ITGβ6 Facilitates Skeletal Muscle Development by Maintaining the Properties and Cytoskeleton Stability of Satellite Cells
Abstract
:1. Introduction
2. Materials and Methods
3. Results
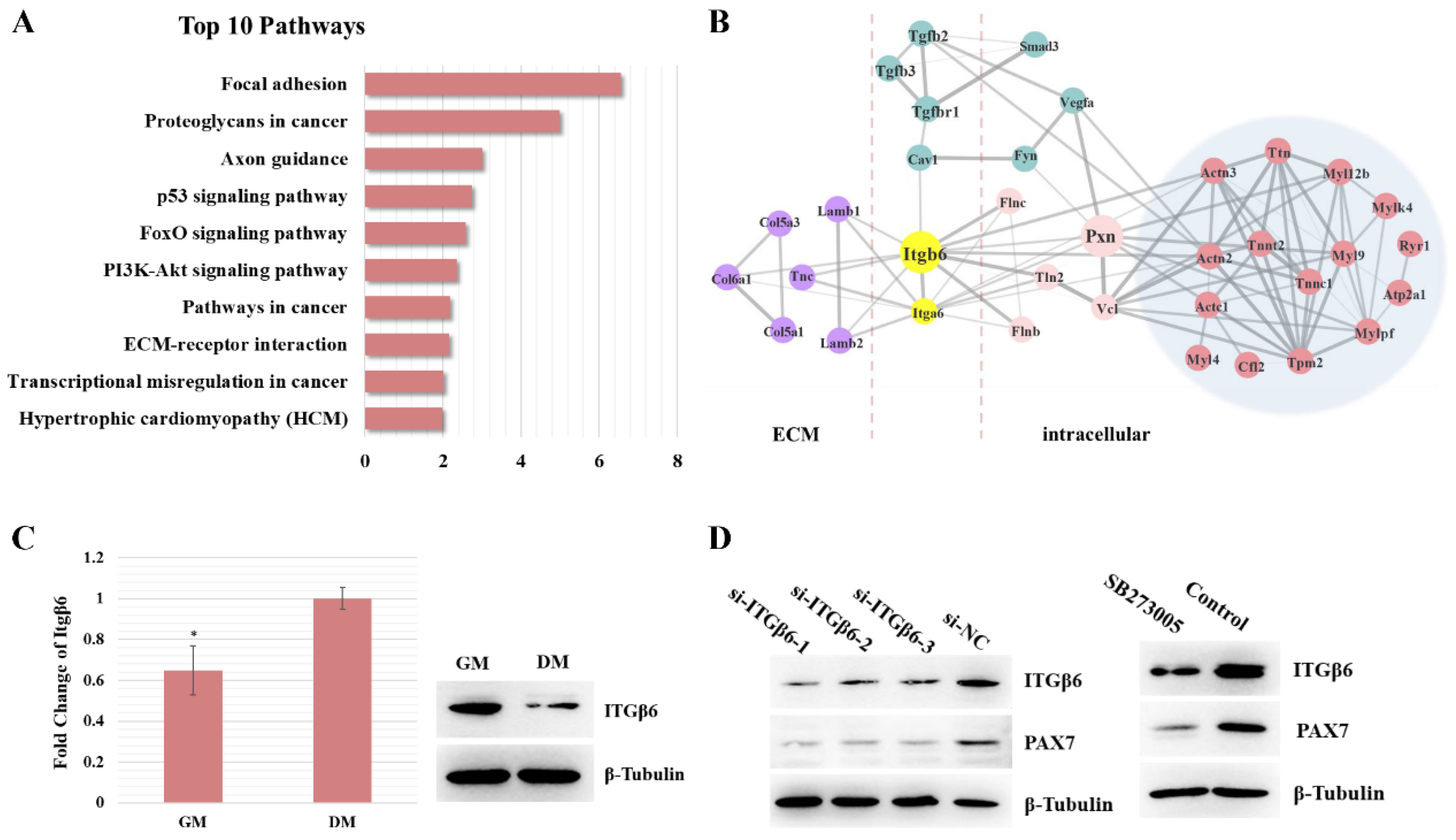
3.1. Itgβ6 Is One of the Key Node Genes during Skeletal Muscle Development
3.2. ITGβ6 Is Necessary for the Differentiation of Satellite Cells
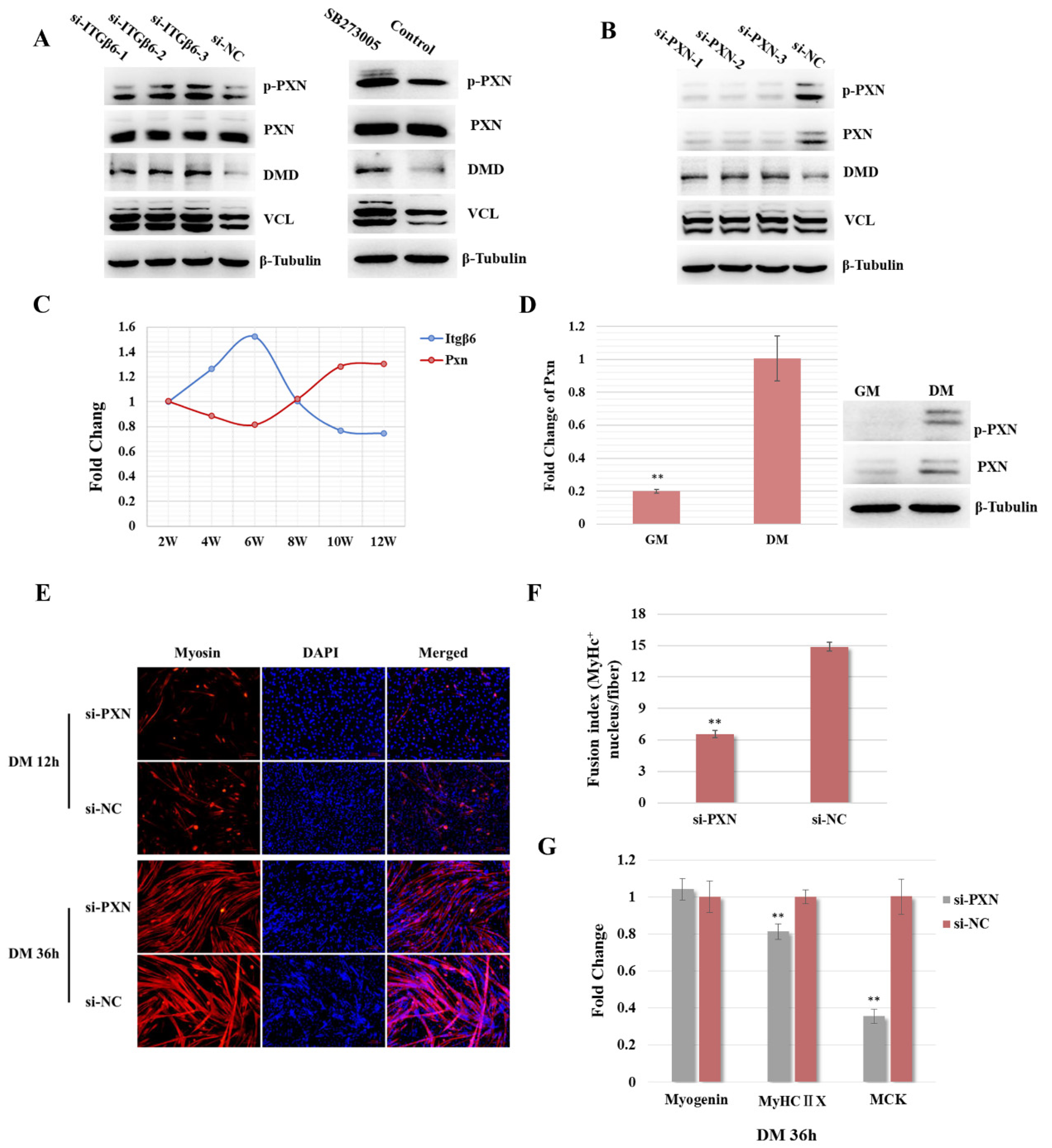
3.3. ITGβ6 Is Required to Maintain the Membrane Integrity and Cytoskeleton Stability of Satellite Cells
3.4. ITGβ6 Regulates the Differentiation of Satellite Cells through Affecting the Expression of Cytoskeleton Proteins
3.5. ITGβ6 Regulates the Expression of β-Laminin, a Component Protein of ECM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Guitart, M.; Lloreta, J.; García, L.M.; Barreiro, E. Muscle regeneration potential and satellite cell activation profile during recovery following hindlimb immobilization in mice. J. Cell. Physiol. 2018, 233, 4360–4372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lepper, C.; Conway, S.J.; Fan, C.-M. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature 2009, 460, 627–631. [Google Scholar] [CrossRef] [Green Version]
- Mauro, A. Satellite cell of skeletal muscle fibers. J. Cell Biol. 1961, 9, 493–495. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, Y.; Zhang, H. Extracellular matrix: An important regulator of cell functions and skeletal muscle development. Cell Biosci. 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Yehezkely, R.G.; Zaffryar-Eilot, S.; Kaganovsky, A.; Malka, N.F.; Aviram, R.; Livneh, I.; Hasson, P. Intracellular Role for the Matrix-Modifying Enzyme Lox in Regulating Transcription Factor Subcellular Localization and Activity in Muscle Regeneration. Dev. Cell 2020, 53, 406–417.e5. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; So, K.K.; Li, Y.; Li, Y.; Yuan, J.; Ding, Y.; Chen, F.; Huang, Y.; Liu, J.; Lee, W.; et al. Elevated H3K27ac in aged skeletal muscle leads to increase in extracellular matrix and fibrogenic conversion of muscle satellite cells. Aging Cell 2019, 18, e12996. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.-X.; Wu, B.-B.; Gong, L.; An, C.-R.; Lin, J.-X.; Li, Q.-K.; Jiang, D.-M.; Jin, K.-X.; Mechakra, A.; Bunpetch, V.; et al. Dissecting cell diversity and connectivity in skeletal muscle for myogenesis. Cell Death Dis. 2019, 10, 427. [Google Scholar] [CrossRef]
- Huang, Y.-L.; Liang, C.-Y.; Ritz, D.; Coelho, R.; Septiadi, D.; Estermann, M.; Cumin, C.; Rimmer, N.; Schötzau, A.; López, M.N.; et al. Collagen-rich omentum is a premetastatic niche for integrin α2-mediated peritoneal metastasis. eLife 2020, 9, e59442. [Google Scholar] [CrossRef] [PubMed]
- Bui, T.; Rennhack, J.; Mok, S.; Ling, C.; Perez, M.; Roccamo, J.; Andrechek, E.R.; Moraes, C.; Muller, W.J. Functional Redundancy between β1 and β3 Integrin in Activating the IR/Akt/mTORC1 Signaling Axis to Promote ErbB2-Driven Breast Cancer. Cell Rep. 2019, 29, 589–602.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Llacua, L.A.; Faas, M.M.; de Vos, P. Extracellular matrix molecules and their potential contribution to the function of transplanted pancreatic islets. Diabetologia 2018, 61, 1261–1272. [Google Scholar] [CrossRef] [Green Version]
- Sanes, J.R. The Basement Membrane/Basal Lamina of Skeletal Muscle. J. Biol. Chem. 2003, 278, 12601–12604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McClure, M.J.; Ramey, A.N.; Rashid, M.; Boyan, B.D.; Schwartz, Z. Integrin-α7 signaling regulates connexin 43, M-cadherin, and myoblast fusion. Am. J. Physiol. Physiol. 2019, 316, C876–C887. [Google Scholar] [CrossRef] [PubMed]
- Bayer, M.L.; Svensson, R.B.; Schjerling, P.; Williams, A.S.; Wasserman, D.H.; Kjaer, M. Influence of the integrin alpha-1 subunit and its relationship with high-fat diet upon extracellular matrix synthesis in skeletal muscle and tendon. Cell Tissue Res. 2020, 381, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Rayagiri, S.S.; Ranaldi, D.; Raven, A.; Azhar, N.I.F.M.; Lefebvre, O.; Zammit, P.; Borycki, A.-G. Basal lamina remodeling at the skeletal muscle stem cell niche mediates stem cell self-renewal. Nat. Commun. 2018, 9, 1075. [Google Scholar] [CrossRef] [Green Version]
- Baghdadi, M.B.; Castel, D.; Machado, L.; Fukada, S.-I.; Birk, D.E.; Relaix, F.; Tajbakhsh, S.; Mourikis, P. Reciprocal signalling by Notch–Collagen V–CALCR retains muscle stem cells in their niche. Nature 2018, 557, 714–718. [Google Scholar] [CrossRef]
- Seko, D.; Fujita, R.; Kitajima, Y.; Nakamura, K.; Imai, Y.; Ono, Y. Estrogen Receptor β Controls Muscle Growth and Regeneration in Young Female Mice. Stem Cell Rep. 2020, 15, 577–586. [Google Scholar] [CrossRef]
- Bae, H.; Hong, K.Y.; Lee, C.-K.; Jang, C.; Lee, S.-J.; Choe, K.; Offermanns, S.; He, Y.; Lee, H.J.; Koh, G.Y. Angiopoietin-2–integrin α5β1 signaling enhances vascular fatty acid transport and prevents ectopic lipid-induced insulin resistance. Nat. Commun. 2020, 11, 2980. [Google Scholar] [CrossRef]
- Magli, A.; Incitti, T.; Kiley, J.; Swanson, S.A.; Darabi, R.; Rinaldi, F.; Selvaraj, S.; Yamamoto, A.; Tolar, J.; Yuan, C.; et al. PAX7 Targets, CD54, Integrin α9β1, and SDC2, Allow Isolation of Human ESC/iPSC-Derived Myogenic Progenitors. Cell Rep. 2017, 19, 2867–2877. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Xu, Y.; Zhang, L.; Wang, S.; Yin, B.; Zhao, S.; Li, X. Synergistic effects of TGFβ2, WNT9a, and FGFR4 signals attenuate satellite cell differentiation during skeletal muscle development. Aging Cell 2018, 17, e12788. [Google Scholar] [CrossRef]
- Velleman, S.G. Recent Developments in Breast Muscle Myopathies Associated with Growth in Poultry. Annu. Rev. Anim. Biosci. 2019, 7, 289–308. [Google Scholar] [CrossRef]
- Pines, M.; Das, R.; Ellis, S.J.; Morin, A.; Czerniecki, S.; Yuan, L.; Klose, M.; Coombs, D.; Tanentzapf, G. Mechanical force regulates integrin turnover in Drosophila in vivo. Nature 2012, 14, 935–943. [Google Scholar] [CrossRef] [PubMed]
- Wood, A.J.; Cohen, N.; Joshi, V.; Li, M.; Costin, A.; Hersey, L.; A McKaige, E.; Manneken, J.D.; Sonntag, C.; Miles, L.B.; et al. RGD inhibition of itgb1 ameliorates laminin-α2-deficient zebrafish fibre pathology. Hum. Mol. Genet. 2018, 28, 1403–1413. [Google Scholar] [CrossRef] [PubMed]
- Heller, K.N.; Montgomery, C.L.; Janssen, P.M.; Clark, K.R.; Mendell, J.R.; Rodino-Klapac, L.R. AAV-mediated Overexpression of Human α7 Integrin Leads to Histological and Functional Improvement in Dystrophic Mice. Mol. Ther. 2013, 21, 520–525. [Google Scholar] [CrossRef] [Green Version]
- Ducceschi, M.; Clifton, L.G.; Stimpson, S.A.; Billin, A.N. Post-transcriptional regulation of ITGB6 protein levels in damaged skeletal muscle. Histochem. J. 2014, 45, 329–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, J.; Wang, S.; Zhou, J.; Tan, B.; Li, Z.; Zheng, E.; Cai, G.; Wu, Z.; Hong, L.; Gu, T. ITGB6 inhibits the proliferation of porcine skeletal muscle satellite cells. Cell Biol. Int. 2021, 46, 96–105. [Google Scholar] [CrossRef]
- Droguett, R.; Cabello-Verrugio, C.; Santander, C.; Brandan, E. TGF-β receptors, in a Smad-independent manner, are required for terminal skeletal muscle differentiation. Exp. Cell Res. 2010, 316, 2487–2503. [Google Scholar] [CrossRef]
- Yin, H.; Zhao, J.; He, H.; Chen, Y.; Wang, Y.; Li, D.; Zhu, Q. Gga-miR-3525 Targets PDLIM3 through the MAPK Signaling Pathway to Regulate the Proliferation and Differentiation of Skeletal Muscle Satellite Cells. Int. J. Mol. Sci. 2020, 21, 5573. [Google Scholar] [CrossRef]
- Gros, J.; Manceau, M.; Thomé, V.; Marcelle, C. A common somitic origin for embryonic muscle progenitors and satellite cells. Nature 2005, 435, 954–958. [Google Scholar] [CrossRef]
- Bhagavati, S.; Song, X.; Siddiqui, M.A.Q. RNAi inhibition of Pax3/7 expression leads to markedly decreased expression of muscle determination genes. Mol. Cell. Biochem. 2007, 302, 257–262. [Google Scholar] [CrossRef]
- Oustanina, S.; Hause, G.; Braun, T. Pax7 directs postnatal renewal and propagation of myogenic satellite cells but not their specification. EMBO J. 2004, 23, 3430–3439. [Google Scholar] [CrossRef] [Green Version]
- Relaix, F.; Rocancourt, D.; Mansouri, A.; Buckingham, M. A Pax3/Pax7-dependent population of skeletal muscle progenitor cells. Nat. 2005, 435, 948–953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seale, P.; Sabourin, L.A.; Girgis-Gabardo, A.; Mansouri, A.; Gruss, P.; Rudnicki, M.A. Pax7 Is Required for the Specification of Myogenic Satellite Cells. Cell 2000, 102, 777–786. [Google Scholar] [CrossRef] [Green Version]
- Schubert, F.; Tremblay, P.; Mansouri, A.; Faisst, A.M.; Kammandel, B.; Lumsden, A.; Gruss, P.; Dietrich, S. Early mesodermal phenotypes insplotch suggest a role forPax3 in the formation of epithelial somites. Dev. Dyn. 2001, 222, 506–521. [Google Scholar] [CrossRef] [Green Version]
- Petäjäniemi, N.; Korhonen, M.; Kortesmaa, J.; Tryggvason, K.; Sekiguchi, K.; Fujiwara, H.; Sorokin, L.; Thornell, L.-E.; Wondimu, Z.; Assefa, D.; et al. Localization of Laminin α4-Chain in Developing and Adult Human Tissues. J. Histochem. Cytochem. 2002, 50, 1113–1130. [Google Scholar] [CrossRef] [Green Version]
- Hall, T.E.; Bryson-Richardson, R.J.; Berger, S.; Jacoby, A.S.; Cole, N.J.; Hollway, G.E.; Berger, J.; Currie, P.D. The zebrafish candyfloss mutant implicates extracellular matrix adhesion failure in laminin α2-deficient congenital muscular dystrophy. Proc. Natl. Acad. Sci. USA 2007, 104, 7092–7097. [Google Scholar] [CrossRef] [Green Version]
- Gupta, V.A.; Kawahara, G.; Myers, J.A.; Chen, A.T.; Hall, T.E.; Manzini, M.C.; Currie, P.D.; Zhou, Y.; Zon, L.I.; Kunkel, L.M.; et al. A Splice Site Mutation in Laminin-α2 Results in a Severe Muscular Dystrophy and Growth Abnormalities in Zebrafish. PLoS ONE 2012, 7, e43794. [Google Scholar] [CrossRef]
- Burridge, K.; E Turner, C.; Romer, L.H. Tyrosine phosphorylation of paxillin and pp125FAK accompanies cell adhesion to extracellular matrix: A role in cytoskeletal assembly. J. Cell Biol. 1992, 119, 893–903. [Google Scholar] [CrossRef]
- Alvarez, K.; Fadic, R.; Brandan, E. Augmented synthesis and differential localization of heparan sulfate proteoglycans in Duchenne muscular dystrophy. J. Cell. Biochem. 2002, 85, 703–713. [Google Scholar] [CrossRef]
- Norwood, F.L.; Sutherland-Smith, A.J.; Keep, N.; Kendrick-Jones, J. The structure of the N-terminal actin-binding domain of human dystrophin and how mutations in this domain may cause Duchenne or Becker muscular dystrophy. Structure 2000, 8, 481–491. [Google Scholar] [CrossRef] [Green Version]
- Fabbrizio, E.; Bonetkerrache, A.; Limas, F.; Hugon, G.; Mornet, D. Dystrophin, the Protein That Promotes Membrane Resistance. Biochem. Biophys. Res. Commun. 1995, 213, 295–301. [Google Scholar] [CrossRef]
- Boujemaa-Paterski, R.; Martins, B.; Eibauer, M.; Beales, C.T.; Geiger, B.; Medalia, O. Talin-activated vinculin interacts with branched actin networks to initiate bundles. eLife 2020, 9, e53990. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Lin, L.; Sun, K.; Zhang, T.; Chen, W.; Li, L.; Xie, Y.; Wu, C.; Wei, Z.; Yu, C. Complex structures of Rsu1 and PINCH1 reveal a regulatory mechanism of the ILK/PINCH/Parvin complex for F-actin dynamics. eLife 2021, 10, e64395. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Liu, Y.; Li, C.; Zhang, W. ITGβ6 Facilitates Skeletal Muscle Development by Maintaining the Properties and Cytoskeleton Stability of Satellite Cells. Life 2022, 12, 926. https://doi.org/10.3390/life12070926
Zhang H, Liu Y, Li C, Zhang W. ITGβ6 Facilitates Skeletal Muscle Development by Maintaining the Properties and Cytoskeleton Stability of Satellite Cells. Life. 2022; 12(7):926. https://doi.org/10.3390/life12070926
Chicago/Turabian StyleZhang, Hong, Yuan Liu, Cencen Li, and Weiya Zhang. 2022. "ITGβ6 Facilitates Skeletal Muscle Development by Maintaining the Properties and Cytoskeleton Stability of Satellite Cells" Life 12, no. 7: 926. https://doi.org/10.3390/life12070926




