RNA-Seq Analysis Demonstrates Different Strategies Employed by Tiger Nuts (Cyperus esculentus L.) in Response to Drought Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Analysis of Physiological Indicators
2.3. RNA-Seq Analysis
2.4. QRT-PCR Analysis
2.5. Validation of the Physiological Indicators in Enriched Pathways
3. Results
3.1. The Physiological Trends under Stress
3.2. RNA-Seq Analysis
3.3. Enrichment Analysis
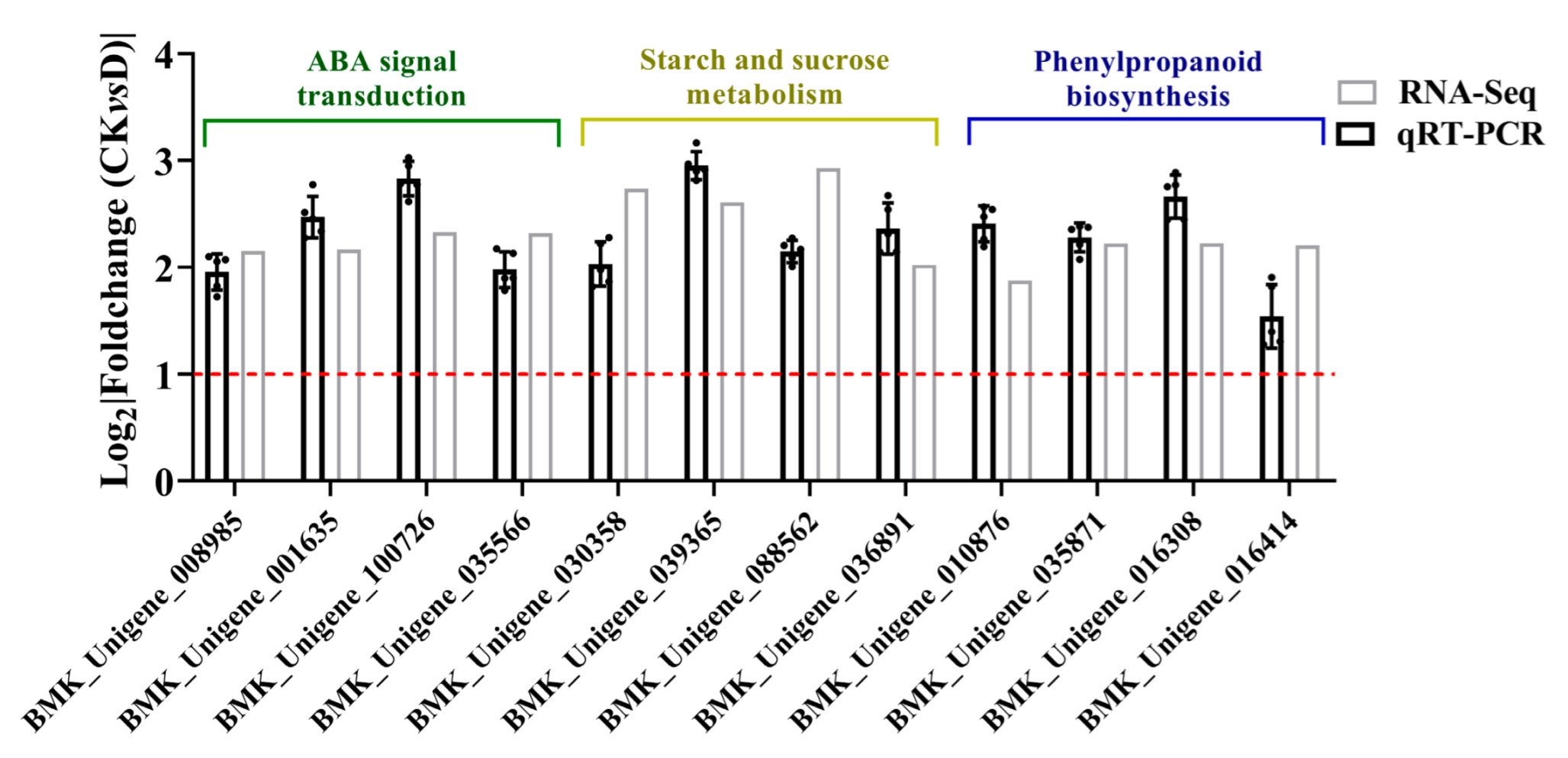
3.4. QRT-PCR Analysis
3.5. Enriched Pathways Analysis
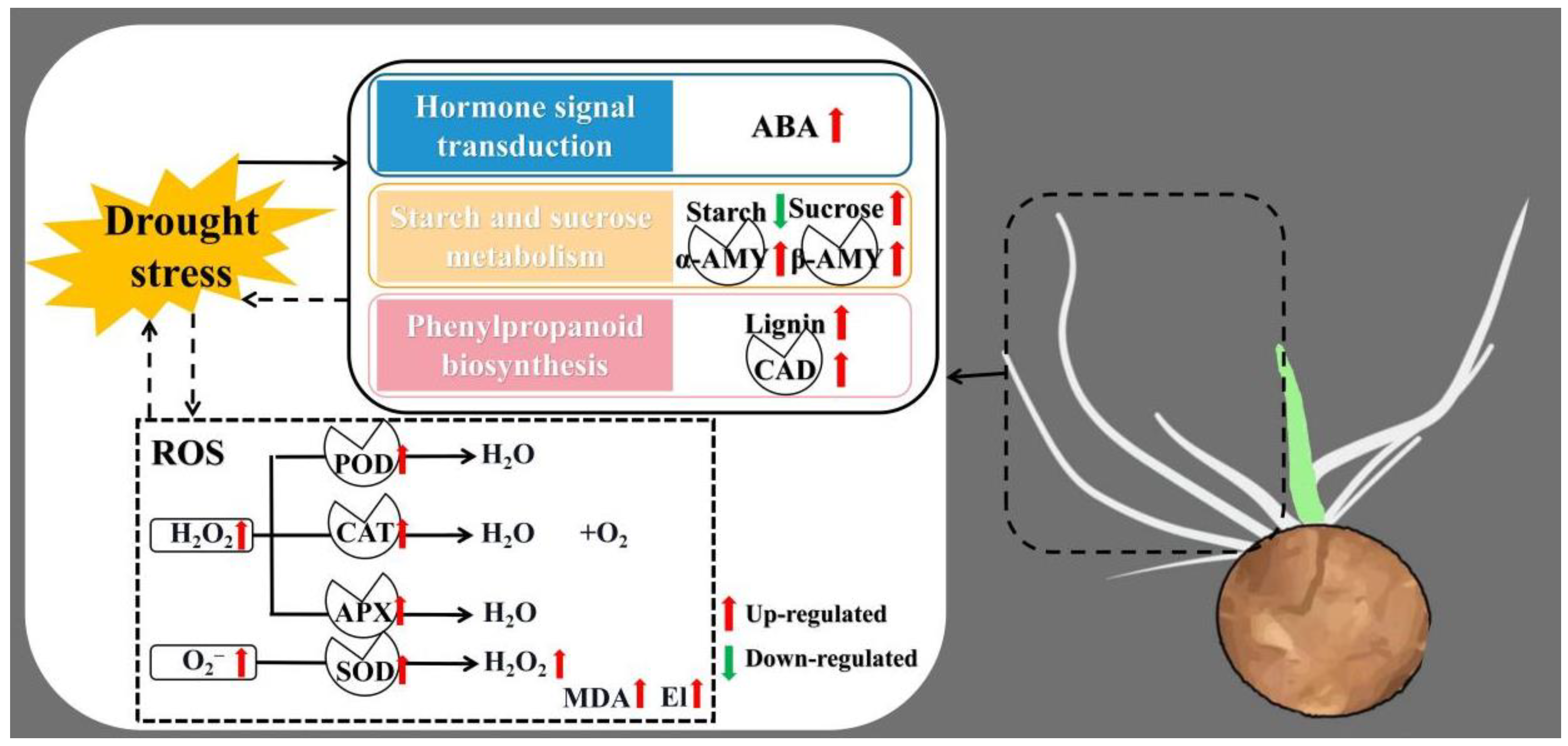
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yang, X.; Niu, L.; Zhang, Y.; Ren, W.; Yang, C.; Yang, J.; Xing, G.; Zhong, X.; Zhang, J.; Slaski, J.; et al. Morpho-Agronomic and Biochemical Characterization of Accessions of Tiger Nut (Cyperus esculentus) Grown in the North Temperate Zone of China. Plants 2022, 11, 923. [Google Scholar] [CrossRef] [PubMed]
- De Castro, O.; Gargiulo, R.; Del Guacchio, E.; Caputo, P.; De Luca, P. A molecular survey concerning the origin of Cyperus esculentus (Cyperaceae, Poales): Two sides of the same coin (weed vs. crop). Ann. Bot. 2015, 115, 733–745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zohari, D. The origin and early spread of agriculture in the Old World. In Developments in Agricultural and Managed Forest Ecology; Elsevier: Amsterdam, The Netherlands, 1986; Volume 16, pp. 3–20. [Google Scholar]
- Maduka, N.; Ire, F. Tigernut plant and useful application of tigernut tubers (Cyperus esculentus)—A review. Curr. J. Appl. Sci. Technol 2018, 29, 1–23. [Google Scholar] [CrossRef]
- Bado, S.; Bazongo, P.; Son, G.; Kyaw, M.T.; Forster, B.P.; Nielen, S.; Lykke, A.M.; Ouédraogo, A.; Bassolé, I.H. Physicochemical Characteristics and Composition of Three Morphotypes of Cyperus esculentus Tubers and Tuber Oils. J. Anal. Methods Chem. 2015, 2015, 673547. [Google Scholar] [CrossRef] [Green Version]
- Adekanmi, O.K.; Oluwatooyin, O.F.; Yemisi, A.A.; Yemisi, A. Influence of processing techniques on the nutrients and antinutrients of tigernut (Cyperus esculentus L.). World J. Dairy Food Sci. 2009, 4, 88–93. [Google Scholar]
- Chukwuma, E.R.; Obioma, N.; Christopher, O.I. The phytochemical composition and some biochemical effects of Nigerian tigernut (Cyperus esculentus L.) tuber. Pak. J. Nutr. 2010, 9, 709–715. [Google Scholar] [CrossRef] [Green Version]
- Adejuyitan, J. Tigernut processing: Its food uses and health benefits. Am. J. Food Technol. 2011, 6, 197–201. [Google Scholar] [CrossRef] [Green Version]
- Defelice, M.S.W.T. Yellow Nutsedge Cyperus esculentus L.—Snack Food of the Gods1. Weed Technol. 2002, 16, 901–907. [Google Scholar] [CrossRef]
- Yu, Y.; Lu, X.; Zhang, T.; Zhao, C.; Guan, S.; Pu, Y.; Gao, F. Tiger Nut (Cyperus esculentus L.): Nutrition, Processing, Function and Applications. Foods 2022, 11, 601. [Google Scholar] [CrossRef]
- Chourasiya, D.; Agnihotri, R.; Prakash, A.; Pal, K.K.; Sharma, M.P. Bioprotection of Soybean Plants from Drought Stress by Application of Bacterial and Fungal Endophytes. In Root Biology; Springer: Berlin/Heidelberg, Germany, 2018; pp. 281–301. [Google Scholar]
- Dubey, A.; Kumar, A.; Malla, M.A.; Chowdhary, K.; Singh, G.; Gudasalamani Ravikanth, H.; Sharma, S.; Saati-Santamaria, Z.; Menéndez, E.; Dames, J.F. Approaches for the amelioration of adverse effects of drought stress on crop plants. Front. Biosci. 2021, 26, 928–947. [Google Scholar]
- Ahmad, P.; Hameed, A.; Abd-Allah, E.F.; Sheikh, S.A.; Wani, M.R.; Rasool, S.; Jamsheed, S.; Kumar, A. Biochemical and molecular approaches for drought tolerance in plants. In Physiological Mechanisms and Adaptation Strategies in Plants under Changing Environment; Springer: Berlin/Heidelberg, Germany, 2014; pp. 1–29. [Google Scholar]
- Hashem, A.; Kumar, A.; Al-Dbass, A.M.; Alqarawi, A.A.; Al-Arjani, A.-B.F.; Singh, G.; Farooq, M.; Abd_Allah, E.F. Arbuscular mycorrhizal fungi and biochar improves drought tolerance in chickpea. Saudi J. Biol. Sci. 2019, 26, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Dubey, A.; Kumar, A.; Khan, M.L. Role of biostimulants for enhancing abiotic stress tolerance in Fabaceae plants. In The Plant Family Fabaceae; Springer: Berlin/Heidelberg, Germany, 2020; pp. 223–236. [Google Scholar]
- Goswami, A.; Banerjee, R.; Raha, S. Drought resistance in rice seedlings conferred by seed priming. Protoplasma 2013, 250, 1115–1129. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Geng, J.; Du, Y.; Zhao, Q.; Zhang, W.; Fang, Q.; Yin, Z.; Li, J.; Yuan, X.; Fan, Y. Heat shock transcription factor (Hsf) gene family in common bean (Phaseolus vulgaris): Genome-wide identification, phylogeny, evolutionary expansion and expression analyses at the sprout stage under abiotic stress. BMC Plant Biol. 2022, 22, 33. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S.P. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 2014, 202, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Ali, F.; Bano, A.; Fazal, A. Recent methods of drought stress tolerance in plants. Plant Growth Regul. 2017, 82, 363–375. [Google Scholar] [CrossRef]
- Zhang, Q.; Li, M.; Xia, C.Y.; Zhang, W.J.; Yin, Z.G.; Zhang, Y.L.; Fang, Q.X.; Liu, Y.C.; Zhang, M.Y.; Zhang, W.H. Transcriptome-based analysis of salt-related genes during the sprout stage of common bean (Phaseolus vulgaris) under salt stress conditions. Biotechnol. Biotechnol. Equip. 2021, 35, 1086–1098. [Google Scholar] [CrossRef]
- Bashir, K.; Matsui, A.; Rasheed, S.; Seki, M. Recent advances in the characterization of plant transcriptomes in response to drought, salinity, heat, and cold stress. F1000Research 2019, 8, 658. [Google Scholar] [CrossRef] [Green Version]
- Hanada, K.; Higuchi-Takeuchi, M.; Okamoto, M.; Yoshizumi, T.; Shimizu, M.; Nakaminami, K.; Nishi, R.; Ohashi, C.; Iida, K.; Tanaka, M. Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc. Natl. Acad. Sci. USA 2013, 110, 2395–2400. [Google Scholar] [CrossRef] [Green Version]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef]
- Krizek, B.A.; Bequette, C.J.; Xu, K.; Blakley, I.C.; Fu, Z.Q.; Stratmann, J.W.; Loraine, A.E. RNA-Seq links the transcription factors AINTEGUMENTA and AINTEGUMENTA-LIKE6 to cell wall remodeling and plant defense pathways. Plant Physiol. 2016, 171, 2069–2084. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Pan, Z.; Chen, L.; Dai, Y.; Wan, J.; Ye, H.; Nguyen, H.T.; Zhang, G.; Chen, H. Analysis of whole transcriptome RNA-seq data reveals many alternative splicing events in soybean roots under drought stress conditions. Genes 2020, 11, 1520. [Google Scholar] [CrossRef]
- Mahdavi Mashaki, K.; Garg, V.; Nasrollahnezhad Ghomi, A.A.; Kudapa, H.; Chitikineni, A.; Zaynali Nezhad, K.; Yamchi, A.; Soltanloo, H.; Varshney, R.K.; Thudi, M. RNA-Seq analysis revealed genes associated with drought stress response in kabuli chickpea (Cicer arietinum L.). PLoS ONE 2018, 13, e0199774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morgil, H.; Tardu, M.; Cevahir, G.; Kavakli, İ.H. Comparative RNA-seq analysis of the drought-sensitive lentil (Lens culinaris) root and leaf under short-and long-term water deficits. Funct. Integr. Genom. 2019, 19, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Wang, S.; Paetzold, L.; Wang, Z.; Hui, K.; Rudd, J.C.; Xue, Q.; Ibrahim, A.M.; Metz, R.; Johnson, C.D. RNA-seq analysis reveals different drought tolerance mechanisms in two broadly adapted wheat cultivars ‘TAM 111’ and ‘TAM 112’. Sci. Rep. 2021, 11, 4301. [Google Scholar] [CrossRef] [PubMed]
- Rezayian, M.; Niknam, V.; Ebrahimzadeh, H. Penconazole and calcium ameliorate drought stress in canola by upregulating the antioxidative enzymes. Funct. Plant Biol. 2020, 47, 825–839. [Google Scholar] [CrossRef] [PubMed]
- Sutinen, M.L.; Palta, J.P.; Reich, P.B. Seasonal differences in freezing stress resistance of needles of Pinus nigra and Pinus resinosa: Evaluation of the electrolyte leakage method. Tree Physiol. 1992, 11, 241–254. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.P.; Choudhuri, M.A. Implications of water stress-induced changes in the levels of endogenous ascorbic acid and hydrogen peroxide in Vigna seedlings. Physiol. Plant. 1983, 58, 166–170. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Gene Ontology Consortium. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, W.-J.; Yin, Z.-G.; Li, W.-J.; Xia, C.-Y.; Sun, H.-Y.; Yang, Y.-M.; Wu, H.-B.; Zhang, S.; Zhao, H.-H. Genome-wide identification reveals the potential functions of the bZIP gene family in common bean (Phaseolus vulgaris) in response to salt stress during the sprouting stage. J. Plant Growth Regul. 2021, 1–16. [Google Scholar] [CrossRef]
- Du, Y.; Zhao, Q.; Chen, L.; Yao, X.; Zhang, H.; Wu, J.; Xie, F. Effect of Drought Stress during Soybean R2-R6 Growth Stages on Sucrose Metabolism in Leaf and Seed. Int. J. Mol. Sci. 2020, 21, 618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goffner, D.; Joffroy, I.; Grima-Pettenati, J.; Halpin, C.; Knight, M.E.; Schuch, W.; Boudet, A.M. Purification and characterization of isoforms of cinnamyl alcohol dehydrogenase from Eucalyptus xylem. Planta 1992, 188, 48–53. [Google Scholar] [CrossRef]
- Ma, Q.; Wu, C.; Liang, S.; Yuan, Y.; Liu, C.; Liu, J.; Feng, B. The alkali tolerance of broomcorn millet (Panicum miliaceum L.) at the germination and seedling stage: The case of 296 broomcorn millet genotypes. Front. Plant Sci. 2021, 12, 711429. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Lu, X.; Zhang, H.; Luo, D.; Zhang, A.; Sun, M.; Wu, Q.; Wang, X.; Huang, L. Transcriptome Reveals the Dynamic Response Mechanism of Pearl Millet Roots under Drought Stress. Genes 2021, 12, 1988. [Google Scholar] [CrossRef]
- Ahmad, A.; Aslam, Z.; Naz, M.; Hussain, S.; Javed, T.; Aslam, S.; Raza, A.; Ali, H.M.; Siddiqui, M.H.; Salem, M.Z. Exogenous salicylic acid-induced drought stress tolerance in wheat (Triticum aestivum L.) grown under hydroponic culture. PLoS ONE 2021, 16, e0260556. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Su, X.; Guo, Y.; Sun, F.; Qu, Y.; Chen, Q. Cotton seedling drought tolerance is improved via salt preconditioning. Protoplasma 2021, 258, 263–277. [Google Scholar] [CrossRef]
- Jones, C.; De Vega, J.; Worthington, M.; Thomas, A.; Gasior, D.; Harper, J.; Doonan, J.; Fu, Y.; Bosch, M.; Corke, F. A comparison of differential gene expression in response to the onset of water stress between three hybrid Brachiaria genotypes. Front. Plant Sci. 2021, 12, 393. [Google Scholar] [CrossRef]
- Ishibashi, Y.; Yuasa, T.; Iwaya-Inoue, M. Mechanisms of maturation and germination in crop seeds exposed to environmental stresses with a focus on nutrients, water status, and reactive oxygen species. Adv. Exp. Med. Biol. 2018, 1081, 233–257. [Google Scholar]
- Wu, L.; Chang, Y.; Wang, L.; Wu, J.; Wang, S. Genetic dissection of drought resistance based on root traits at the bud stage in common bean. TAG Theor. Appl. Genet. Theor. Angew. Genet. 2021, 134, 1047–1061. [Google Scholar] [CrossRef]
- Kim, Y.; Chung, Y.S.; Lee, E.; Tripathi, P.; Heo, S.; Kim, K.-H. Root response to drought stress in rice (Oryza sativa L.). Int. J. Mol. Sci. 2020, 21, 1513. [Google Scholar] [CrossRef] [Green Version]
- Dubey, A.; Saiyam, D.; Kumar, A.; Hashem, A.; Abd_Allah, E.F.; Khan, M.L. Bacterial root endophytes: Characterization of their competence and plant growth promotion in soybean (Glycine max (L.) Merr.) under drought stress. Int. J. Environ. Res. Public Health 2021, 18, 931. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Li, X.; Zou, J.; Ren, J.; Jin, C.; Zhang, H.; Yu, H.; Jin, H. Comparative transcriptome analysis of genes involved in the drought stress response of two peanut (Arachis hypogaea L.) varieties. BMC Plant Biol. 2021, 21, 64. [Google Scholar] [CrossRef]
- Chakhchar, A.; Chaguer, N.; Ferradous, A.; Filali-Maltouf, A.; El Modafar, C. Root system response in Argania spinosa plants under drought stress and recovery. Plant Signal. Behav. 2018, 13, e1489669. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, M.N.; Léon, J.; Naz, A.A.; Ballvora, A. Genetics and genomics of root system variation in adaptation to drought stress in cereal crops. J. Exp. Bot. 2021, 72, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Dev Sharma, A.; Kumar, S.; Singh, P. Expression analysis of a stress-modulated transcript in drought tolerant and susceptible cultivars of sorghum (Sorghum bicolor). J. Plant Physiol. 2006, 163, 570–576. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Qiao, X.; Tian, Z.; Zhang, X.; Zou, X.; Cheng, Y.; Lu, G.; Zeng, L.; Fu, G.; Ding, X.; et al. Proteomic Analysis of Rapeseed Root Response to Waterlogging Stress. Plants 2018, 7, 71. [Google Scholar] [CrossRef] [Green Version]
- Song, H.; Huang, Y.; Gu, B. QTL-Seq identifies quantitative trait loci of relative electrical conductivity associated with heat tolerance in bottle gourd (Lagenaria siceraria). PLoS ONE 2020, 15, e0227663. [Google Scholar] [CrossRef]
- Guo, Y.; Zhao, S.; Zhu, C.; Chang, X.; Yue, C.; Wang, Z.; Lin, Y.; Lai, Z. Identification of drought-responsive miRNAs and physiological characterization of tea plant (Camellia sinensis L.) under drought stress. BMC Plant Biol. 2017, 17, 211. [Google Scholar] [CrossRef] [Green Version]
- Hamurcu, M.; Khan, M.K.; Pandey, A.; Ozdemir, C.; Avsaroglu, Z.Z.; Elbasan, F.; Omay, A.H.; Gezgin, S. Nitric oxide regulates watermelon (Citrullus lanatus) responses to drought stress. 3 Biotech 2020, 10, 494. [Google Scholar] [CrossRef]
- Mubarakshina, M.M.; Ivanov, B.N.; Naydov, I.A.; Hillier, W.; Badger, M.R.; Krieger-Liszkay, A. Production and diffusion of chloroplastic H2O2 and its implication to signalling. J. Exp. Bot. 2010, 61, 3577–3587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittler, R. ROS Are Good. Trends Plant Sci. 2016, 22, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashwini, N.; Sajeevan, R.S.; Udayakumar, M.; Nataraja, K.N. Identification of splice variant of OsGBF1 in Oryza sativa ssp. indica genotypes under salinity stress. 3 Biotech 2018, 8, 345. [Google Scholar] [CrossRef] [PubMed]
- Dos Reis, M.V.; Rouhana, L.V.; Sadeque, A.; Koga, L.; Clough, S.J.; Calla, B.; Paiva, P.D.O.; Korban, S.S. Genome-wide expression of low temperature response genes in Rosa hybrida L. Plant Physiol. Biochem. PPB 2020, 146, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, M.; Turkyilmaz Unal, B.; García-Caparrós, P.; Khursheed, A.; Gul, A.; Hasanuzzaman, M. Osmoregulation and its actions during the drought stress in plants. Physiol. Plant. 2021, 172, 1321–1335. [Google Scholar] [CrossRef]
- Ng, L.M.; Melcher, K.; Teh, B.T.; Xu, H.E. Abscisic acid perception and signaling: Structural mechanisms and applications. Acta Pharmacol. Sin. 2014, 35, 567–584. [Google Scholar] [CrossRef] [Green Version]
- Long, H.; Zheng, Z.; Zhang, Y.; Xing, P.; Wan, X.; Zheng, Y.; Li, L. An abscisic acid (ABA) homeostasis regulated by its production, catabolism and transport in peanut leaves in response to drought stress. PLoS ONE 2019, 14, e0213963. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Seng, S.; Li, D.; Zhang, F.; Liu, Y.; Yao, T.; Liang, J.; Yi, M.; Wu, J. Antagonism between abscisic acid and gibberellin regulates starch synthesis and corm development in Gladiolus hybridus. Hortic. Res. 2021, 8, 155. [Google Scholar] [CrossRef]
- Lv, X.; Ding, Y.; Long, M.; Liang, W.; Gu, X.; Liu, Y.; Wen, X. Effect of Foliar Application of Various Nitrogen Forms on Starch Accumulation and Grain Filling of Wheat (Triticum aestivum L.) Under Drought Stress. Front. Plant Sci. 2021, 12, 645379. [Google Scholar] [CrossRef]
- Mathan, J.; Singh, A.; Ranjan, A. Sucrose transport in response to drought and salt stress involves ABA-mediated induction of OsSWEET13 and OsSWEET15 in rice. Physiol. Plant. 2021, 171, 620–637. [Google Scholar] [CrossRef]
- Guo, J.; Qu, L.; Hu, Y.; Lu, W.; Lu, D. Proteomics reveals the effects of drought stress on the kernel development and starch formation of waxy maize. BMC Plant Biol. 2021, 21, 434. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Wang, Y.; Xie, H.; Qiu, C.; Zhang, S.; Xiao, J.; Li, H.; Chen, L.; Li, X.; Ding, Z. Drought stress triggers proteomic changes involving lignin, flavonoids and fatty acids in tea plants. Sci. Rep. 2020, 10, 15504. [Google Scholar] [CrossRef] [PubMed]
- Mandoulakani, B.A.; Eyvazpour, E.; Ghadimzadeh, M. The effect of drought stress on the expression of key genes involved in the biosynthesis of phenylpropanoids and essential oil components in basil (Ocimum basilicum L.). Phytochemistry 2017, 139, 1–7. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Jin, G.; Luo, X.; Chen, C.; Li, W.; Zhu, G. Transcriptome analysis and transcription factors responsive to drought stress in Hibiscus cannabinus. PeerJ 2020, 8, e8470. [Google Scholar] [CrossRef] [Green Version]
- De Vega, J.J.; Teshome, A.; Klaas, M.; Grant, J.; Finnan, J.; Barth, S. Physiological and transcriptional response to drought stress among bioenergy grass Miscanthus species. Biotechnol. Biofuels 2021, 14, 60. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of Phenylpropanoid Pathway and the Role of Polyphenols in Plants under Abiotic Stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [Green Version]
- Lamin-Samu, A.T.; Farghal, M.; Ali, M.; Lu, G. Morpho-Physiological and Transcriptome Changes in Tomato Anthers of Different Developmental Stages under Drought Stress. Cells 2021, 10, 1809. [Google Scholar] [CrossRef]







| Groups | Total DEGs | Up DEGs | Down DEGs |
|---|---|---|---|
| CK vs. D | 1760 | 854 | 906 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mu, Z.; Wei, Z.; Liu, J.; Cheng, Y.; Song, Y.; Yao, H.; Yuan, X.; Wang, S.; Gu, Y.; Zhong, J.; et al. RNA-Seq Analysis Demonstrates Different Strategies Employed by Tiger Nuts (Cyperus esculentus L.) in Response to Drought Stress. Life 2022, 12, 1051. https://doi.org/10.3390/life12071051
Mu Z, Wei Z, Liu J, Cheng Y, Song Y, Yao H, Yuan X, Wang S, Gu Y, Zhong J, et al. RNA-Seq Analysis Demonstrates Different Strategies Employed by Tiger Nuts (Cyperus esculentus L.) in Response to Drought Stress. Life. 2022; 12(7):1051. https://doi.org/10.3390/life12071051
Chicago/Turabian StyleMu, Zhongsheng, Zunmiao Wei, Jiayao Liu, Yan Cheng, Yu Song, Hongbing Yao, Xiankai Yuan, Shukun Wang, Yanhua Gu, Jingwen Zhong, and et al. 2022. "RNA-Seq Analysis Demonstrates Different Strategies Employed by Tiger Nuts (Cyperus esculentus L.) in Response to Drought Stress" Life 12, no. 7: 1051. https://doi.org/10.3390/life12071051





