] [CrossRef]
- Daye, M.; Klepac-Ceraj, V.; Pajusalu, M.; Rowland, S.; Farrell-Sherman, A.; Beukes, N.; Tamura, N.; Fournier, G.; Bosak, T. Light-Driven anaerobic microbial oxidation of manganese. Nature 2019, 576, 311–314. [Google Scholar] [CrossRef]
- Seto, Y.; Ohtsuka, M. ReciPro: Free and Open-Source Multipurpose Crystallographic Software Integrating a Crystal Model Database and Viewer, Diffraction and Microscopy Simulators, and Diffraction Analysis Tools. J. Appl. Crystallogr. 2022, 55, 397–410. [Google Scholar] [CrossRef]
- Tsukamoto, Y.; Nonaka, K.; Ishida, A.; Kakegawa, T. Geochemical and mineralogical studies of ca. 12 Ma hydrothermal manganese-rich rocks in the Hokuroku district in Japan. Ore Geol. Rev. 2020, 121, 103539. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Piazza, A.; Casalini, L.C.; Pacini, V.A.; Sanguinetti, G.; Ottado, J.; Gottig, N. Environmental Bacteria Involved in Manganese(II) Oxidation and Removal from Groundwater. Front. Microbiol. 2019, 10, 119. [Google Scholar] [CrossRef] [Green Version]
- Marcus, D.N.; Pinto, A.; Anantharaman, K.; Ruberg, S.A.; Kramer, E.L.; Raskin, L.; Dick, G.J. Diverse manganese(II)-oxidizing bacteria are prevalent in drinking water systems. Environ. Microbiol. Rep. 2017, 9, 120–128. [Google Scholar] [CrossRef]
- Francis, C.A.; Tebo, B.M. Enzymatic Manganese(II) oxidation by metabolically dormant spores of diverse Bacillus Species. Appl. Environ. Microbiol. 2002, 68, 874–880. [Google Scholar] [CrossRef] [Green Version]
- Templeton, A.S.; Staudigel, H.; Tebo, B.M. Diverse Mn(II)-oxidizing bacteria isolated from submarine basalts at Loihi eamount. Geomicrobiol. J. 2005, 22, 127–139. [Google Scholar] [CrossRef]
- Carmichael, M.J.; Carmichael, S.K.; Santelli, C.M.; Strom, A.; Bräuer, S.L. Mn(II)-oxidizing bacteria are abundant and environmentally relevant members of ferromanganese deposits in caves of the upper tennessee river basin. Geomicrobiol. J. 2013, 30, 779–800. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, Y.; Qin, Z.; Luo, P.; Ma, Z.; Tan, M.; Kang, H.; Huang, Z. A novel manganese oxidizing bacterium-Aeromonas hydrophila strain DS02: Mn(II) oxidization and biogenic Mn oxides generation. J. Hazard. Mater. 2019, 367, 539–545. [Google Scholar] [CrossRef]
- Yang, W.; Zhang, Z.; Zhang, Z.; Chen, H.; Liu, J.; Ali, M.; Liu, F.; Li, L. Population Structure of Manganese-Oxidizing Bacteria in Stratified Soils and Properties of Manganese Oxide Aggregates under Manganese-Complex Medium Enrichment. PLoS ONE 2013, 8, e73778. [Google Scholar] [CrossRef] [Green Version]
- Caspi, R.; Haygood, M.G.; Tebo, B.M. Unusual ribulose-1,5-bisphosphate carboxylase/oxygenase genes from a marine manganese-oxidizing bacterium. Microbiology 1996, 142, 2549–2559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caspi, R.; Tebo, B.M.; Haygood, M.G. c-type cytochromes and manganese oxidation in Pseudomonas putida MnB1. Appl. Environ. Microbiol. 1998, 64, 3549–3555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cerrato, J.M.; Falkinham, J.O.; Dietrich, A.M.; Knocke, W.R.; McKinney, C.W.; Pruden, A. Manganese-oxidizing and -reducing microorganisms isolated from biofilms in chlorinated drinking water systems. Water Res. 2010, 44, 3935–3945. [Google Scholar] [CrossRef] [PubMed]
- Santelli, C.M.; Chaput, D.L.; Hansel, C.M. Microbial Communities Promoting Mn(II) Oxidation in Ashumet Pond, a Historically Polluted Freshwater Pond Undergoing Remediation. Geomicrobiol. J. 2014, 31, 605–616. [Google Scholar] [CrossRef]
- Wang, X.; Wiens, M.; Divekar, M.; Grebenjuk, V.A.; Schröder, H.C.; Batel, R.; Müller, W.E.G. I Isolation and characterization of a Mn(II)-oxidizing Bacillus strain from the demosponge Suberites domuncula. Mar. Drugs 2011, 9, 1–28. [Google Scholar] [CrossRef] [Green Version]
- Rajasabapathy, R.; Mohandass, C.; Dastager, S.G.; Liu, Q.; Li, W.J.; Colaço, A. Citreicella manganoxidans sp. nov., a novel manganese oxidizing bacterium isolated from a shallow water hydrothermal vent in Espalamaca (Azores). Antonie van Leeuwenhoek 2015, 108, 1433–1439. [Google Scholar] [CrossRef]
- Mayhew, L.E.; Swanner, E.D.; Martin, A.P.; Templeton, A.S. Phylogenetic relationships and functional genes: Distribution of a gene (mnxG) encoding a putative manganese-oxidizing enzyme in Bacillus species. Appl. Environ. Microbiol. 2008, 74, 7265–7271. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Liu, Y.; Wang, Y.; Dai, X.; Zhang, X.H. Celeribacter manganoxidans sp. Nov., A manganese-oxidizing bacterium isolated from deep-sea sediment of a polymetallic nodule province. Int. J. Syst. Evol. Microbiol. 2015, 65, 4180–4185. [Google Scholar] [CrossRef]
- Sujith, P.P.; Mourya, B.S.; Krishnamurthi, S.; Meena, R.M.; Loka Bharathi, P.A. Mobilization of manganese by basalt associated Mn(II)-oxidizing bacteria from the Indian Ridge System. Chemosphere 2014, 95, 486–495. [Google Scholar] [CrossRef]
- Corstjens, P.L.A.M.; Devrind, J.P.M.; Goosen, T.; de Vrind-de Jong, E.W. Identification and molecular analysis of the leptothrix discophora SS-1 mofA gene, a gene putatively encoding a manganese-oxidizing protein with copper domains. Geomicrobiol. J. 1997, 14, 91–108. [Google Scholar] [CrossRef]
- Brouwers, G.J.; Corstjens, P.L.A.M.; De Vrind, J.P.M.; Verkamman, A.; De Kuyper, M.; De Vrind-De Jong, E.W. Stimulation of Mn2+ oxidation in leptothrix discophora SS-1 by Cu2+ and sequence analysis of the region flanking the gene encoding putative multicopper oxidase mofA. Geomicrobiol. J. 2000, 17, 25–33. [Google Scholar]
- Liao, S.; Zhou, J.; Wang, H.; Chen, X.; Wang, H.; Wang, G. Arsenite Oxidation Using Biogenic Manganese Oxides Produced by a Deep-Sea Manganese-Oxidizing Bacterium, Marinobacter sp. MnI7-9. Geomicrobiol. J. 2013, 30, 150–159. [Google Scholar] [CrossRef]
- Sly, L.I.; Vullapa, A.; Hodgkinson, M.C. Pedomicrobium manganicum from rinking-Water Distribution Systems with Manganese-Related “Dirty Water” Problems. Syst. Appl. Microbiol. 1988, 11, 75–184. [Google Scholar] [CrossRef]
- Bräuer, S.L.; Adams, C.; Kranzler, K.; Murphy, D.; Xu, M.; Zuber, P.; Simon, H.M.; Baptista, A.M.; Tebo, B.M. Culturable Rhodobacter and Shewanella species are abundant in estuarine turbidity maxima of the Columbia River. Environ. Microbiol. 2011, 13, 589–603. [Google Scholar] [CrossRef]
- Hansel, C.M.; Francis, C.A. Coupled photochemical and enzymatic Mn(II) oxidation pathways of a planktonic Roseobacter-like bacterium. Appl. Environ. Microbiol. 2006, 72, 3543–3549. [Google Scholar] [CrossRef] [Green Version]
- Mariner, R.; Johnson, D.B.; Hallberg, K.B. Characterisation of an attenuation system for the remediation of Mn(II) contaminated waters. Hydrometallurgy 2008, 94, 100–104. [Google Scholar] [CrossRef]
- Wright, M.H.; Farooqui, S.M.; White, A.R.; Greene, A.C. Production of manganese oxide nanoparticles by Shewanella Species. Appl. Environ. Microbiol. 2016, 82, 5402–5409. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Liu, Y.; Shi, X.; Wang, Y.; Zheng, Y.; Dai, X.; Zhang, X.H. Xuhuaishuia manganoxidans Gen. Nov., Sp. Nov., a manganese-oxidizing bacterium isolated from deep-sea sediments from the Pacific polymetallic nodule province. Int. J. Syst. Evol. Microbiol. 2016, 66, 1521–1526. [Google Scholar] [CrossRef] [Green Version]
- Uritskiy, G.V.; DiRuggiero, J.; Taylor, J. MetaWRAP—A flexible pipeline for genome-resolved metagenomic data analysis. Microbiome 2018, 6, 158. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [Green Version]
- Kang, D.D.; Li, F.; Kirton, E.; Thomas, A.; Egan, R.; An, H.; Wang, Z. MetaBAT 2: An adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 2019, 7, e7359. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 2016, 32, 605–607. [Google Scholar] [CrossRef] [PubMed]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Loman, N.J.; Andersson, A.F.; Quince, C. CONCOCT: Clustering CONtigs on COverage and ComposiTion. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowers, R.M.; Kyrpides, N.C.; Stepanauskas, R.; Harmon-Smith, M.; Doud, D.; Reddy, T.B.K.; Schulz, F.; Jarett, J.; Rivers, A.R.; Eloe-Fadrosh, E.A.; et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017, 35, 725–731. [Google Scholar] [CrossRef] [Green Version]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Chaumeil, P.A.; Rinke, C.; Mussig, A.J.; Hugenholtz, P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020, 38, 1079–1086. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Deng, L.; Huang, L.; Guo, S.; Liu, F.; He, J. Catalytic oxidation of manganese(II) by multicopper oxidase CueO and characterization of the biogenic Mn oxide. Water Res. 2014, 56, 304–313. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2018, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Takami, H.; Taniguchi, T.; Arai, W.; Takemoto, K.; Moriya, Y.; Goto, S. An automated system for evaluation of the potential functionome: MAPLE Version 2.1.0. DNA Res. 2016, 23, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Tanizawa, Y.; Fujisawa, T.; Kaminuma, E.; Nakamura, Y.; Arita, M. DFAST and DAGA: Web-based integrated genome annotation tools and resources. Biosci. Microbiota Food Health 2016, 35, 173–184. [Google Scholar] [CrossRef] [Green Version]
- Tanizawa, Y.; Fujisawa, T.; Nakamura, Y. DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics 2018, 34, 1037–1039. [Google Scholar] [CrossRef] [Green Version]
- Villalobos, M.; Toner, B.; Bargar, J.; Sposito, G. Characterization of the manganese oxide produced by Pseudomonas putida strain MnB1. Geochim. Cosmochim. Acta 2003, 67, 2649–2662. [Google Scholar] [CrossRef]
- Feng, X.H.; Zhu, M.; Ginder-Vogel, M.; Ni, C.; Parikh, S.J.; Sparks, D.L. Formation of nano-crystalline todorokite from biogenic Mn oxides. Geochim. Cosmochim. Acta 2010, 74, 3232–3245. [Google Scholar] [CrossRef]
- Su, J.; Bao, P.; Bai, T.; Deng, L.; Wu, H.; Liu, F.; He, J. CotA, a Multicopper Oxidase from Bacillus pumilus WH4, Exhibits Manganese-Oxidase Activity. PLoS ONE 2013, 8, e60573. [Google Scholar] [CrossRef] [Green Version]
- Bargar, J.R.; Fuller, C.C.; Marcus, M.A.; Brearley, A.J.; Perez De la Rosa, M.; Webb, S.M.; Caldwell, W.A. Structural characterization of terrestrial microbial Mn oxides from Pinal Creek, AZ. Geochim. Cosmochim. Acta 2009, 73, 889–910. [Google Scholar] [CrossRef] [Green Version]
- Learman, D.R.; Wankel, S.D.; Webb, S.M.; Martinez, N.; Madden, A.S.; Hansel, C.M. Coupled biotic-abiotic Mn(II) oxidation pathway mediates the formation and structural evolution of biogenic Mn oxides. Geochim. Cosmochim. Acta 2011, 75, 6048–6063. [Google Scholar] [CrossRef]
- Burns, R.G.; Burns, V.M. Authigenic todorokite and phillipsite inside deep-sea manganese nodules. Am. Miner. 1978, 63, 827–831. [Google Scholar]
- Takahashi, Y.; Manceau, A.; Geoffroy, N.; Marcus, M.A.; Usui, A. Chemical and structural control of the partitioning of Co, Ce, and Pb in marine ferromanganese oxides. Geochim. Cosmochim. Acta 2007, 71, 984–1008. [Google Scholar] [CrossRef] [Green Version]
- Mellin, T.A.; Lei, G. Stabilization of 10Å-manganates by interlayer cations and hydrothermal treatment: Implications for the mineralogy of marine manganese concretions. Mar. Geol. 1993, 115, 67–83. [Google Scholar] [CrossRef]
- Moy, Y.P.; Neilan, B.A.; Foster, L.J.R.; Madgwick, J.C.; Rogers, P.L. Screening, identification and kinetic characterization of a bacterium for Mn(II) uptake and oxidation. Biotechnol. Lett 2003, 25, 1407–1413. [Google Scholar] [CrossRef] [PubMed]
- Mansor, M.; Berti, D.; Hochella, M.F.; Murayama, M.; Xu, J. Phase, Morphology, elemental composition, and formation mechanisms of biogenic and abiogenic Fe-Cu-sulfide nanoparticles: A comparative study on their occurrences under anoxic conditions. Am. Mineral. 2019, 104, 703–717. [Google Scholar] [CrossRef]
- Goldberg, E.D. Marine geochemistry. I. Chemical scavengers of the sea. J. Geol. 1954, 62, 249–265. [Google Scholar] [CrossRef]
- Hein, J.R.; Conrad, T.A.; Frank, M.; Christl, M.; Sager, W.W. Copper-nickel-rich, amalgamated ferromanganese crust-nodule deposits from Shatsky Rise, NW Pacific. Geochem. Geophys. Geosyst. 2012, 13, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Moreau, J.W.; Weber, P.K.; Martin, M.C.; Gilbert, B.; Hutcheon, I.D.; Banfield, J.F. Extracellular proteins limit the dispersal of biogenic nanoparticles. Science 2007, 316, 1600–1603. [Google Scholar] [CrossRef] [Green Version]
- Estes, E.R.; Andeer, P.F.; Nordlund, D.; Wankel, S.D.; Hansel, C.M. Biogenic manganese oxides as reservoirs of organic carbon and proteins in terrestrial and marine environments. Geobiology 2017, 15, 158–172. [Google Scholar] [CrossRef] [Green Version]
- Johnson, K.; Purvis, G.; Lopez-Capel, E.; Peacock, C.; Gray, N.; Wagner, T.; März, C.; Bowen, L.; Ojeda, J.; Finlay, N.; et al. Towards a mechanistic understanding of carbon stabilization in manganese oxides. Nat. Commun. 2015, 6, 7628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schippers, A.; Neretin, L.N.; Lavik, G.; Leipe, T.; Pollehne, F. Manganese(II) oxidation driven by lateral oxygen intrusions in the Western Black Sea. Geochim. Cosmochim. Acta 2005, 69, 2241–2252. [Google Scholar] [CrossRef]
- Clement, B.G.; Luther, G.W.; Tebo, B.M. Rapid, oxygen-dependent microbial Mn(II) oxidation kinetics at sub-micromolar oxygen concentrations in the Black Sea suboxic zone. Geochim. Cosmochim. Acta 2009, 73, 1878–1889. [Google Scholar] [CrossRef]
- Wang, X.; Schröder, H.C.; Wiens, M.; Schloßmacher, U.; Müller, W.E.G. Manganese/polymetallic nodules: Micro-structural characterization of exolithobiontic- and endolithobiontic microbial biofilms by scanning electron microscopy. Micron 2009, 40, 350–358. [Google Scholar] [CrossRef]
- Antony, R.; Sujith, P.P.; Fernandes, S.O.; Verma, P.; Khedekar, V.D.; Loka Bharathi, P.A. Cobalt immobilization by manganese oxidizing bacteria from the Indian Ridge System. Curr. Microbiol. 2011, 62, 840–849. [Google Scholar] [CrossRef]
- Cowen, J.P.; Bertram, M.A.; Baker, E.T.; Feely, R.A.; Massoth, G.J.; Summit, M. Geomicrobial transformation of manganese in Gorda Ridge Event Plumes. Deep-Sea Res. Part. II 1998, 45, 2713–2737. [Google Scholar] [CrossRef]
- Cowen, J.P.; Wilcox, S.M. The association of iron and manganese with bacteria on marine macroparticulate material. Science 1984, 224, 1340–1342. [Google Scholar] [CrossRef]
- Moffett, J.W. The importance of microbial Mn oxidation in the upper ocean: A comparison of the Sargasso Sea and Equatorial Pacific. Deep-Sea Res. Part. I 1997, 44, 1277–1291. [Google Scholar] [CrossRef]
- Northup, D.E.; Barns, S.M.; Yu, L.E.; Spilde, M.N.; Schelble, R.T.; Dano, K.E.; Crossey, L.J.; Connolly, C.A.; Boston, P.J.; Natvig, D.O.; et al. Diverse microbial communities inhabiting ferromanganese deposits in Lechuguilla and Spider Caves. Environ. Microbiol. 2003, 5, 1071–1086. [Google Scholar] [CrossRef]
- Hungate, B.; Danin, A.; Pellerin, N.B.; Stemmler, J.; Kjellander, P.; Adams, J.B.; Staley, J.T. Characterization of manganese-oxidizing (MnII-MnIV) bacteria from Negev Desert Rock Varnish: Implications in Desert Varnish Formation. Can. J. Microbiol. 1987, 33, 939–943. [Google Scholar] [CrossRef]
- Cox, T.L.; Sly, L.I. Phylogenetic relationships and uncertain taxonomy of Pedomicrobium Species. Int. J. Syst. Bacteriol. 1997, 47, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Larsen, E.I.; Sly, L.I.; McEwan, A.G. Manganese(II) adsorption and oxidation by whole cells and a membrane fraction of Pedomicrobium Sp. ACM 3067. Arch. Microbiol. 1999, 171, 257–264. [Google Scholar] [CrossRef]
- Francis, C.A.; Casciotti, K.L.; Tebo, B.M. Localization of Mn(II)-oxidizing activity and the putative multicopper oxidase, MnxG, to the exosporium of the marine Bacillus sp. Strain SG-1. Arch. Microbiol. 2002, 178, 450–456. [Google Scholar] [CrossRef] [PubMed]
- Brouwers, G.J.; De Vrind, J.P.M.; Corstjens, P.L.A.M.; Cornelis, P.; Baysse, C.; De Vrind-De Jong, E.W. CumA, a gene encoding a multicopper oxidase, is involved in Mn2+ oxidation in Pseudomonas putida GB-1. Appl. Environ. Microbiol. 1999, 65, 1762–1768. [Google Scholar] [CrossRef] [Green Version]
- Ishida, K.; Tsukamoto, Y.; Horitani, M.; Ogawa, T.; Tanaka, Y. Biochemical properties of CumA multicopper oxidase from plant pathogen, Pseudomonas syringae. Biosci. Biotechnol. Biochem. 2021, 85, 1995–2002. [Google Scholar] [CrossRef] [PubMed]
- Geszvain, K.; Smesrud, L.; Tebo, B.M. Identification of a third Mn(II) oxidase enzyme in Pseudomonas putida GB-1. Appl. Environ. Microbiol. 2016, 82, 3774–3782. [Google Scholar] [CrossRef] [Green Version]
- Liang, J.; Bai, Y.; Men, Y.; Qu, J. Microbe-microbe interactions trigger Mn(II)-oxidizing gene expression. ISME J. 2017, 11, 67–77. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsukamoto, Y.; Kakegawa, T. Mineralogical and Genomic Constraints on the Origin of Microbial Mn Oxide Formation in Complexed Microbial Community at the Terrestrial Hot Spring. Life 2022, 12, 816. https://doi.org/10.3390/life12060816
Tsukamoto Y, Kakegawa T. Mineralogical and Genomic Constraints on the Origin of Microbial Mn Oxide Formation in Complexed Microbial Community at the Terrestrial Hot Spring. Life. 2022; 12(6):816. https://doi.org/10.3390/life12060816
Chicago/Turabian StyleTsukamoto, Yuya, and Takeshi Kakegawa. 2022. "Mineralogical and Genomic Constraints on the Origin of Microbial Mn Oxide Formation in Complexed Microbial Community at the Terrestrial Hot Spring" Life 12, no. 6: 816. https://doi.org/10.3390/life12060816





