Validation of AmpliSeq NGS Panel for BRCA1 and BRCA2 Variant Detection in Canine Formalin-Fixed Paraffin-Embedded Mammary Tumors
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Selection
2.2. DNA Extraction
2.3. Sequencing and Data Analysis
3. Results
3.1. DNA Quality
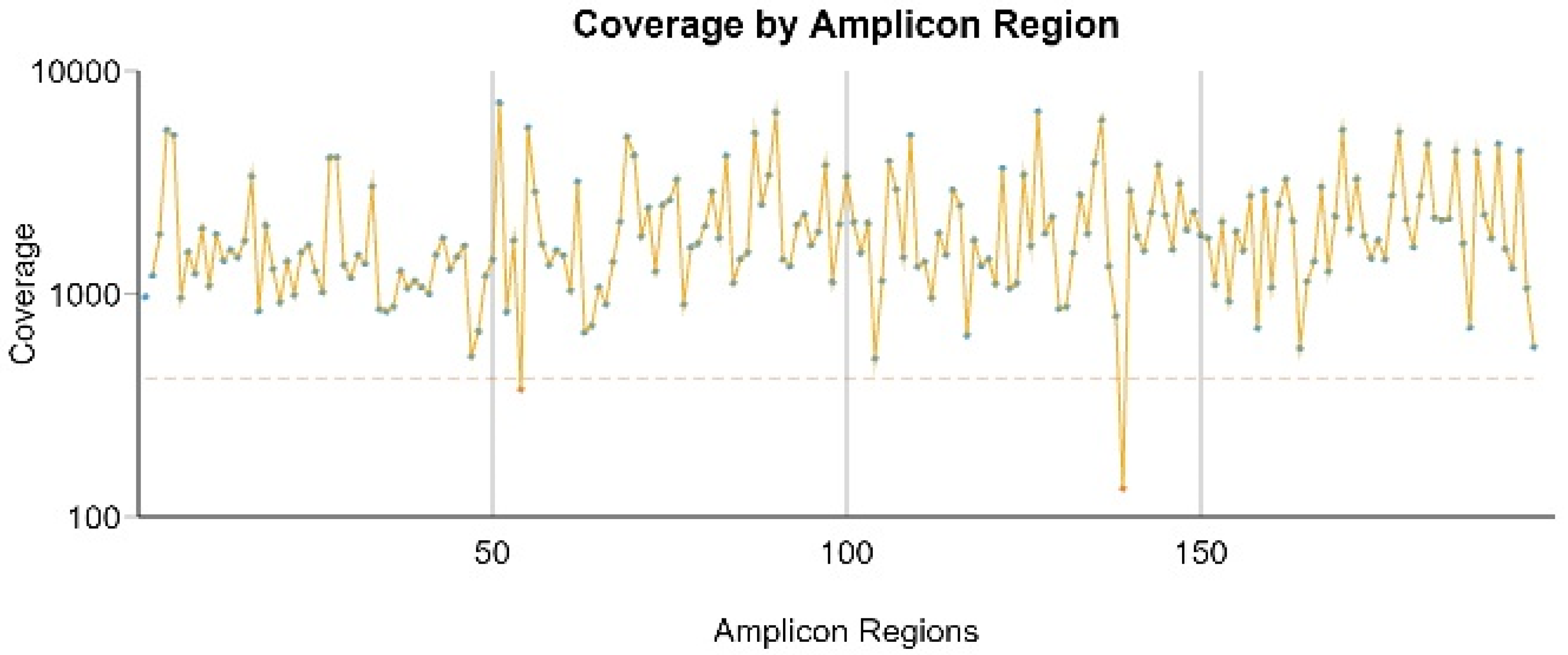
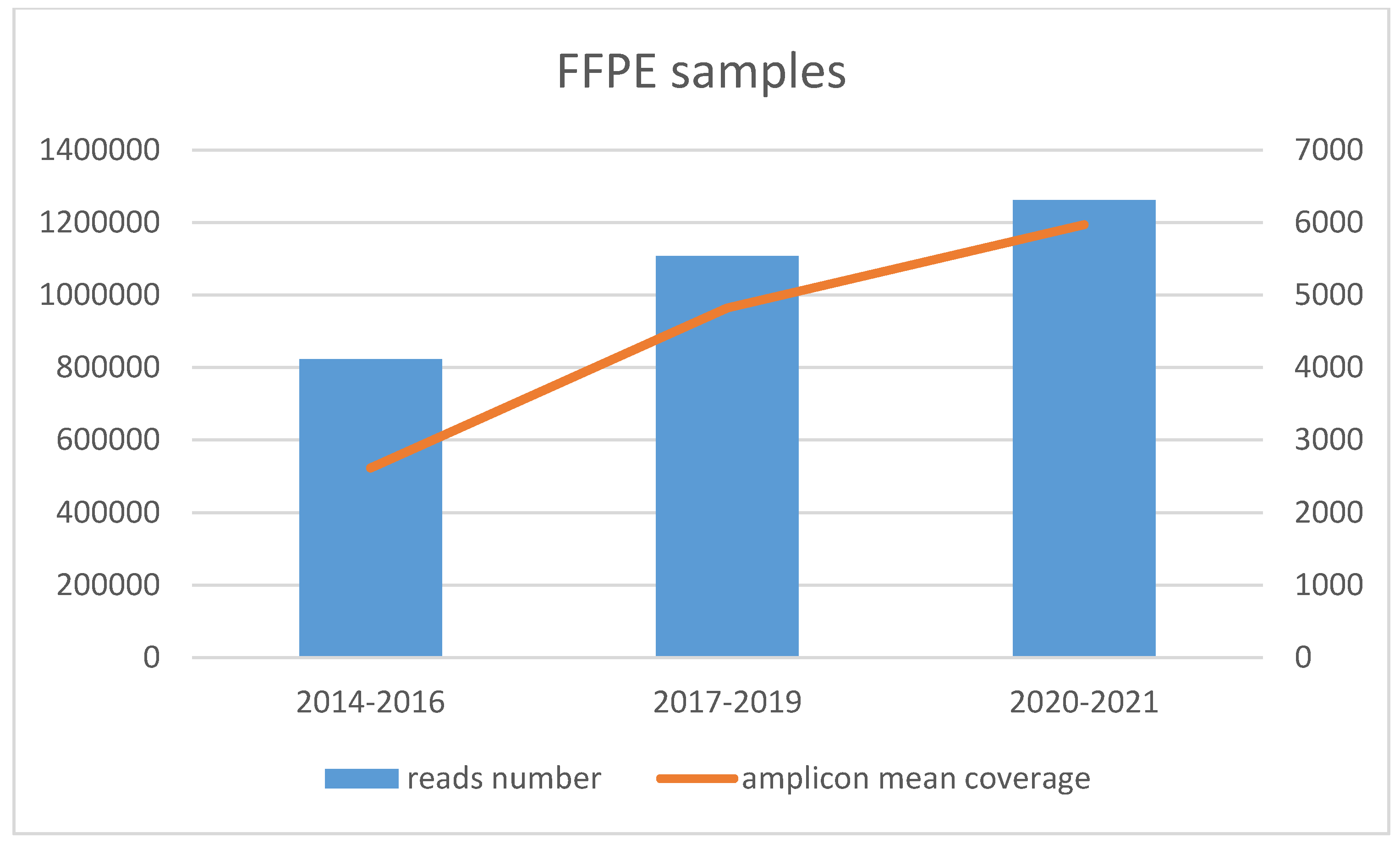
3.2. Amplicon Coverage and Variants Found
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scully, R.; Livingston, D.M. In search of the tumor-suppressor functions of BRCA1 and BRCA2. Nature 2000, 408, 429–432. [Google Scholar] [CrossRef] [PubMed]
- Szabo, C.I.; Wagner, L.A.; Francisco, L.V.; Roach, J.C.; Argonza, R.; King, M.C.; Ostrander, E.A. Human, canine and murine BRCA1 genes: Sequence comparison among species. Hum. Mol. Genet. 1996, 5, 1289–1298. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, F.; Peña, L.; Ibisch, C.; Loussouarn, D.; Gama, A.; Rieder, N.; Belousov, A.; Campone, M.; Abadie, J. Canine invasive mammary carcinomas as models of human breast cancer. Part 1: Natural history and prognostic factors. Breast Cancer Res. Treat. 2018, 167, 635–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shafiee, R.; Javanbakht, J.; Atyabi, N.; Kheradmand, P.; Kheradmand, D.; Bahrami, A.; Daraei, H.; Khadivar, F. Diagnosis, classification and grading of canine mammary tumours as a model to study human breast cancer: A Clinico-Cytohistopathological study with environmental factors influencing public health and medicine. Cancer Cell Int. 2013, 13, 79. [Google Scholar] [CrossRef] [Green Version]
- Feliubadaló, L.; Lopez-Doriga, A.; Castellsagué, E.; del Valle, J.; Menéndez, M.; Tornero, E.; Montes, E.; Cuesta, R.; Gómez, C.; Campos, O.; et al. Next-generation sequencing meets genetic diagnostics: Development of a comprehensive workflow for the analysis of BRCA1 and BRCA2 genes. Eur. J. Hum. Genet. 2013, 21, 864–870. [Google Scholar] [CrossRef]
- Strom, C.M.; Rivera, S.; Elzinga, C.; Angeloni, T.; Rosenthal, S.H.; Goos-Root, D.; Siaw, M.; Platt, J.; Braastadt, C.; Cheng, L.; et al. Development and Validation of a Next Generation Sequencing Assay for BRCA1 and BRCA2 Variants for the clinical Laboratory. PLoS ONE 2015, 10, e0136419. [Google Scholar] [CrossRef] [Green Version]
- Nicolussi, A.; Belardinilli, F.; Mahdavian, Y.; Colicchia, V.; D’Inzeo, S.; Petroni, M.; Zani, M.; Ferraro, S.; Valentini, V.; Ottini, L.; et al. Next-generation sequencing of BRCA1 and BRCA2 genes for rapid detection of germline mutations in hereditary breast/ovarian cancer. PeerJ 2019, 7, e6661. [Google Scholar] [CrossRef] [Green Version]
- Mercatante Carrick, D.; Mehaffey, M.G.; Sachs, M.C.; Altekruse, S.; Camalier, C.; Chuaqui, R.; Cozen, W.; Das, B.; Hernandez, B.Y.; Lih, C.J.; et al. Robustness of Next Generation Sequencing on Older Formalin-Fixed Paraffin-Embedded Tissue. PLoS ONE 2015, 10, e0127353. [Google Scholar]
- Einaga, N.; Yoshida, A.; Noda, H.; Suemitsu, M.; Nakayama, Y.; Sakurada, A.; Kawaji, Y.; Yamaguchi, H.; Sasaki, Y.; Tokino, T.; et al. Assessment of the quality of DNA from various formalin-fixed paraffin-embedded (FFPE) tissues and the use of this DNA for next-generation sequencing (NGS) with no artifactual mutation. PLoS ONE 2017, 12, e0176280. [Google Scholar] [CrossRef]
- He, Y.; Taylor, L.T.; Dimitrov, K.D.; Butt, S.L.; Stanton, J.B.; Goraichuk, I.V.; Fenton, H.; Poulson, R.; Zhang, J.; Brown, C.C.; et al. Whole-genome sequencing of genotype VI Newcastle disease viruses from formalin-fixed paraffin-embedded tissues from wild pigeons reveals continuous evolution and previously unrecognized genetic diversity in the US. Virol. J. 2018, 15, 9. [Google Scholar] [CrossRef] [Green Version]
- Govoni, V.M.; Da Silva, T.C.; Guerra, J.M.; Pereira, I.V.A.; Queiroga, F.L.; Cogliati, B. Genetic variants of BRCA1 and BRCA2 genes in cats with mammary gland carcinoma. Vet. Comp. Oncol. 2021, 19, 404–408. [Google Scholar] [PubMed]
- Engingler, S.O.; Akış, I.; Toydemir, T.S.F.; Oztabak, K.; Haktanir, D.; Gündüz, M.C.; Kırşan, I.; Fırat, I. Genetic variations of BRCA1 and BRCA2 genes in dogs with mammary tumours. Vet. Res. Commun. 2014, 38, 21–27. [Google Scholar]
- Ozmen, O.; Kul, S.; Risvanli, A.; Ozalp, G.; Sabuncu, A.; Kul, O. Somatic SNPs of the BRCA2 gene at the fragments encoding RAD51 binding sites of canine mammary tumors. Vet. Comp. Oncol. 2017, 15, 1479–1486. [Google Scholar] [CrossRef] [PubMed]
- Thumser-Henner, P.; Nytko, K.J.; Rohrer Bley, C. Mutations of BRCA2 in canine mammary tumors and their targeting potential in clinical therapy. BMC Vet. Res. 2020, 16, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldschmidt, M.; Peña, L.; Rasotto, R.; Zappulli, V. Classification and grading of canine mammary tumors. Vet. Pathol. 2021, 48, 117–131. [Google Scholar] [CrossRef]
- Peña, L.; De Andrés, P.J.; Clemente, M.; Cuesta, P.; Pérez-Alenza, M.D. Prognostic value of histological grading in non-inflammatory canine mammary carcinomas in a prospective study with two-year follow-up: Relationship with clinical and histological characteristics. Vet. Pathol. 2013, 50, 94–105. [Google Scholar] [CrossRef]
- Bhoyar, R.C.; Jain, A.; Sehgal, P.; Divakar, M.K.; Sharma, D.; Imran, M. High throughput detection and genetic epidemiology of SARS-CoV-2 using COVIDSeq next-generation sequencing. PLoS ONE 2021, 16, e0247115. [Google Scholar] [CrossRef]
- Ogiso-Tanaka, E.; Shimizu, T.; Hajika, M.; Kaga, A.; Ishimoto, M. Highly multiplexed AmpliSeq technology identifies novel variation of flowering time-related genes in soybean (Glycine Max). DNA Res. 2019, 26, 243–260. [Google Scholar] [CrossRef] [Green Version]
- Tariq, H.; Gul, A.; Zubair, M.; Jaffer, S.R.; Zafar, N.; Sadaf, G. Targetedet Next-generation Sequencing for Reliable Detection of Genetic Status in Breast Cancer. J. Coll. Physicians Surg. Pak. 2021, 30, 837–840. [Google Scholar]
- Jennings, L.J.; Arcila, M.E.; Corless, C.; Kamel-Reid, S.; Lubin, I.M.; Pfeifer, J.; Temple-Smolkin, R.L.; Voelkerding, K.V.; Nikiforova, M.N. Guidelines for Validation of Next-Generation Sequencing-Based Oncology Panels: A Joint Consensus Recommendation of the Association for Molecular Pathology and College of American Pathologists. J. Mol. Diagn. 2017, 19, 341–365. [Google Scholar] [CrossRef] [Green Version]
- Lu, K.H.; Wood, M.E.; Daniels, M.; Burke, C.; Ford, J.; Kauff, N.D.; Kohlmann, W.; Lindor, N.M.; Mulvey, T.M.; Robinson, L.; et al. American Society of Clinical Oncology Expert Statement: Collection and Use of a Cancer Family History for Oncology Providers. J. Clin. Oncol. 2014, 32, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Maués, T.; El-Jaick, K.B.; Costa, F.B.; Araujo, G.E.F.; Soares, M.V.G.; Moreira, A.S.; Ferreira, M.L.G.; Ferreira, A.M.R. Common germline haplotypes and genotypes identified in BRCA2 exon 11 of dogs with mammary tumours and histopathological analyses. Vet. Comp. Oncol. 2018, 16, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Davies, A.A.; Masson, J.Y.; McIlwraith, M.J.; Stasiak, A.Z.; Stasiak, A.; Venkitaraman, A.R.; West, S.C. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol. Cell 2001, 7, 273–282. [Google Scholar] [CrossRef]
- Mizuta, R.; La Salle, J.M.; Cheng, H.L.; Shinohara, A.; Ogawa, H.; Copeland, N.; Jenkins, N.A.; Lalande, M.; Alt, F.W. RAB22 and RAB163/mouse BRCA2: Proteins that specifically interact with the RAD51 protein. Proc. Natl. Acad. Sci. USA 1997, 94, 6927–6932. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bork, P.; Blomberg, N.; Nilges, M. Internal repeats in the BRCA2 protein sequence. Nat. Genet. 1996, 13, 22–23. [Google Scholar] [CrossRef]
- Wong, A.K.; Pero, R.; Ormonde, P.A.; Tavitigian, S.V.; Bartel, P.L. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene BRCA2. J. Biol. Chem. 1997, 272, 31941–33194. [Google Scholar] [CrossRef] [Green Version]
- Yoshikawa, Y.; Morimatsu, M.; Ochiai, K.; Nagano, M.; Yamane, Y.; Tomizawa, N.; Sasaki, N.; Hashizume, K.J. Analysis of genetic variations in the exon 27 region of the canine BRCA2 locus. Vet. Med. Sci. 2005, 67, 1013–1017. [Google Scholar] [CrossRef] [Green Version]

| Dog ID | Year of Collection | Sex and Hormonal Status | Breed | Age (Years) | Tissue | Histologic Type † | Grade of Malignancy ‡ |
|---|---|---|---|---|---|---|---|
| 1 § | 2019 | Fi | Yorkshire terrier | 8 | Mammary | Tubular carcinoma | I |
| 2 § | 2019 | Fi | Maremma sheepdog | 11 | Mammary | Intraductal papillary carcinoma (cystic type) | I |
| 3 § | 2020 | Fi | Mixed breed | 10 | Mammary | Tubular carcinoma | III |
| 4 § | 2020 | Fi | Mixed breed | 10 | Mammary | Solid carcinoma | I |
| 5 | 2020 | Fi | Golden retriever | 9 | Mammary | Intraductal papillary carcinoma (cystic type) | I |
| 6 | 2021 | Fi | Pinscher | 15 | Mammary | Tubulopapillary carcinoma | I |
| 7 § | 2021 | Fi | Mixed breed | 12 | Mammary | Anaplastic carcinoma | II |
| 8 § | 2021 | F- | Pinscher | 17 | Mammary | Tubular carcinoma, with foci of necrosis | II |
| 9 § | 2021 | Fi | German shepherd | 12 | Mammary | Ductal carcinoma | I |
| 10 | 2016 | Mi | Mixed breed | 13 | Mammary | Intraductal carcinoma | I |
| 11 | 2020 | Mi | Mixed breed | 13 | Mammary | Tubulopapillary carcinoma, with foci of necrosis | II |
| 12 | 2018 | Fi | German shepherd | 6 | Mammary | Anaplastic carcinoma with multifocal aspects of intraductal papillary carcinoma and foci of necrosis | III |
| 13 | 2016 | Fn | German shepherd | 5 | Mammary | Tubular carcinoma | I |
| 14 | 2016 | Fn | Mixed breed | 4 | Mammary | Complex carcinoma | I |
| 15 | 2016 | Fn | Mixed breed | 5 | Mammary | Intraductal papillary carcinoma (cystic type), with focal areas of squamous metaplasia | III |
| 16 | 2015 | Fn | American pit bull terrier | 11 | Mammary | Solid carcinoma with foci of necrosis | II |
| 17 | 2015 | Fn | Poodle | 4 | Mammary | Tubular carcinoma | II |
| 18 | 2015 | Fn | Siberian husky | - | Mammary | Solid carcinoma | II |
| 19 | 2015 | Fn | Golden retriever | 3 | Mammary | Tubulopapillary carcinoma | I |
| 20 | 2014 | Fi | German shepherd | 9 | Mammary | Intraductal papillary carcinoma | II |
| 21 | 2021 | Fi | Italian Cane Corso | 4 | Mammary | Normal tissue | n/a |
| 22 | 2021 | F- | German shepherd | 4 | Mammary | Normal tissue | n/a |
| Parameters | FFPE | Blood | Fresh Tissue | ||
|---|---|---|---|---|---|
| Old | Medium | Recent | |||
| DIN | 1.5–2.2 | 2.3–4.2 | 3.4–6.2 | 8.3–9.1 | 7.7–8.2 |
| Concentration (ng/µL) | 8.2–19.6 | 26.2–34.5 | 12.8–24.8 | 12.0–28.6 | 16.3–31.0 |
| N° Tot | N° Exonic Variants | N° Intronic Variants | |
|---|---|---|---|
| BRCA1 variants | 16 | 12 | 4 |
| BRCA2 variants | 26 | 23 | 3 |
| Gene | Chromosome (Dog) | Exon/Intron | Nucleotide Change § | Protein Change | Human Variant at Corresponding Position § | Human Clinical Classification |
|---|---|---|---|---|---|---|
| BRCA1 | chr 9 | INT 2 | c.81-26delC | - | c.81-26delC | Not referenced |
| BRCA1 | chr 9 | INT 2 | c.81-18delC | - | c.81-12delC | Benign/Likely benign |
| BRCA1 | chr 9 | INT 3 | c.135-43delA | - | c.135-43delA | Not referenced |
| BRCA1 | chr 9 | EX 5 | c.169G>A | p.(Gly57Arg) | c.169G>A; p.(Gly57Arg) | Not referenced |
| BRCA1 | chr 9 | EX 10 | c.606A>G | p.(Asp200Gly) | c.602A>G; p.(Asp201Gly) | Not referenced |
| BRCA1 | chr 9 | EX 10 | c.611A>G | p.(Lys202Glu) | c.607G>A; p.(Glu203Lys) | Not referenced |
| BRCA1 | chr 9 | EX 11 | c.723G>A | p.(Gly239Ser) | c.720A>G; p.(Gln240=) | Not referenced |
| BRCA1 | chr 9 | EX 11 | c.1315C>T | p.(Ala436Val) | c.1316C>T; p.(Ala439Val) | Not referenced |
| BRCA1 | chr 9 | EX 11 | c.1337A>C | p.(Arg443Ser) | c.1338A>C; p.(Arg446Ser) | Not referenced |
| BRCA1 | chr 9 | EX 11 | c.3628A>G | p.(Thr1207Ala) | c.3616G>A; p.(Ala1206Thr) | Not referenced |
| BRCA1 | chr 9 | EX 11 | c.3963G>A | p.(Ser1318=) | c.3951G>A; p.(Leu1317=) | Likely benign |
| BRCA1 | chr 9 | EX 13 | c.4219C>T | p.(Thr1403=) | c.4197C>T; p.(Thr1399=) | Not referenced |
| BRCA1 | chr 9 | INT 13 | c.4381-19C>T | - | c.4358-16C>T | Not referenced |
| BRCA1 | chr 9 | EX 16 | c.4777C>T | p.(Ser1588Pro) | c.4754C>T; p.(Pro1585Leu) | Not referenced |
| BRCA1 | chr 9 | EX 19 | c.5203G>A | p.(Arg1729Lys) | c.5177G>A; p.(Arg1726Lys) | Not referenced |
| BRCA1 | chr 9 | EX 24 | c.5525 G>A | p.(Ala1869Thr) | c.5581A>G; p.(Ser1861Gly) | Not referenced |
| BRCA2 | chr 25 | EX 4 | c.308T>C | p.(Ile103Thr) | c.326T>C; p.(Val109Ala) | Not referenced |
| BRCA2 | chr 25 | INT 4 | c.408-9_408-8del | - | - | Not found |
| BRCA2 | chr 25 | EX 5 | c.428A>G | p.(His143Pro) | c.449A>G; p.(His150Arg) | Uncertain significance |
| BRCA2 | chr 25 | INT 5 | c.455-40del | - | - | Not found |
| BRCA2 | chr 25 | EX 10 | c.1122C>T | p.(Thr371Ile) | c.1139G>T; p.(Ser380Ile) | Not referenced |
| BRCA2 | chr 25 | EX 10 | c.1168T>G | p.(Cys386Trp) | c.1185G>T; p.(Trp395Cys) | Uncertain significance |
| BRCA2 | chr 25 | EX 11 | c.1937G>A | p.(Glu643Lys) | c.1957G>A; p.(Glu653Lys) | Not referenced |
| BRCA2 | chr 25 | EX 11 | c.2131T>C | p.(His707=) | c.2148G>C; p.(Gln716His) | Uncertain significance |
| BRCA2 | chr 25 | EX 11 | c.2154A>C | p.( Pro715 Gln) | c.2170A>C; p.(Lys724Gln) | Not referenced |
| BRCA2 | chr 25 | EX 11 | c.2164C>A | p.(Ser718=) | c.2181A>C; p.(Ser727=) | Likely benign |
| BRCA2 | chr 25 | EX 11 | c.2193C>T | p.(Ala728Val) | c.2210C>T; p.(Ala737Val) | Not referenced |
| BRCA2 | chr 25 | EX 11 | c.2232A>G | p.(Asp741Ser) | c.2246G>A; p.(Ser749Asn) | Not referenced |
| BRCA2 | chr 25 | EX 11 | c.2269A>C | p.(Lys801Gln) | c.2431A>C; p.(Lys811Gln) | Not referenced |
| BRCA2 | chr 25 | EX 11 | c.4283A>C | p.(Thr1425Pro) | c.4288A>C; p.(Thr1430Pro) | Not referenced |
| BRCA2 | chr 25 | EX 11 | c.4314A>G | p.(Lys1435Arg) | c.4319A>G; p.(Lys1440Arg) | Uncertain significance |
| BRCA2 | chr 25 | EX 11 | c.4147G>A | p.(Val2013Ile) | c.5962G>A; p.(Val1988Ile) | Likely benign |
| BRCA2 | chr 25 | EX 11 | c.4175G>A | p.(Arg2022Lys) | c.5990G>A; p.(Arg1997Lys) | Uncertain significance |
| BRCA2 | chr 25 | EX 11 | c.4198G>A | p.(Asp2030Asn) | c.6013G>A; p.(Asp2005Asn) | Not referenced |
| BRCA2 | chr 25 | EX 11 | c.4204G>A | p.(Ala2032Thr) | c.6019A>G; p.(Thr2007Ala) | Not referenced |
| BRCA2 | chr 25 | EX 12 | c.6952_6954del | p.(Leu2306 o 2307del) | c.6868_6870del; p.(Leu2290del) | Not referenced |
| BRCA2 | chr 25 | EX 12 | c.6966C>T | p.(Phe2310=) | c.6879T>C; p.(Phe2293=) | Not referenced |
| BRCA2 | chr 25 | EX 16 | G>A | p.(Asp2611Asn) | c.7774G>A; p.(Asp2592Asn) | Not referenced |
| BRCA2 | chr 25 | EX 18 | T>C | p.(Ile2683=) | c.7992T>C; p.(Ile2664=) | Likely benign |
| BRCA2 | chr 25 | INT 25 | c.9692+57A>T | - | - | Not found |
| BRCA2 | chr 25 | EX 27 | c.10204_1026insAAA | p.(Met3332_Ile3333insLys) | c.9936_9938dup; p.(Lys3315dup) | Not referenced |
| BRCA2 | chr 25 | EX 27 | c.10450A>C | p.(Thr3405Pro) | c.101111A>C; p.(Thr3371Pro) | Not referenced |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Giacomo, D.; Di Domenico, M.; Defourny, S.V.P.; Malatesta, D.; Di Teodoro, G.; Martino, M.; Viola, A.; D’Alterio, N.; Cammà, C.; Modesto, P.; et al. Validation of AmpliSeq NGS Panel for BRCA1 and BRCA2 Variant Detection in Canine Formalin-Fixed Paraffin-Embedded Mammary Tumors. Life 2022, 12, 851. https://doi.org/10.3390/life12060851
Di Giacomo D, Di Domenico M, Defourny SVP, Malatesta D, Di Teodoro G, Martino M, Viola A, D’Alterio N, Cammà C, Modesto P, et al. Validation of AmpliSeq NGS Panel for BRCA1 and BRCA2 Variant Detection in Canine Formalin-Fixed Paraffin-Embedded Mammary Tumors. Life. 2022; 12(6):851. https://doi.org/10.3390/life12060851
Chicago/Turabian StyleDi Giacomo, Daniela, Marco Di Domenico, Sabrina Vanessa Patrizia Defourny, Daniela Malatesta, Giovanni Di Teodoro, Michele Martino, Antonello Viola, Nicola D’Alterio, Cesare Cammà, Paola Modesto, and et al. 2022. "Validation of AmpliSeq NGS Panel for BRCA1 and BRCA2 Variant Detection in Canine Formalin-Fixed Paraffin-Embedded Mammary Tumors" Life 12, no. 6: 851. https://doi.org/10.3390/life12060851
APA StyleDi Giacomo, D., Di Domenico, M., Defourny, S. V. P., Malatesta, D., Di Teodoro, G., Martino, M., Viola, A., D’Alterio, N., Cammà, C., Modesto, P., & Petrini, A. (2022). Validation of AmpliSeq NGS Panel for BRCA1 and BRCA2 Variant Detection in Canine Formalin-Fixed Paraffin-Embedded Mammary Tumors. Life, 12(6), 851. https://doi.org/10.3390/life12060851






