Structural Analysis of Human Fascin-1: Essential Protein for Actin Filaments Bundling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Making Constructs and the Purification of the Recombinant WT Fascin and Its Mutants
2.2. F-Actin Bundling and Binding Assay
2.3. Data Collection Using Transmission Electron Microscope
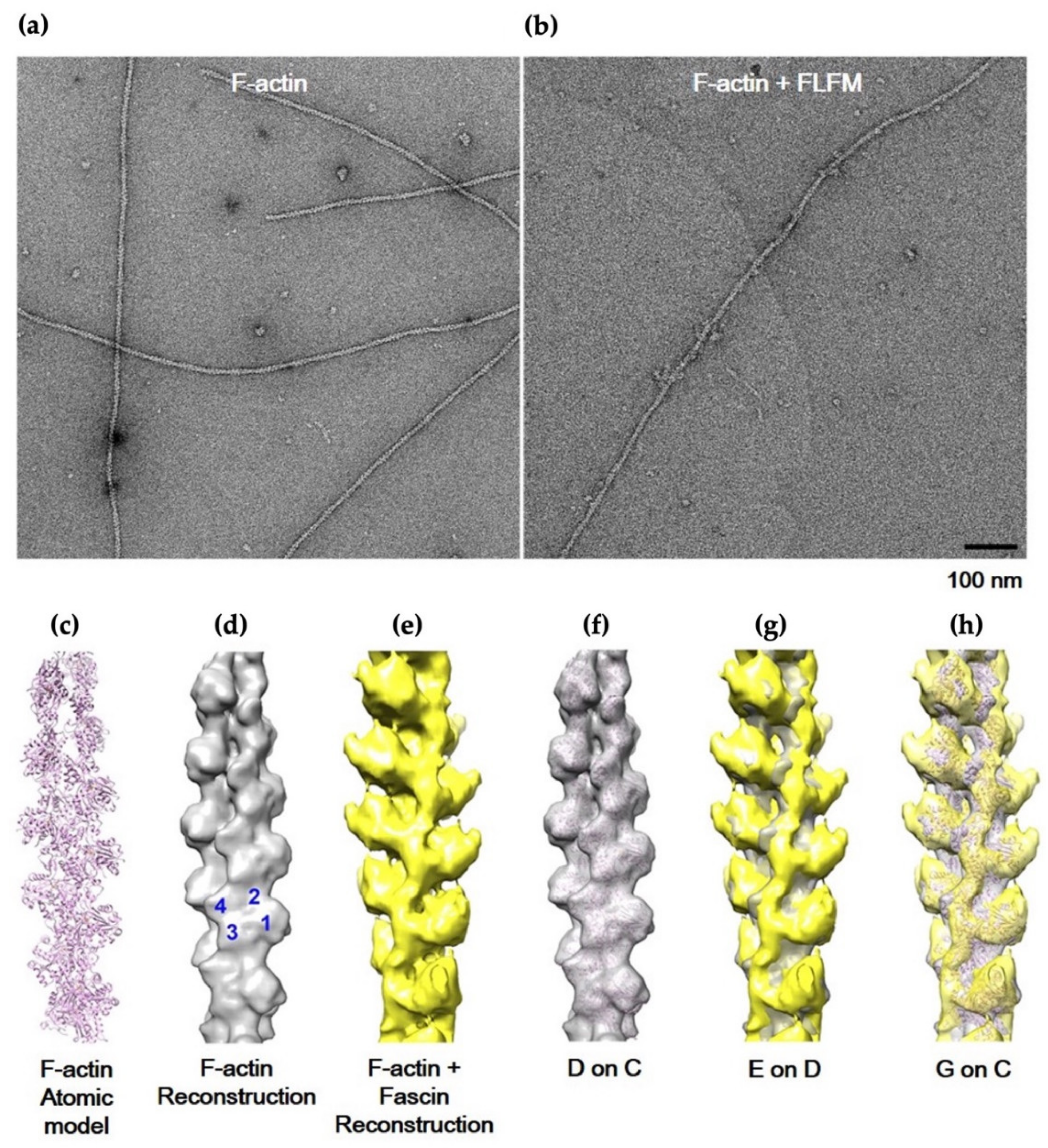
2.4. Image Processing
3. Results
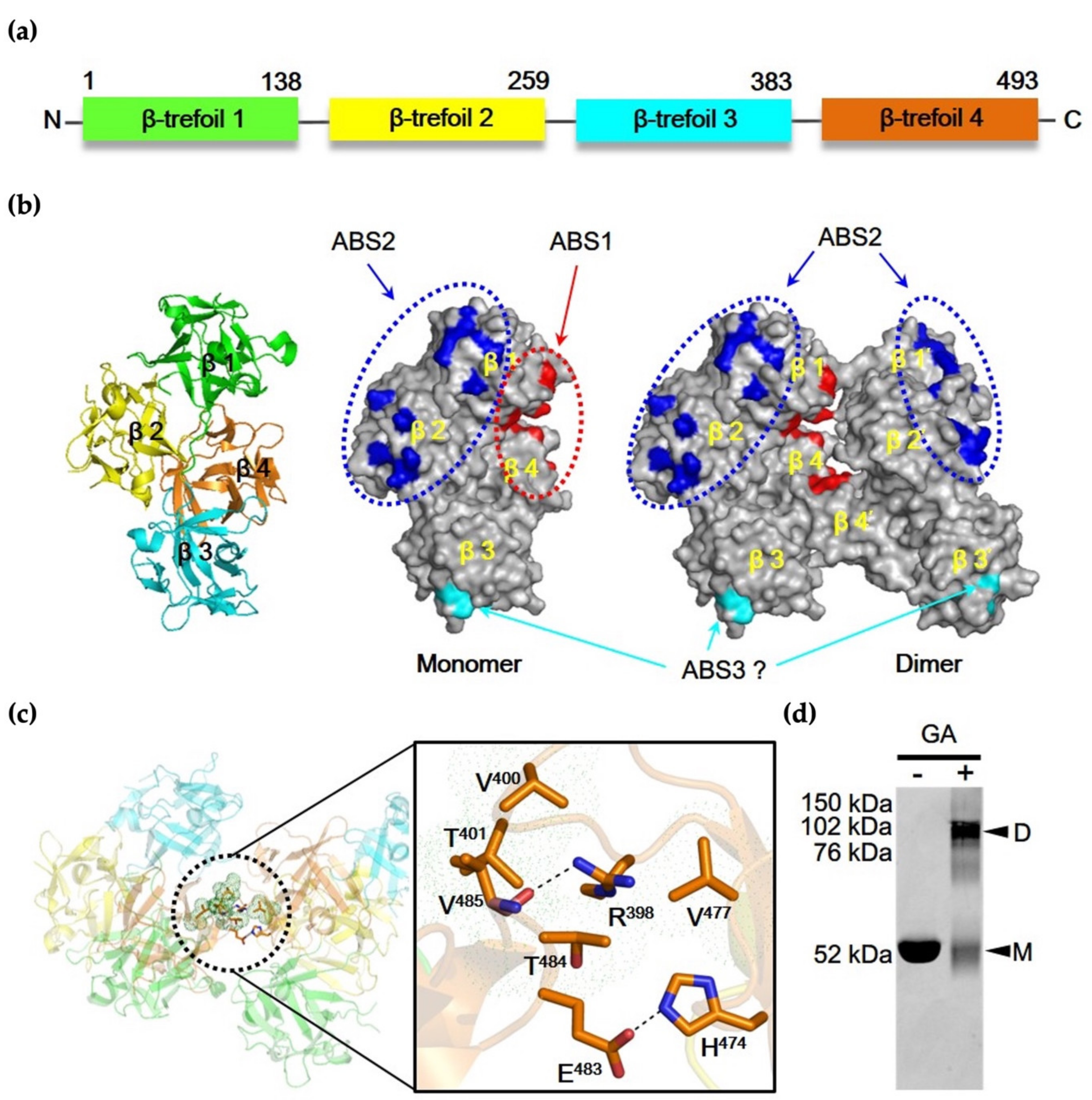
3.1. Two-Major Actin-Binding Sites of Fascin
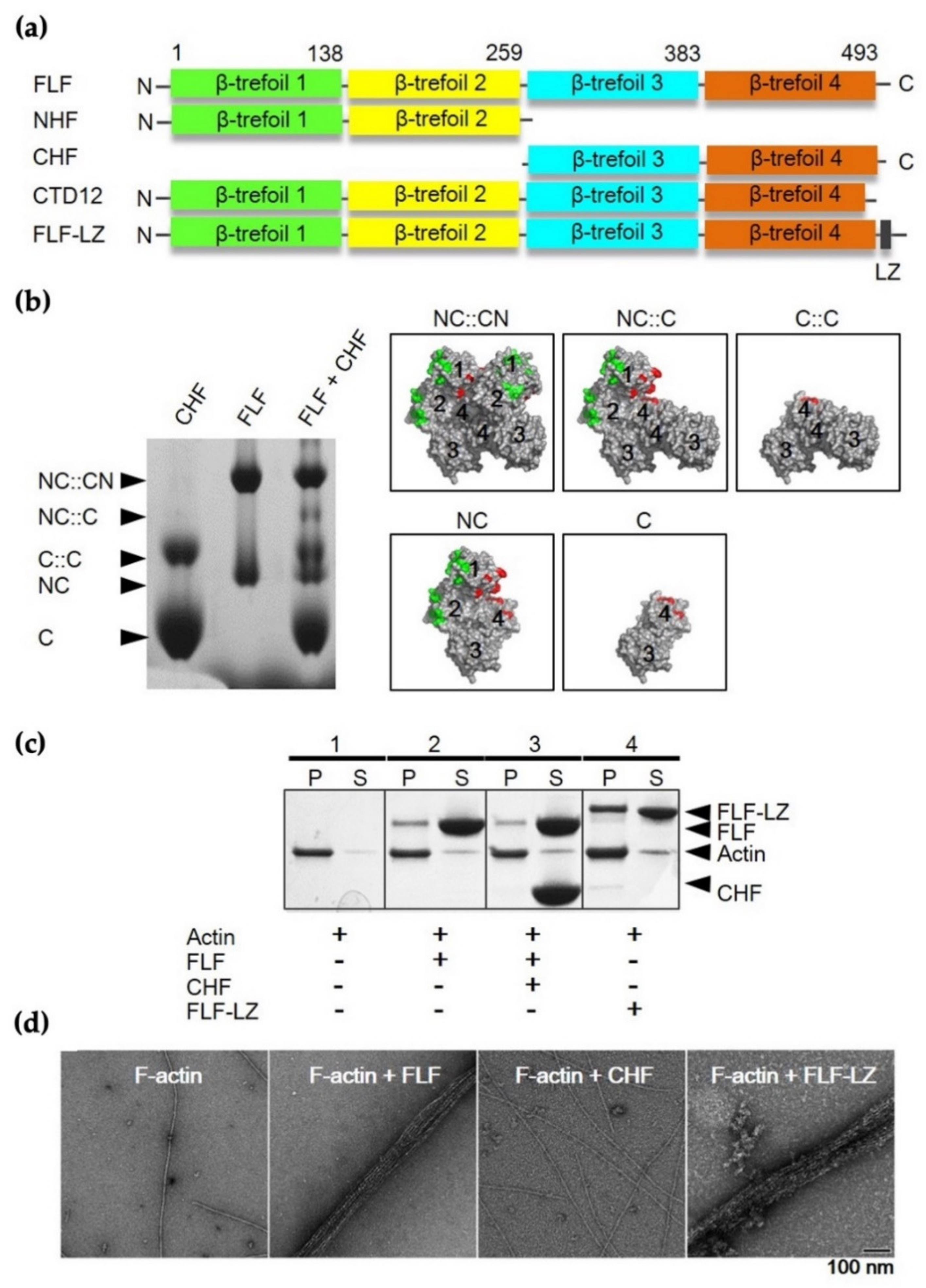
3.2. Actin-Bundling Activity of Fascin and Its Truncated Mutants
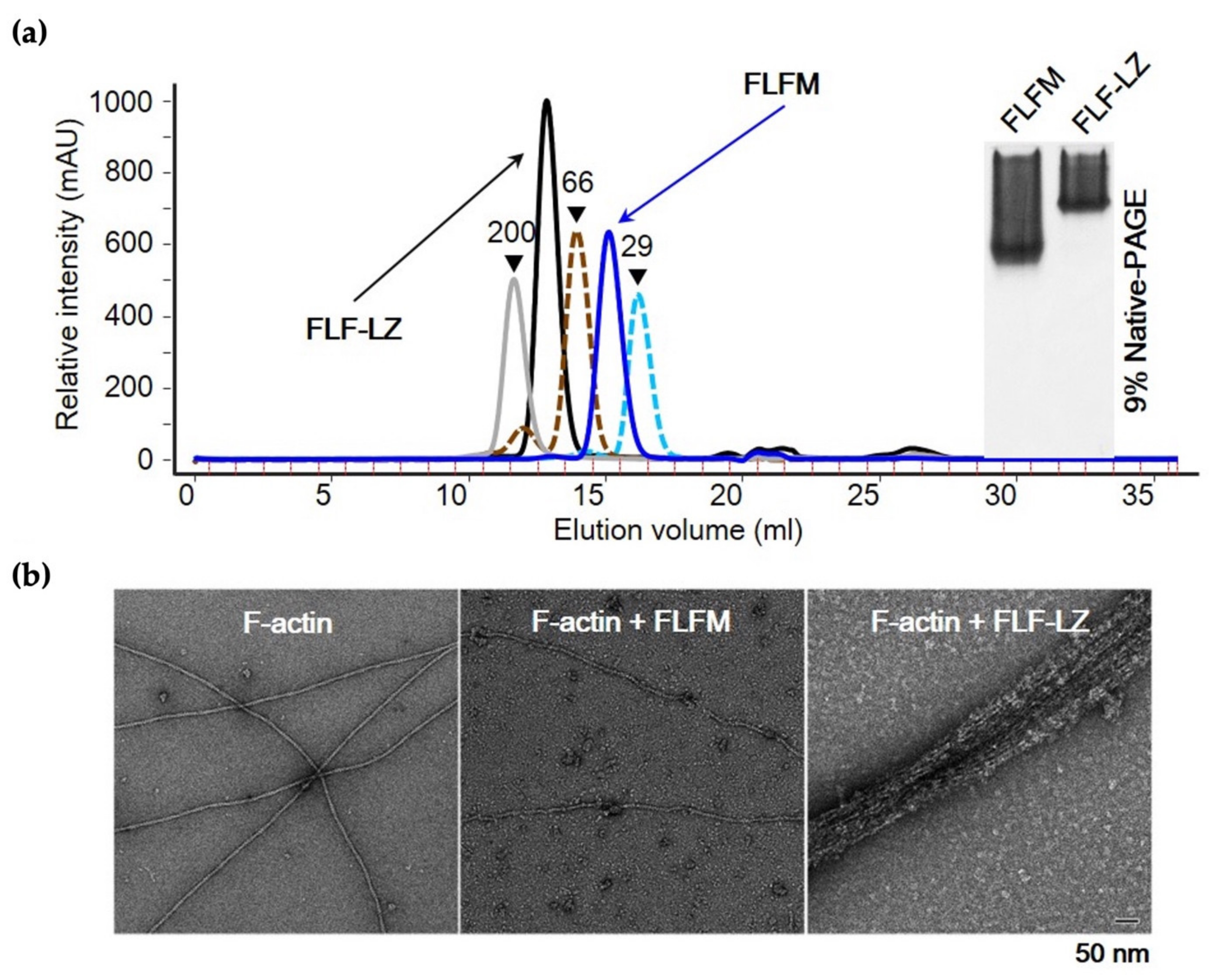
3.3. Effect of Dimerization on Actin-Bundling Activity of Fascin
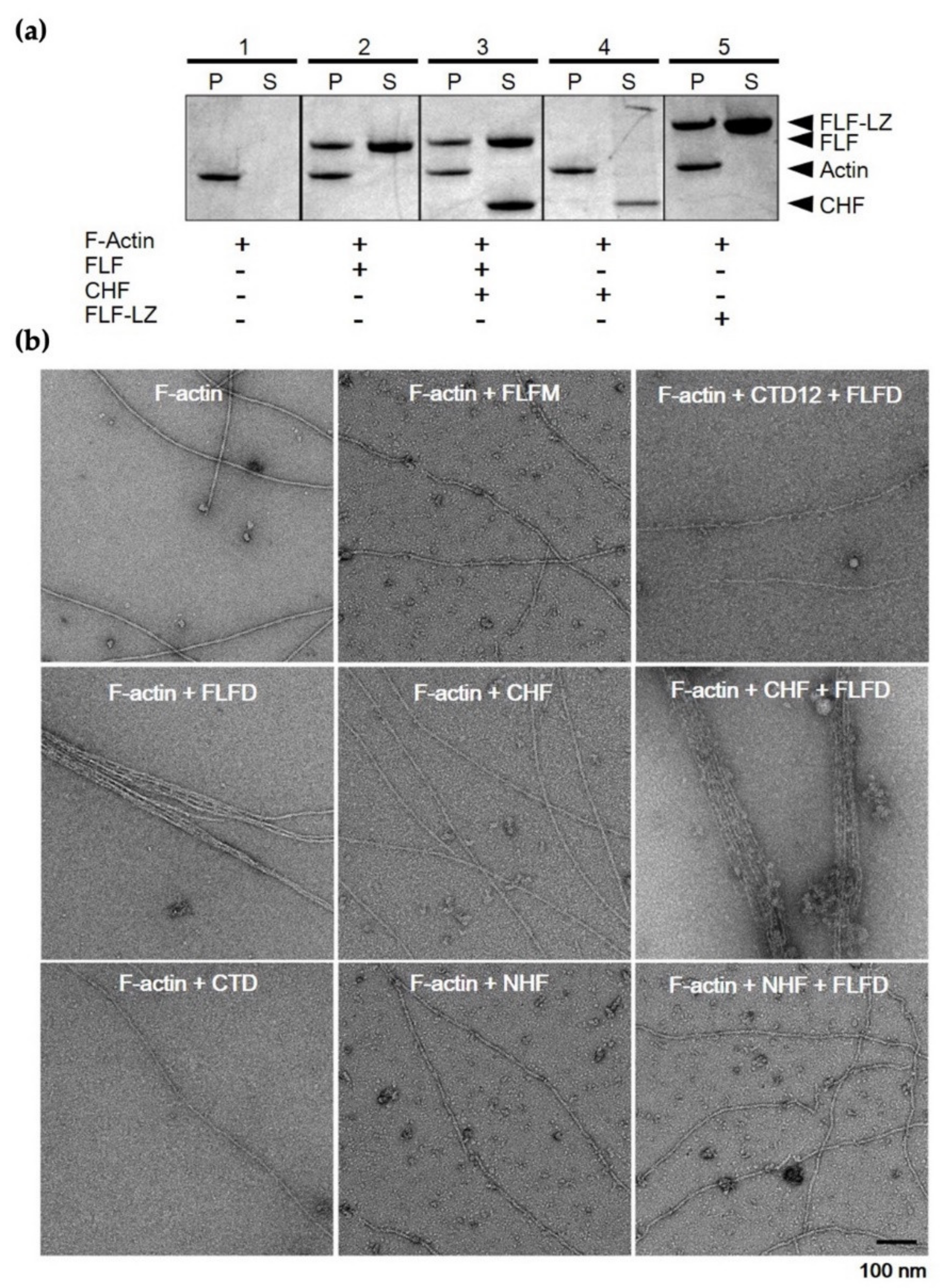
3.4. F-Actin-Binding and -Bundling Models of Fascin
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Conflicts of Interest
References
- Otto, J.J. Actin-bundling proteins. Curr. Opin. Cell Biol. 1994, 6, 105–109. [Google Scholar] [CrossRef]
- Chung, J.M.; Kim, H.-U.; Kim, G.J.; Jeoung, D.; Jung, H.S. The actin bundling activity of actin bundling protein 34 is inhibited by calcium binding to the EF2. Biochem. Biophys. Res. Commun. 2018, 503, 1836–1840. [Google Scholar] [CrossRef]
- Bryan, J.; Kane, R.E. Actin gelation in sea urchin egg extracts. Methods Cell Biol. 1982, 25, 175–199. [Google Scholar]
- Vignjevic, D.; Kojima, S.-I.; Aratyn, Y.; Danciu, O.; Svitkina, T.; Borisy, G.G. Role of fascin in filopodial protrusion. J. Cell Biol. 2006, 174, 863–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machesky, L.M.; Li, A. Fascin: Invasive filopodia promoting metastasis. Commun. Integr. Biol. 2010, 3, 263–270. [Google Scholar] [CrossRef]
- Ono, S.; Yamakita, Y.; Yamashiro, S.; Matsudaira, P.T.; Gnarra, J.; Obinata, T.; Matsumura, F. Identification of an actin binding region and a protein kinase C phosphorylation site on human fascin. J. Biol. Chem. 1997, 272, 2527–2533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, R.A.; Herrera-Sosa, H.; Otto, J.; Bryan, J. Cloning and expression of a murine fascin homolog from mouse brain. J. Biol. Chem. 1995, 270, 10764–10770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, Y.; Hayashi, K.; Shirao, T.; Ishikawa, R.; Kohama, K. Inhibition by drebrin of the actin-bundling activity of brain fascin, a protein localized in filopodia of growth cones. J. Neurochem. 2002, 66, 980–988. [Google Scholar] [CrossRef]
- Yamashiro-Matsumura, S.; Matsumura, F. Purification and characterization of an F-actin-bundling 55-kilodalton protein from HeLa cells. J. Biol. Chem. 1985, 260, 5087–5097. [Google Scholar] [CrossRef]
- Yamakita, Y.; Ono, S.; Matsumura, F.; Yamashiro, S. Phosphorylation of human fascin inhibits its actin binding and bundling activities. J. Biol. Chem. 1996, 271, 12632–12638. [Google Scholar] [CrossRef] [Green Version]
- Weiner, M.P.; Costa, G.L.; Schoettlin, W.; Cline, J.; Mathur, E.; Bauer, J.C. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 1994, 151, 119–123. [Google Scholar] [CrossRef]
- O’Shea, E.K.; Klemm, J.D.; Kim, P.S.; Alber, T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 1991, 254, 539–544. [Google Scholar] [CrossRef] [Green Version]
- Sato, O.; White, H.D.; Inoue, A.; Belknap, B.; Ikebe, R.; Ikebe, M. Human deafness mutation of myosin VI (C442Y) accelerates the ADP dissociation rate. J. Biol. Chem. 2004, 279, 28844–28854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fechheimer, M.; Taylor, D.L. Isolation and characterization of a 30,000-dalton calcium-sensitive actin cross-linking protein from Dictyostelium discoideum. J. Biol. Chem. 1984, 259, 4514–4520. [Google Scholar] [CrossRef]
- Fechheimer, M. The Dictyostelium discoideum 30,000-dalton protein is an actin filament-bundling protein that is selectively present in filopodia. J. Cell Biol. 1987, 104, 1539–1551. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.S.; Burgess, S.A.; Billington, N.; Colegrave, M.; Patel, H.; Chalovich, J.M.; Chantler, P.D.; Knight, P.J. Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc. Natl. Acad. Sci. USA 2008, 105, 6022–6026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egelman, E.H. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy 2000, 85, 225–234. [Google Scholar] [CrossRef]
- Jung, H.S.; Craig, R. Ca2+-induced tropomyosin movement in scallop striated muscle thin filaments. J. Mol. Biol. 2008, 383, 512–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kremer, J.R.; Mastronarde, D.N.; Mc Intosh, J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996, 116, 71–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bag, S.; Prentice, M.; Liang, M.; Warren, M.; Choudhury, K.R. Classification of polyhedral shapes from individual anisotropically resolved cryo-electron tomography reconstructions. BMC Bioinform. 2016, 17, 234. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera? A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sedeh, R.S.; Fedorov, A.A.; Fedorov, E.V.; Ono, S.; Matsumura, F.; Almo, S.C.; Bathe, M. Structure, evolutionary conservation, and conformational dynamics of homo sapiens fascin-1, an f-actin crosslinking protein. J. Mol. Biol. 2010, 400, 589–604. [Google Scholar] [CrossRef]
- Jansen, S.; Collins, A.; Yang, C.; Rebowski, G.; Svitkina, T.; Dominguez, R. Mechanism of actin filament bundling by fascin. J. Biol. Chem. 2011, 286, 30087–30096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Yang, S.; Jakoncic, J.; Zhang, J.J.; Huang, X.-Y. Migrastatin analogues target fascin to block tumour metastasis. Nature 2010, 464, 1062–1066, 1.8 Angstrom human fascin 1 crystal structure 10.2210/pdb3LLP/pdb. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Huang, F.-K.; Huang, J.; Chen, S.; Jakoncic, J.; Leo-Macias, A.; Diaz-Avalos, R.; Chen, L.; Zhang, J.J.; Huang, X.-Y. Molecular mechanism of fascin function in filopodial formation. J. Biol. Chem. 2013, 288, 274–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, J.C.; Clelland, J.D.; Collett, G.D.; Matsumura, F.; Yamashiro, S.; Zhang, L. Cell-matrix adhesions differentially regulate fascin phosphorylation. Mol. Biol. Cell 1999, 10, 4177–4190. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Machesky, L.M. Fascin1 in carcinomas: Its regulation and prognostic value. Int. J. Cancer 2014, 137, 2534–2544. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, Y.; Kim, D.J.; Adams, J.C. The roles of fascins in health and disease. J. Pathol. 2011, 224, 289–300. [Google Scholar] [CrossRef]
- Lamb, M.C.; Tootle, T.L. Fascin in Cell Migration: More Than an Actin Bundling Protein. Biology 2020, 9, 403. [Google Scholar] [CrossRef]
- Alburquerque-González, B.; Bernabé-García, M.; Montoro-García, S.; Rodrigues, P.C.; Sanz, J.R.; López-Calderón, F.F.; Luque, I.; Nicolás, F.J.; Cayuela, M.L.; Salo, T.; et al. New role of the antidepressant imipramine as a Fascin1 inhibitor in colorectal cancer cells. Exp. Mol. Med. 2020, 52, 281–292. [Google Scholar] [CrossRef] [Green Version]
- Miao, Q.; Hill, M.C.; Chen, F.; Mo, Q.; Ku, A.T.; Ramos, C.; Sock, E.; Lefebvre, V.; Nguyen, H. SOX11 and SOX4 drive the reactivation of an embryonic gene program during murine wound repair. Nat. Commun. 2019, 10, 4042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castaño, E.M.; Maarouf, C.L.; Wu, T.; Leal, M.; Whiteside, C.M.; Lue, L.-F.; Kokjohn, T.A.; Sabbagh, M.N.; Beach, T.G.; Roher, A.E. Alzheimer disease periventricular white matter lesions exhibit specific proteomic profile alterations. Neurochem. Int. 2012, 62, 145–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryu, M.-J.; Lee, C.; Kim, J.; Shin, H.-S.; Yu, M.-H. Proteomic analysis of stargazer mutant mouse neuronal proteins involved in absence seizure. J. Neurochem. 2007, 104, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Lin-Jones, J.; Burnside, B. Retina-specific protein fascin 2 is an actin cross-linker associated with actin bundles in photoreceptor inner segments and calycal processes. Investig. Opthalmol. Vis. Sci. 2007, 48, 1380–1388. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chung, J.M.; Sato, O.; Ikebe, R.; Lee, S.; Ikebe, M.; Jung, H.S. Structural Analysis of Human Fascin-1: Essential Protein for Actin Filaments Bundling. Life 2022, 12, 843. https://doi.org/10.3390/life12060843
Chung JM, Sato O, Ikebe R, Lee S, Ikebe M, Jung HS. Structural Analysis of Human Fascin-1: Essential Protein for Actin Filaments Bundling. Life. 2022; 12(6):843. https://doi.org/10.3390/life12060843
Chicago/Turabian StyleChung, Jeong Min, Osamu Sato, Reiko Ikebe, Sangmin Lee, Mitsuo Ikebe, and Hyun Suk Jung. 2022. "Structural Analysis of Human Fascin-1: Essential Protein for Actin Filaments Bundling" Life 12, no. 6: 843. https://doi.org/10.3390/life12060843
APA StyleChung, J. M., Sato, O., Ikebe, R., Lee, S., Ikebe, M., & Jung, H. S. (2022). Structural Analysis of Human Fascin-1: Essential Protein for Actin Filaments Bundling. Life, 12(6), 843. https://doi.org/10.3390/life12060843






