Strain Development in Microalgal Biotechnology—Random Mutagenesis Techniques
Abstract
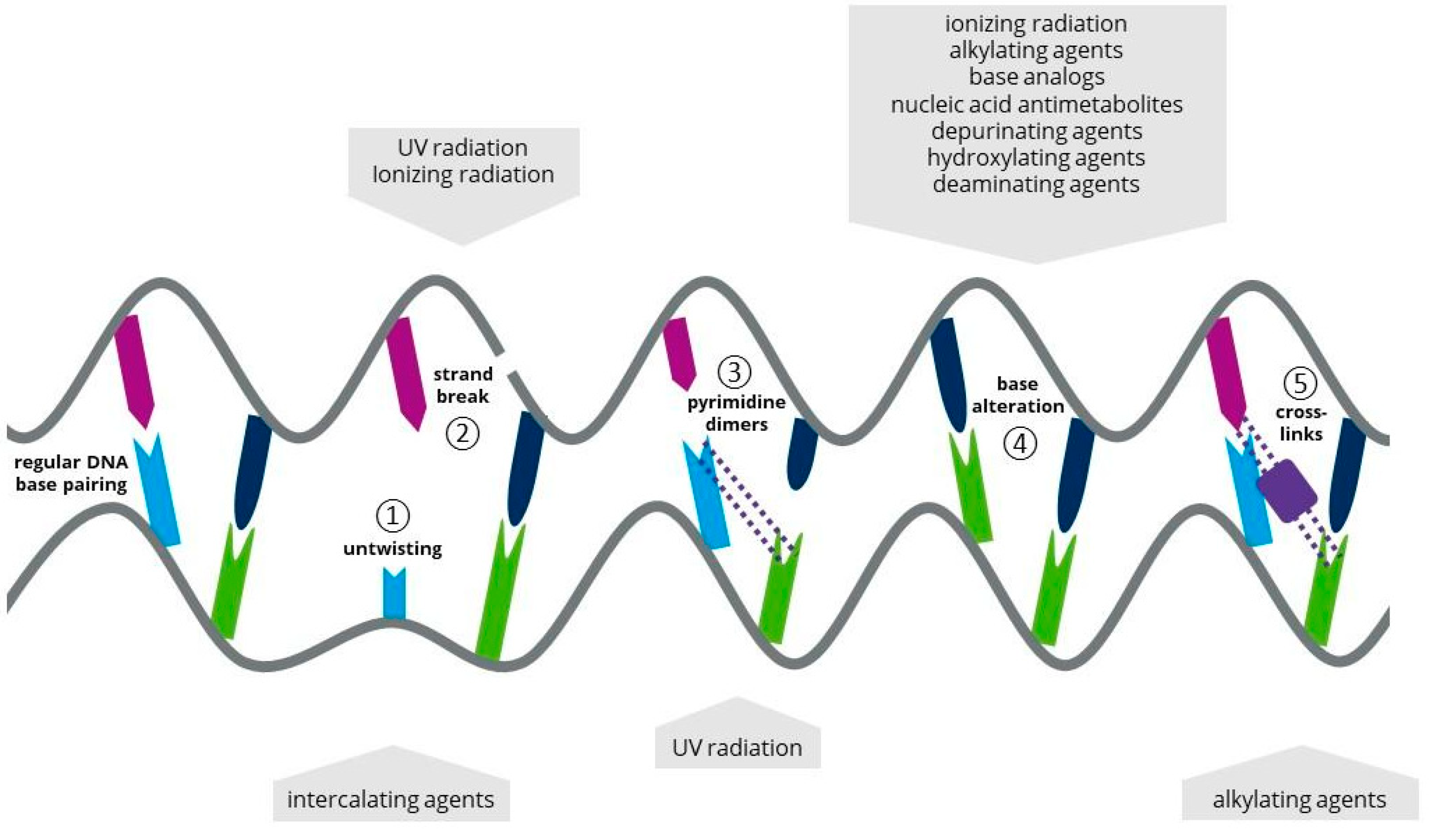
1. Introduction
2. Mutagens Applied to Microalgae for Random Mutagenesis
2.1. Physical Mutagens in Microalgal Biotechnology
2.1.1. Ultraviolet Light
2.1.2. Ionizing Radiation
2.1.3. Atmospheric and Room Temperature Plasma
2.1.4. Laser Radiation
| Mutagen | Method, Exposure Time, Source, Distance, Recovery Time | Reference Microalgae | Mutation Results | References | ||
|---|---|---|---|---|---|---|
| Mutated trait | WT * | M ** | ||||
| UV | UV 18 W, for 13 min, 15 cm, 24 h darkness | Chlorella vulgaris Y-019 | neutral lipid accumulation [g/g dry wt] | 0.11 | 0.26 | [36] |
| UV-C | UV-C 253.7 nm, 30-W, 3–30 min, 9 cm, 24 h darkness | Chlorella sp. | protein content [g/L] | 0.0242 | 0.0688 | [37] |
| UV-C 254 nm 1.4 mW/cm2 for 60 s, 15 cm, 16 h darkness | Chlorella vulgaris | fatty acids 16:0;18:0, 20:0 [% of total fatty acids] | 27.9; 3.9; 11.9 | 47.4; 5.9; 19.9 | [68] | |
| UV-C 254 nm, 15 W, (Vilber–Lourmat, France), for 30–180 s, 5 cm, 24 h darkness | natural isolates of photosynthetic microorganism | lipid content though Nile red autofluorescence; with fluorescence emission | 35; 1081 | 983; 89,770 | [38] | |
| UV-C 40,000 μJ/cm, 254 nm, overnight darkness | Scenedesmus obliquus | trans-fatty acid productivity [g/(L·d)] | 0.095 | 0.112 | [69] | |
| UV-C 254 nm 340 mW cm2, for 3–32 min, 13.5 cm, 24 h darkness | Isochrysis affinis galbana | total fatty acid [g/g dry wt] | 0.262 | 0.409 | [40] | |
| UV-C, for 1–10 min, 40 cm, overnight darkness | Chlorella vulgaris | lipid content [g/g] | 0.58 | 0.75 | [35] | |
| Gamma irradiation | 10 doses of irradiation 50–7000 kGy, 60Co gamma ray irradiator, room temperature | Scenedesmus sp. | lipid productivity [g/L·d] | 0.0648 | 0.097 | [70] |
| ARTP | He RF power 100 W, plasma temperature 25–35 °C, for 20; 40; 60 and 80 s, 2 mm | Spirulina platensis | Carbohydrates productivity [g/L·d] | 0.0157 | 0.026 | [59] |
| He RF power 100 W, plasma temperature 25–35 °C, 20–60 s, 2 mm | Chlamydomonas reinhardtii | H2 production [mL/L] | ~16.1 | 84.1 | [71] | |
| He RF power 150 W, for 100 s | Crypthecodinium cohnii | biomass concentration [g dry wt/L] | 3.60 | 4.24 | [72] | |
| Heavy ion beam | 12 C6+ ion beam 31 keVµm−1 160 Gy, | Nannochloropsis oceanica | lipid productivity [g/L·d] | 0.211 | 0.295 | [73] |
| 12 C6+ ion beam, 90 Gy | Desmodesmus sp. | lipid productivity [g/L·d] | 0.247 | 0.298 | [74] | |
| Low-energy ion beam implementation | N+ ion beam chamber pressure 10−2 Pa Dose of implantation 0.3–3.3·1015 ions cm−2 s−1 | Chlorella pyrenoidosa | lipid productivity [g/ L·d]; Lipid content [g/g dry wt] | 47.7; 0.337 | 64.4; 0.446 | [75] |
| laser radiation | He–Ne laser 808 nm, 6 W, 4 min, 24 h darkness | C. pyrenoidesa | lipid content [g/g dry wt] | 0.354 | 0.780 | [66] |
| Nd:YAG laser 1064 nm, 40 mW 8 min, 24 h darkness | Chlorella vulgaris | lipid content [g/g dry wt] | 0.315 | 0.525 | [66] | |
| Nd:YAG laser 1064 nm, 40 mW 2 min, 24 h darkness | Chlorella pacifica | lipid content [g L−1] | 0.033 | 0.088 | [76] | |
| semiconductor laser 632 nm, 40 mW, 4 min, 24 h darkness | Chlorella pacifica | lipid content [g L−1] | 0.033 | 0.077 | [76] | |
2.2. Chemical Mutagens in Microalgal Biotechnology
2.2.1. Alkylating Agents as a Chemical Mutagen
2.2.2. Base Analogs (BAs) as a Chemical Mutagen
2.2.3. Antimetabolites (AMs) as a Chemical Mutagen
2.2.4. Intercalating Agents (IAs) as a Chemical Mutagen
2.2.5. Other Approaches for Chemical Mutagenesis
| Mutagen | Mutagen Concentration, Time of Exposure | Reference Microalgae | Mutation Results | References | ||
|---|---|---|---|---|---|---|
| Mutated trait | WT * | M ** | ||||
| EMS | EMS 0.1–1.2 M for 60 min | Nannochloropsis sp. | fatty acid methyl esters [g/g of dry wt] | 0.123 | 0.238 | [101] |
| EMS 0.4–1 g/L for 60–120 min | Haematococcus pluvialis | total carotenoid; Astaxanthin [g/g of dry wt] | 0.02; 0.005 | 0.02; 0.019 | [102] | |
| EMS 300 mM for 60 min | Chlorella vulgaris | protein content [g/g of dry wt] | 0.353 | 0.455 | [34] | |
| EMS 0.2–0.4 M for 2 h in darkness | Chlorella vulgaris | violaxanthin [mg/L culture] | 1.64 | 5.23 | [103] | |
| EMS 0.1–0.2 M | Phaeodactylum tricornutum | total carotenoids [g/g dry wt] | 0.009 | 0.011 | [104] | |
| EMS 0.2 M for 2 h in the dark | Dunaliella tertiolecta | Zeaxanthin [μg/106·cells] | 0.131 | 0.359 | [105] | |
| EMS 20–40 µL/mL for 2 h | Chlamydomonas reinhardtii | fatty acid methyl esters yield [%] | 6.53 | 7.56 | [106] | |
| EMS 0.2 M for 2 h in the dark | Dunaliella salina | carotenoid synthesis [Mol Car/Mol Chl] | 0.99 | 1.24 | [107] | |
| EMS 100 μ mol mL−1, for 30 min | Chlorella sp. | lipid content [g/g of dry wt]; productivity [g/(L·d)] | 0.247; 0.1536 | 0.356; 0.2487 | [108] | |
| EMS 0.4 M, for 60 min | Coelastrum sp. | Astaxanthin content [g/L] | 0.0145 | 0.0283 | [109] | |
| EMS + UV | UV + EMS 25 mM for 60 min | Chlorella vulgaris | lipid content [%] | 100 | 167 | [85] |
| UV 5–240 s, 245 nm + EMS 0.24 mol/L for 30 min | Nannochloropsis salina | fatty acid methyl ester [g/g of dry wt] | 0.175 | 0.787 | [110] | |
| MNNG | MNNG 0.1 mM for 60 min | Haematococcus pluvialis | Total carotenoid content [g/L] | ~0.067 | 0.089 | [80] |
| MNNG 5 µg/mL for 60 min | Chlorella sp. | max. growth rate under alkaline conditions [ d−1] | 0.064 | 0.554 | [111] | |
| MNNG 0.02 mol/L for 60 min | Nannochloropsis oceanica | Total lipid content [g/g] Lipid productivity [g/(L·d)] | 0.241; 0.0065 | 0.299; 0.0086 | [33] | |
| MNNG 0.1–0.2 M | Phaeodactylum tricornutum | total carotenoids [g/g dry wt] | 0.009 | 0.011 | [104] | |
| MNNG 0.2 mg/mL | Chlorella sorokiniana | Lutein content [g/L] | 0.025 | 0.042 | [83] | |
| MNNG 0.25–0.5 mM | Botryosphaerella sp. | lipid [g dry wt/(m2 day)]; biomass productivity [g dry wt/(m2·day)] | 1.0; 3.2 | 1.9; 5.4 | [84] | |
| NMU | NMU 5 mM for 60–90 min | Nannochloropsis oculata | Total fatty acid [g/g dry wt] | 0.0634 | 0.0762 | [82] |
| DES + UV | UV 7–11 min 254 nm + DES 0.1–1.5% (V/V) 40 min | Haematococcus pluvialis | astaxanthin content [mg/L] | ~0.031 | ~0.089 | [81] |
| 5BU | 5BU 1 mM for 48 h | Chlamydomonas reinhardtii | O2 tolerance [%] | 100 | 1400 | [112] |
| 5′FDU | 5′FDU 0.25 and 0.50 mM for 1 week | Chlorella vulgaris | fatty acids 16:0; 18:0; 20:0 [% of total fatty acids] | 27.9; 3.9; 11.9 | 46.9; 5.5; 18.5 | [68] |
| Acriflavin | Acriflavin 2–8 μg/mL for 1–3 d in darkness | Chlamydomonas reinhardtii zyklo | Loss of respiratory rate [nmol O2/(min·107 cells)] through loss of mitochondrial DNA | 23.2 | 3.7 | [100] |
3. Further Approaches in Random Mutagenesis
4. Overview of High-Throughput Screening Methods and Techniques for Strain Selection
4.1. Screening Approaches on a Quantitative Basis
4.2. Screening Approaches on a Qualitative Basis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments

Conflicts of Interest
References
- Sydney, E.B.; Novak, A.C.; de Carvalho, J.C.; Soccol, C.R. Chapter—Balance and Carbon Fixation of Industrially Important Algae. In Biofuels from Algae; Pandey, A., Lee, D.-J., Chisti, Y., Soccol, C.R., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 67–84. ISBN 978-0-444-59558-4. [Google Scholar]
- Perez-Lopez, P.; González-García, S.; Jeffryes, C.; Agathos, S.; McHugh, E.; Walsh, D.; Murray, P.; Moane, S.; Feijoo, G.; Moreira, M.T. Life cycle assessment of the production of the red antioxidant carotenoid astaxanthin by microalgae: From lab to pilot scale. J. Clean. Prod. 2014, 64, 332–344. [Google Scholar] [CrossRef]
- León, R.; Martin, M.; Vigara, J.; Vilchez, C.; Vega, J.M. Microalgae mediated photoproduction of β-carotene in aqueous–organic two phase systems. Biomol. Eng. 2003, 20, 177–182. [Google Scholar] [CrossRef]
- Chen, C.-Y.; Jesisca; Hsieh, C.; Lee, D.-J.; Chang, C.-H.; Chang, J.-S. Production, extraction and stabilization of lutein from microalga Chlorella sorokiniana MB-1. Bioresour. Technol. 2015, 200, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Khandual, S.; Sanchez, E.O.L.; Andrews, H.E.; de la Rosa, J.D.P. Phycocyanin content and nutritional profile of Arthrospira platensis from Mexico: Efficient extraction process and stability evaluation of phycocyanin. BMC Chem. 2021, 15, 24. [Google Scholar] [CrossRef]
- De Jesus Raposo, M.F.; De Morais, A.M.B.; Santos Costa de Morais, R.M. Marine Polysaccharides from Algae with Potential Biomedical Applications. Mar. Drugs 2015, 13, 2967–3028. [Google Scholar] [CrossRef] [PubMed]
- Baianova, I.; Trubachev, I.N. Comparative evaluation of the vitamin composition of unicellular algae and higher plants grown under artificial conditions. Prikl. Biokhim. Mikrobiol. 1981, 17, 400–407. [Google Scholar]
- Brányiková, I.; Maršálková, B.; Doucha, J.; Brányik, T.; Bišová, K.; Zachleder, V.; Vítová, M. Microalgae-novel highly efficient starch producers. Biotechnol. Bioeng. 2010, 108, 766–776. [Google Scholar] [CrossRef] [PubMed]
- Bárcenas-Pérez, D.; Lukeš, M.; Hrouzek, P.; Kubáč, D.; Kopecký, J.; Kaštánek, P.; Cheel, J. A biorefinery approach to obtain docosahexaenoic acid and docosapentaenoic acid n-6 from Schizochytrium using high performance countercurrent chromatography. Algal Res. 2021, 55, 102241. [Google Scholar] [CrossRef]
- Abdo, S.; Ali, G.; El-Baz, F. Potential Production of Omega Fatty Acids from Microalgae. Int. J. Pharm. Sci. Rev. Res. 2015, 34, 210–215. [Google Scholar]
- Maltsev, Y.; Maltseva, K. Fatty acids of microalgae: Diversity and applications. Rev. Environ. Sci. Bio/Technol. 2021, 20, 515–547. [Google Scholar] [CrossRef]
- Loh, S.H.; Chen, M.K.; Fauzi, N.S.; Aziz, A.; Cha, T.S. Enhanced fatty acid methyl esters recovery through a simple and rapid direct transesterification of freshly harvested biomass of Chlorella vulgaris and Messastrum gracile. Sci. Rep. 2021, 11, 2720. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-L. (Ed.) Hb25_Springer Handbook of Marine Biotechnology; Springer: Berlin/Heidelberg, Germany, 2015; ISBN 978-3-642-53970-1. [Google Scholar]
- Woolston, B.M.; Edgar, S.; Stephanopoulos, G. Metabolic Engineering: Past and Future. Annu. Rev. Chem. Biomol. Eng. 2013, 4, 259–288. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Park, J.-H.; Bhattacharya, D.; Yoon, H.S. Applications of next-generation sequencing to unravelling the evolutionary history of algae. Int. J. Syst. Evol. Microbiol. 2014, 64, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Cao, X.; Wang, Y.; Zhu, Z.; Zhang, H.; Xue, S.; Tian, J. A Method for Microalgae Proteomics Analysis Based on Modified Filter-Aided Sample Preparation. Appl. Biochem. Biotechnol. 2017, 183, 923–930. [Google Scholar] [CrossRef]
- Chen, T.; Zhao, Q.; Wang, L.; Xu, Y.; Wei, W. Comparative Metabolomic Analysis of the Green Microalga Chlorella sorokiniana Cultivated in the Single Culture and a Consortium with Bacteria for Wastewater Remediation. Appl. Biochem. Biotechnol. 2017, 183, 1062–1075. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.; Xu, G.; Zhang, X.; Shi, J.; Xu, Z. Integration of ARTP mutagenesis with biosensor-mediated high-throughput screening to improve l-serine yield in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2018, 102, 5939–5951. [Google Scholar] [CrossRef]
- Ma, F.; Chung, M.T.; Yao, Y.; Nidetz, R.; Lee, L.M.; Liu, A.P.; Feng, Y.; Kurabayashi, K.; Yang, G.-Y. Efficient molecular evolution to generate enantioselective enzymes using a dual-channel microfluidic droplet screening platform. Nat. Commun. 2018, 9, 1–18. [Google Scholar] [CrossRef]
- Acevedo-Rocha, C.G.; Agudo, R.; Reetz, M.T. Directed evolution of stereoselective enzymes based on genetic selection as opposed to screening systems. J. Biotechnol. 2014, 191, 3–10. [Google Scholar] [CrossRef]
- Graham, P.J.; Riordon, J.; Sinton, D. Microalgae on display: A microfluidic pixel-based irradiance assay for photosynthetic growth. Lab Chip 2015, 15, 3116–3124. [Google Scholar] [CrossRef]
- Morschett, H.; Loomba, V.; Huber, G.; Wiechert, W.; Von Lieres, E.; Oldiges, M. Laboratory-scale photobiotechnology—current trends and future perspectives. FEMS Microbiol. Lett. 2018, 365, fnx238. [Google Scholar] [CrossRef]
- Morschett, H.; Schiprowski, D.; Müller, C.; Mertens, K.; Felden, P.; Meyer, J.; Wiechert, W.; Oldiges, M. Design and validation of a parallelized micro-photobioreactor enabling phototrophic bioprocess development at elevated throughput. Biotechnol. Bioeng. 2017, 114, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Weiss, T.L.; Thapa, H.R.; Devarenne, T.P.; Han, A. A microfluidic photobioreactor array demonstrating high-throughput screening for microalgal oil production. Lab Chip 2014, 14, 1415–1425. [Google Scholar] [CrossRef] [PubMed]
- Rowlands, R. Industrial strain improvement: Mutagenesis and random screening procedures. Enzym. Microb. Technol. 1984, 6, 3–10. [Google Scholar] [CrossRef]
- Khromov-Borisov, N.N. Naming the mutagenic nucleic acid base analogs: The Galatea syndrome. Mutat. Res. Mol. Mech. Mutagen. 1997, 379, 95–103. [Google Scholar] [CrossRef]
- Azin, M.; Noroozi, E. Random mutagenesis and use of 2-deoxy-D-glucose as an antimetabolite for selection of α-amylase-overproducing mutants of Aspergillus oryzae. World J. Microbiol. Biotechnol. 2001, 17, 747–750. [Google Scholar] [CrossRef]
- Buysschaert, B.; Byloos, B.; Leys, N.; Van Houdt, R.; Boon, N. Reevaluating multicolor flow cytometry to assess microbial viability. Appl. Microbiol. Biotechnol. 2016, 100, 9037–9051. [Google Scholar] [CrossRef]
- Elisabeth, B.; Rayen, F.; Behnam, T. Microalgae culture quality indicators: A review. Crit. Rev. Biotechnol. 2021, 41, 457–473. [Google Scholar] [CrossRef]
- Krujatz, F.; Lode, A.; Brüggemeier, S.; Schütz, K.; Kramer, J.; Bley, T.; Gelinsky, M.; Weber, J. Green bioprinting: Viability and growth analysis of microalgae immobilized in 3D-plotted hydrogels versus suspension cultures. Eng. Life Sci. 2015, 15, 678–688. [Google Scholar] [CrossRef]
- Bernaerts, T.M.M.; Gheysen, L.; Foubert, I.; Hendrickx, M.E.; Van Loey, A. Evaluating microalgal cell disruption upon ultra high pressure homogenization. Algal Res. 2019, 42, 101616. [Google Scholar] [CrossRef]
- Nescerecka, A.; Hammes, F.; Juhna, T. A pipeline for developing and testing staining protocols for flow cytometry, demonstrated with SYBR Green I and propidium iodide viability staining. J. Microbiol. Methods 2016, 131, 172–180. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, L.; Yang, G.; Han, J.; Thomsen, L.; Pan, K. Breeding 3 elite strains of Nannochloropsis oceanica by nitrosoguanidine mutagenesis and robust screening. Algal Res. 2016, 19, 104–108. [Google Scholar] [CrossRef]
- Schüler, L.M.; de Morais, E.G.; Dos Santos, M.; Machado, A.; Carvalho, B.; Carneiro, M.; Maia, I.B.; Soares, M.; Duarte, P.; Barros, A.; et al. Isolation and Characterization of Novel Chlorella Vulgaris Mutants with Low Chlorophyll and Improved Protein Contents for Food Applications. Front. Bioeng. Biotechnol. 2020, 8, 469. [Google Scholar] [CrossRef] [PubMed]
- Carino, J.D.; Vital, P.G. Characterization of isolated UV-C-irradiated mutants of microalga Chlorella vulgaris for future biofuel application. Environ. Dev. Sustain. 2022, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Li, Y.; Fei, X. Effects of Selective Medium on Lipid Accumulation of Chlorellas and Screening of High Lipid Mutants through Ultraviolet Mutagenesis. Afr. J. Agric. Res. 2011, 6, 3768–3774. [Google Scholar]
- Liu, S.; Zhao, Y.; Liu, L.; Ao, X.; Ma, L.; Wu, M.; Ma, F. Improving Cell Growth and Lipid Accumulation in Green Microalgae Chlorella sp. via UV Irradiation. Appl. Biochem. Biotechnol. 2015, 175, 3507–3518. [Google Scholar] [CrossRef]
- Ardelean, A.V.; Ardelean, I.I.; Sicuia-Boiu, O.A.; Cornea, P. Random- Mutagenesis in Photosynthetic Microorganisms Further Selected with Respect to Increased Lipid Content. Agric. Life Life Agric. Conf. Proc. 2018, 1, 501–507. [Google Scholar] [CrossRef][Green Version]
- De Jaeger, L.; Verbeek, R.E.; Draaisma, R.B.; Martens, D.E.; Springer, J.; Eggink, G.; Wijffels, R.H. Superior triacylglycerol (TAG) accumulation in starchless mutants of Scenedesmus obliquus: (I) mutant generation and characterization. Biotechnol. Biofuels 2014, 7, 69. [Google Scholar] [CrossRef]
- Bougaran, G.; Rouxel, C.; Dubois, N.; Kaas, R.; Grouas, S.; Lukomska, E.; Le Coz, J.-R.; Cadoret, J.-P. Enhancement of neutral lipid productivity in the microalga Isochrysis affinis Galbana (T-Iso) by a mutation-selection procedure. Biotechnol. Bioeng. 2012, 109, 2737–2745. [Google Scholar] [CrossRef]
- Yamamoto, J.; Plaza, P.; Brettel, K. Repair of (6-4) Lesions in DNA by (6-4) Photolyase: 20 Years of Quest for the Photoreaction Mechanism. Photochem. Photobiol. 2017, 93, 51–66. [Google Scholar] [CrossRef]
- Ness, R.W.; Morgan, A.D.; Colegrave, N.; Keightley, P.D. Estimate of the Spontaneous Mutation Rate in Chlamydomonas reinhardtii. Genetics 2012, 192, 1447–1454. [Google Scholar] [CrossRef]
- Krasovec, M.; Sanchez-Brosseau, S.; Grimsley, N.; Piganeau, G. Spontaneous mutation rate as a source of diversity for improving desirable traits in cultured microalgae. Algal Res. 2018, 35, 85–90. [Google Scholar] [CrossRef]
- García-Villada, L.; López-Rodas, V.; Bañares-España, E.; Flores-Moya, A.; Agrelo, M.; Martín-Otero, L.; Costas, E. Evolution of microalgae in highly stressing environments: An experimental model analyzing the rapid adaptation ofdictyosphaerium chlorelloides (chlorophyceae) from sensitivity to resistance against 2,4,6-trinitrotoluene by rare preselective mutations1. J. Phycol. 2002, 38, 1074–1081. [Google Scholar] [CrossRef]
- Park, E.-J.; Choi, J.-I. Resistance and Proteomic Response of Microalgae to Ionizing Irradiation. Biotechnol. Bioprocess Eng. 2018, 23, 704–709. [Google Scholar] [CrossRef]
- Chen, L.; Deng, S.; De Philippis, R.; Tian, W.; Wu, H.; Wang, J. UV-B resistance as a criterion for the selection of desert microalgae to be utilized for inoculating desert soils. J. Appl. Phycol. 2012, 25, 1009–1015. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Deng, S.; de Philippis, R.; Tian, W.; Wu, H.; Wang, J. Molecular Mechanisms of Ultraviolet Radiation-Induced DNA Damage and Repair. J. Nucleic Acids 2010, 2010, 592980. [Google Scholar] [CrossRef]
- Tillich, U.M.; Lehmann, S.; Schulze, K.; Dühring, U.; Frohme, M. The Optimal Mutagen Dosage to Induce Point-Mutations in Synechocystis sp. PCC6803 and Its Application to Promote Temperature Tolerance. PLoS ONE 2012, 7, e49467. [Google Scholar] [CrossRef]
- Holzinger, A.; Lütz, C. Algae and UV irradiation: Effects on ultrastructure and related metabolic functions. Micron 2006, 37, 190–207. [Google Scholar] [CrossRef]
- Rastogi, R.P.; Sinha, R.P.; Moh, S.H.; Lee, T.K.; Kottuparambil, S.; Kim, Y.-J.; Rhee, J.-S.; Choi, E.-M.; Brown, M.; Häder, D.-P.; et al. Ultraviolet radiation and cyanobacteria. J. Photochem. Photobiol. B Biol. 2014, 141, 154–169. [Google Scholar] [CrossRef]
- Graw, J. Genetik; Springer: Berlin/Heidelberg, Germany, 2010; ISBN 978-3-642-04998-9. [Google Scholar]
- Pfeifer, G.P.; You, Y.-H.; Besaratinia, A. Mutations induced by ultraviolet light. Mutat. Res. Mol. Mech. Mutagen. 2005, 571, 19–31. [Google Scholar] [CrossRef]
- Yi, Z.; Xu, M.; Magnusdottir, M.; Zhang, Y.; Brynjolfsson, S.; Fu, W. Photo-Oxidative Stress-Driven Mutagenesis and Adaptive Evolution on the Marine Diatom Phaeodactylum tricornutum for Enhanced Carotenoid Accumulation. Mar. Drugs 2015, 13, 6138–6151. [Google Scholar] [CrossRef]
- Sikder, S.; Biswas, P.; Hazra, P.; Akhtar, S.; Chattopadhyay, A.; Badigannavar, A.M.; D’Souza, S.F. Induction of mutation in tomato (Solanum lycopersicum L.) by gamma irradiation and EMS. Indian J. Genet. Plant Breed. 2013, 73, 392. [Google Scholar] [CrossRef]
- Min, J.; Lee, C.W.; Gu, M.B. Gamma-radiation dose-rate effects on DNA damage and toxicity in bacterial cells. Radiat. Environ. Biophys. 2003, 42, 189–192. [Google Scholar] [CrossRef]
- Klug, W.S.; Cummings, M.R.; Spencer, C.A.; Palladino, M.A. Concepts of Genetics, 11th ed.; Person Education Limited: Harlow, UK, 2016. [Google Scholar]
- Gomes, T.; Xie, L.; Brede, D.; Lind, O.-C.; Solhaug, K.A.; Salbu, B.; Tollefsen, K.E. Sensitivity of the green algae Chlamydomonas reinhardtii to gamma radiation: Photosynthetic performance and ROS formation. Aquat. Toxicol. 2017, 183, 1–10. [Google Scholar] [CrossRef]
- Senthamilselvi, D.; Kalaiselvi, T. Gamma ray mutants of oleaginous microalga Chlorella sp. KM504965 with enhanced biomass and lipid for biofuel production. Biomass-Convers. Biorefinery 2022, 1–17. [Google Scholar] [CrossRef]
- Fang, M.; Jin, L.; Zhang, C.; Tan, Y.; Jiang, P.; Ge, N.; Li, H.; Xing, X. Rapid Mutation of Spirulina platensis by a New Mutagenesis System of Atmospheric and Room Temperature Plasmas (ARTP) and Generation of a Mutant Library with Diverse Phenotypes. PLoS ONE 2013, 8, e77046. [Google Scholar] [CrossRef]
- Fridman, G.; Brooks, A.D.; Balasubramanian, M.; Fridman, A.; Gutsol, A.; Vasilets, V.N.; Ayan, H.; Friedman, G. Comparison of Direct and Indirect Effects of Non-Thermal Atmospheric-Pressure Plasma on Bacteria. Plasma Process. Polym. 2007, 4, 370–375. [Google Scholar] [CrossRef]
- Locke, B.; Sato, M.; Sunka, P.; Hoffmann, M.R.; Chang, J.-S. Electrohydraulic Discharge and Nonthermal Plasma for Water Treatment. Ind. Eng. Chem. Res. 2005, 45, 882–905. [Google Scholar] [CrossRef]
- Gaunt, L.F.; Beggs, C.B.; Georghiou, G. Bactericidal Action of the Reactive Species Produced by Gas-Discharge Nonthermal Plasma at Atmospheric Pressure: A Review. IEEE Trans. Plasma Sci. 2006, 34, 1257–1269. [Google Scholar] [CrossRef]
- Laroussi, M.; Leipold, F. Evaluation of the roles of reactive species, heat, and UV radiation in the inactivation of bacterial cells by air plasmas at atmospheric pressure. Int. J. Mass Spectrom. 2004, 233, 81–86. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.-F.; Li, H.-P.; Wang, L.-Y.; Zhang, C.; Xing, X.-H.; Bao, C.-Y. Atmospheric and room temperature plasma (ARTP) as a new powerful mutagenesis tool. Appl. Microbiol. Biotechnol. 2014, 98, 5387–5396. [Google Scholar] [CrossRef]
- Ouf, S.A.; Alsarrani, A.Q.; Al-Adly, A.A.; Ibrahim, M.K. Evaluation of low-intensity laser radiation on stimulating the cholesterol degrading activity: Part I. Microorganisms isolated from cholesterol-rich materials. Saudi J. Biol. Sci. 2012, 19, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Xing, W.; Zhang, R.; Shao, Q.; Meng, C.; Wang, X.; Wei, Z.; Sun, F.; Wang, C.; Cao, K.; Zhu, B.; et al. Effects of Laser Mutagenesis on Microalgae Production and Lipid Accumulation in Two Economically Important Fresh Chlorella Strains under Heterotrophic Conditions. Agronomy 2021, 11, 961. [Google Scholar] [CrossRef]
- Wang, K.; Lin, B.; Meng, C.; Gao, Z.; Li, Z.; Zhang, H.; Du, H.; Xu, F.; Jiang, X. Screening of three Chlorella mutant strains with high lipid production induced by 3 types of lasers. J. Appl. Phycol. 2020, 32, 1655–1668. [Google Scholar] [CrossRef]
- Anthony, J.; Rangamaran, V.R.; Gopal, D.; Shivasankarasubbiah, K.T.; Thilagam, M.L.J.; Dhassiah, M.P.; Padinjattayil, D.S.M.; Valsalan, V.N.; Manambrakat, V.; Dakshinamurthy, S.; et al. Ultraviolet and 5′Fluorodeoxyuridine Induced Random Mutagenesis in Chlorella vulgaris and Its Impact on Fatty Acid Profile: A New Insight on Lipid-Metabolizing Genes and Structural Characterization of Related Proteins. Mar. Biotechnol. 2014, 17, 66–80. [Google Scholar] [CrossRef] [PubMed]
- Breuer, G.; De Jaeger, L.; Artus, V.P.G.; Martens, D.E.; Springer, J.; Draaisma, R.B.; Eggink, G.; Wijffels, R.H.; Lamers, P.P. Superior triacylglycerol (TAG) accumulation in starchless mutants of Scenedesmus obliquus: (II) evaluation of TAG yield and productivity in controlled photobioreactors. Biotechnol. Biofuels 2014, 7, 70. [Google Scholar] [CrossRef]
- Liu, B.; Ma, C.; Xiao, R.; Xing, D.; Ren, H.; Ren, N. The screening of microalgae mutant strain Scenedesmus sp. Z-4 with a rich lipid content obtained by 60Co γ-ray mutation. RSC Adv. 2015, 5, 52057–52061. [Google Scholar] [CrossRef]
- Ban, S.; Lin, W.; Luo, Z.; Luo, J. Improving hydrogen production of Chlamydomonas reinhardtii by reducing chlorophyll content via atmospheric and room temperature plasma. Bioresour. Technol. 2018, 275, 425–429. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Sun, Z.; Ma, X.; Yang, B.; Jiang, Y.; Wei, D.; Chen, F. Mutation Breeding of Extracellular Polysaccharide-Producing Microalga Crypthecodinium cohnii by a Novel Mutagenesis with Atmospheric and Room Temperature Plasma. Int. J. Mol. Sci. 2015, 16, 8201–8212. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, Z.; Zhu, M.; Yu, C.; Cao, Y.; Zhang, D.; Zhou, G. Increased lipid productivity and TAG content in Nannochloropsis by heavy-ion irradiation mutagenesis. Bioresour. Technol. 2013, 136, 360–367. [Google Scholar] [CrossRef]
- Hu, G.; Fan, Y.; Zhang, L.; Yuan, C.; Wang, J.; Li, W.; Hu, Q.; Li, F.-L. Enhanced Lipid Productivity and Photosynthesis Efficiency in a Desmodesmus sp. Mutant Induced by Heavy Carbon Ions. PLoS ONE 2013, 8, e60700. [Google Scholar] [CrossRef]
- Tu, R.; Jin, W.; Wang, M.; Han, S.; Abomohra, A.E.-F.; Wu, W.-M. Improving of lipid productivity of the biodiesel promising green microalga Chlorella pyrenoidosa via low-energy ion implantation. J. Appl. Phycol. 2016, 28, 2159–2166. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, Z.; Li, Z.; Du, H.; Lin, B.; Cui, M.; Yin, Y.; Lei, F.; Yu, C.; Meng, C. Laser Radiation Induces Growth and Lipid Accumulation in the Seawater Microalga Chlorella pacifica. Energies 2017, 10, 1671. [Google Scholar] [CrossRef]
- Drabløs, F.; Feyzi, E.; Aas, P.A.; Vaagbø, C.B.; Kavli, B.; Bratlie, M.S.; Peña-Diaz, J.; Otterlei, M.; Slupphaug, G.; Krokan, H.E. Alkylation damage in DNA and RNA—Repair mechanisms and medical significance. DNA Repair 2004, 3, 1389–1407. [Google Scholar] [CrossRef] [PubMed]
- Slameňová, D.; Gábelová, A.; Ružeková, L.; Chalupa, I.; Horváthová, E.; Farkašová, T.; Bozsakyová, E.; Štětina, R. Detection of MNNG-induced DNA lesions in mammalian cells; validation of comet assay against DNA unwinding technique, alkaline elution of DNA and chromosomal aberrations. Mutat. Res. Repair 1997, 383, 243–252. [Google Scholar] [CrossRef]
- Engelward, B.P.; Allan, J.M.; Dreslin, A.J.; Kelly, J.D.; Wu, M.M.; Gold, B.; Samson, L.D. A Chemical and Genetic Approach Together Define the Biological Consequences of 3-Methyladenine Lesions in the Mammalian Genome. J. Biol. Chem. 1998, 273, 5412–5418. [Google Scholar] [CrossRef]
- Kamath, B.S.; Vidhyavathi, R.; Sarada, R.; Ravishankar, G. Enhancement of carotenoids by mutation and stress induced carotenogenic genes in Haematococcus pluvialis mutants. Bioresour. Technol. 2008, 99, 8667–8673. [Google Scholar] [CrossRef]
- Wang, N.; Guan, B.; Kong, Q.; Sun, H.; Geng, Z.; Duan, L. Enhancement of astaxanthin production from Haematococcus pluvialis mutants by three-stage mutagenesis breeding. J. Biotechnol. 2016, 236, 71–77. [Google Scholar] [CrossRef]
- Chaturvedi, R.; Uppalapati, S.R.; Alamsjah, M.A.; Fujita, Y. Isolation of quizalofop-resistant mutants of Nannochloropsis oculata (Eustigmatophyceae) with high eicosapentaenoic acid following N-methyl-N-nitrosourea-induced random mutagenesis. J. Appl. Phycol. 2004, 16, 135–144. [Google Scholar] [CrossRef]
- Cordero, B.F.; Obraztsova, I.; Couso, I.; Leon, R.; Vargas, M.A.; Rodriguez, H. Enhancement of Lutein Production in Chlorella sorokiniana (Chorophyta) by Improvement of Culture Conditions and Random Mutagenesis. Mar. Drugs 2011, 9, 1607–1624. [Google Scholar] [CrossRef]
- Nojima, D.; Ishizuka, Y.; Muto, M.; Ujiro, A.; Kodama, F.; Yoshino, T.; Maeda, Y.; Matsunaga, T.; Tanaka, T. Enhancement of Biomass and Lipid Productivities of Water Surface-Floating Microalgae by Chemical Mutagenesis. Mar. Drugs 2017, 15, 151. [Google Scholar] [CrossRef]
- Sarayloo, E.; Simsek, S.; Ünlü, Y.S.; Cevahir, G.; Erkey, C.; Kavakli, I.H. Enhancement of the lipid productivity and fatty acid methyl ester profile of Chlorella vulgaris by two rounds of mutagenesis. Bioresour. Technol. 2018, 250, 764–769. [Google Scholar] [CrossRef] [PubMed]
- Lian, M.; Huang, H.; Ren, L.; Ji, X.; Zhu, J.; Jin, L. Increase of Docosahexaenoic Acid Production by Schizochytrium sp. Through Mutagenesis and Enzyme Assay. Appl. Biochem. Biotechnol. 2009, 162, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Psoda, A.; Kierdaszuk, B.; Pohorille, A.; Geller, M.; Kusmierek, J.T.; Shugar, D. Interaction of the mutagenic base analogs O6-methylguanine and N4-hydroxycytosine with potentially complementary bases. Int. J. Quantum Chem. 1981, 20, 543–554. [Google Scholar] [CrossRef]
- Becket, E.; Tse, L.; Yung, M.; Cosico, A.; Miller, J.H. Polynucleotide Phosphorylase Plays an Important Role in the Generation of Spontaneous Mutations in Escherichia coli. J. Bacteriol. 2012, 194, 5613–5620. [Google Scholar] [CrossRef][Green Version]
- Cupples, C.G.; Miller, J.H. A set of lacZ mutations in Escherichia coli that allow rapid detection of each of the six base substitutions. Proc. Natl. Acad. Sci. USA 1989, 86, 5345–5349. [Google Scholar] [CrossRef] [PubMed]
- Jackson-Grusby, L.; Laird, P.W.; Magge, S.N.; Moeller, B.J.; Jaenisch, R. Mutagenicity of 5-aza-2′-deoxycytidine is mediated by the mammalian DNA methyltransferase. Proc. Natl. Acad. Sci. USA 1997, 94, 4681–4685. [Google Scholar] [CrossRef]
- Ang, J.; Song, L.Y.; D’Souza, S.; Hong, I.L.; Luhar, R.; Yung, M.; Miller, J.H. Mutagen Synergy: Hypermutability Generated by Specific Pairs of Base Analogs. J. Bacteriol. 2016, 198, 2776–2783. [Google Scholar] [CrossRef]
- PubChem Floxuridine|C9H11FN2O5|CID 5790. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/5790 (accessed on 3 June 2020).
- Sibirnyĭ, A.A.; Shavlovskiĭ, G.M.; Goloshchapova, G.V. Mutants of Pichia guilliermondii yeasts with multiple sensitivity to antibiotics and antimetabolites. I. The selection and properties of the mutants. Genetika 1977, 13, 872–879. [Google Scholar]
- Streisinger, G.; Okada, Y.; Emrich, J.; Newton, J.; Tsugita, A.; Terzaghi, E.; Inouye, M. Frameshift Mutations and the Genetic Code. Cold Spring Harb. Symp. Quant. Biol. 1966, 31, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, L.R.; Denny, W.A. Genotoxicity of non-covalent interactions: DNA intercalators. Mutat. Res. Mol. Mech. Mutagen. 2007, 623, 14–23. [Google Scholar] [CrossRef]
- Ferguson, L.R.; Turner, P.M.; Denny, W.A. The mutagenic effects of diacridines and diquinolines in microbial systems. Mutat. Res. Mol. Mech. Mutagen. 1990, 232, 337–343. [Google Scholar] [CrossRef]
- Shafer, R.H.; Waring, M.J. DNA bis-intercalation: Application of theory to the binding of echinomycin to DNA. Biopolymers 1980, 19, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Ephrussi, B. Action de l’ac-Riflavine Sur Res Levures. I. La Mutation “Petite Colonie”. Ann. Inst. Pasteur 1949, 76, 351–367. [Google Scholar]
- Ferguson, L.R.; von Borstel, R. Induction of the cytoplasmic ‘petite’ mutation by chemical and physical agents in Saccharomyces cerevisiae. Mutat. Res. Mol. Mech. Mutagen. 1992, 265, 103–148. [Google Scholar] [CrossRef]
- Matagne, R.F.; Michel-Wolwertz, M.-R.; Munaut, C.; Duyckaerts, C.; Sluse, F. Induction and characterization of mitochondrial DNA mutants in Chlamydomonas reinhardtii. J. Cell Biol. 1989, 108, 1221–1226. [Google Scholar] [CrossRef]
- Doan, T.T.Y.; Obbard, J.P. Enhanced intracellular lipid in Nannochloropsis sp. via random mutagenesis and flow cytometric cell sorting. Algal Res. 2012, 1, 17–21. [Google Scholar] [CrossRef]
- Gómez, P.I.; Inostroza, I.; Pizarro, M.; Pérez, J. From genetic improvement to commercial-scale mass culture of a Chilean strain of the green microalga Haematococcus pluvialis with enhanced productivity of the red ketocarotenoid astaxanthin. AoB Plants 2013, 5, plt026. [Google Scholar] [CrossRef]
- Kim, J.; Kim, M.; Lee, S.; Jin, E. Development of a Chlorella vulgaris mutant by chemical mutagenesis as a producer for natural violaxanthin. Algal Res. 2020, 46, 101790. [Google Scholar] [CrossRef]
- Yi, Z.; Su, Y.; Xu, M.; Bergmann, A.; Ingthorsson, S.; Rolfsson, O.; Salehi-Ashtiani, K.; Brynjolfsson, S.; Fu, W. Chemical Mutagenesis and Fluorescence-Based High-Throughput Screening for Enhanced Accumulation of Carotenoids in a Model Marine Diatom Phaeodactylum tricornutum. Mar. Drugs 2018, 16, 272. [Google Scholar] [CrossRef]
- Kim, M.; Ahn, J.; Jeon, H.; Jin, E. Development of a Dunaliella tertiolecta Strain with Increased Zeaxanthin Content Using Random Mutagenesis. Mar. Drugs 2017, 15, 189. [Google Scholar] [CrossRef]
- Lee, B.; Choi, G.-G.; Choi, Y.-E.; Sung, M.; Park, M.S.; Yang, J.-W. Enhancement of lipid productivity by ethyl methane sulfonate-mediated random mutagenesis and proteomic analysis in Chlamydomonas reinhardtii. Korean J. Chem. Eng. 2014, 31, 1036–1042. [Google Scholar] [CrossRef]
- Jin, E.; Feth, B.; Melis, A. A mutant of the green algaDunaliella salina constitutively accumulates zeaxanthin under all growth conditions. Biotechnol. Bioeng. 2002, 81, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Nayak, M.; Suh, W.I.; Oh, Y.T.; Ryu, A.J.; Jeong, K.J.; Kim, M.; Mohapatra, R.K.; Lee, B.; Chang, Y.K. Directed evolution of Chlorella sp. HS2 towards enhanced lipid accumulation by ethyl methanesulfonate mutagenesis in conjunction with fluorescence-activated cell sorting based screening. Fuel 2022, 316, 123410. [Google Scholar] [CrossRef]
- Tharek, A.; Yahya, A.; Salleh, M.; Jamaluddin, H.; Yoshizaki, S.; Hara, H.; Iwamoto, K.; Suzuki, I.; Mohamad, S.E. Improvement and screening of astaxanthin producing mutants of newly isolated Coelastrum sp. using ethyl methane sulfonate induced mutagenesis technique. Biotechnol. Rep. 2021, 32, e00673. [Google Scholar] [CrossRef] [PubMed]
- Beacham, T.; Macia, V.M.; Rooks, P.; White, D.; Ali, S. Altered lipid accumulation in Nannochloropsis salina CCAP849/3 following EMS and UV induced mutagenesis. Biotechnol. Rep. 2015, 7, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Kuo, C.-M.; Lin, T.-H.; Yang, Y.-C.; Zhang, W.-X.; Lai, J.-T.; Wu, H.-T.; Chang, J.-S.; Lin, C.-S. Ability of an alkali-tolerant mutant strain of the microalga Chlorella sp. AT1 to capture carbon dioxide for increasing carbon dioxide utilization efficiency. Bioresour. Technol. 2017, 244, 243–251. [Google Scholar] [CrossRef] [PubMed]
- Seibert, M.; Flynn, T.Y.; Ghirardi, M.L. Strategies for Improving Oxygen Tolerance of Algal Hydrogen Production. In Biohydrogen II; Elsevier: Amsterdam, The Netherlands, 2001; pp. 67–77. ISBN 978-0-08-043947-1. [Google Scholar]
- Sivaramakrishnan, R.; Incharoensakdi, A. Enhancement of lipid production in Scenedesmus sp. by UV mutagenesis and hydrogen peroxide treatment. Bioresour. Technol. 2017, 235, 366–370. [Google Scholar] [CrossRef]
- Hu, X.; Tang, X.; Bi, Z.; Zhao, Q.; Ren, L. Adaptive evolution of microalgae Schizochytrium sp. under high temperature for efficient production of docosahexaeonic acid. Algal Res. 2021, 54, 102212. [Google Scholar] [CrossRef]
- Mavrommati, M.; Daskalaki, A.; Papanikolaou, S.; Aggelis, G. Adaptive laboratory evolution principles and applications in industrial biotechnology. Biotechnol. Adv. 2021, 54, 107795. [Google Scholar] [CrossRef]
- Dragosits, M.; Mattanovich, D. Adaptive laboratory evolution—Principles and applications for biotechnology. Microb. Cell Fact. 2013, 12, 64. [Google Scholar] [CrossRef]
- Sun, X.-M.; Ren, L.-J.; Bi, Z.-Q.; Ji, X.-J.; Zhao, Q.-Y.; Huang, H. Adaptive evolution of microalgae Schizochytrium sp. under high salinity stress to alleviate oxidative damage and improve lipid biosynthesis. Bioresour. Technol. 2018, 267, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, M.J.; Bonturi, N.; Belouah, I.; Miranda, E.A.; Lahtvee, P.-J. Xylose Metabolism and the Effect of Oxidative Stress on Lipid and Carotenoid Production in Rhodotorula toruloides: Insights for Future Biorefinery. Front. Bioeng. Biotechnol. 2020, 8, 1008. [Google Scholar] [CrossRef] [PubMed]
- Miazek, K.; Iwanek, W.; Remacle, C.; Richel, A.; Goffin, D. Effect of Metals, Metalloids and Metallic Nanoparticles on Microalgae Growth and Industrial Product Biosynthesis: A Review. Int. J. Mol. Sci. 2015, 16, 23929–23969. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Chen, L.; Zhang, W. Chemicals to enhance microalgal growth and accumulation of high-value bioproducts. Front. Microbiol. 2015, 6, 56. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Tan, L.; Yang, B.; Wu, J.; Li, T.; Wu, H.; Wu, H.; Xiang, W. A mutant of seawater Arthrospira platensis with high polysaccharides production induced by space environment and its application potential. Algal Res. 2021, 61, 102562. [Google Scholar] [CrossRef]
- Liu, L.X.; Guo, H.J.; Zhao, L.S.; Wang, J.; Gu, J.Y.; Zhao, S.R. Achievements and Perspectives of Crop Space Breeding in China. Induced Plant Mutations in the Genomics Era; FAO: Rome, Italy, 2009; pp. 213–215. [Google Scholar]
- Han, W.; Li, C.; Miao, X.; Yu, G. A Novel Miniature Culture System to Screen CO2-Sequestering Microalgae. Energies 2012, 5, 4372–4389. [Google Scholar] [CrossRef]
- Linde, T.; Hansen, N.B.; Lübeck, M.; Lübeck, P.S. Fermentation in 24-well plates is an efficient screening platform for filamentous fungi. Lett. Appl. Microbiol. 2014, 59, 224–230. [Google Scholar] [CrossRef]
- Sharma, S.K.; Nelson, D.R.; Abdrabu, R.; Khraiwesh, B.; Jijakli, K.; Arnoux, M.; O’Connor, M.J.; Bahmani, T.; Cai, H.; Khapli, S.; et al. An integrative Raman microscopy-based workflow for rapid in situ analysis of microalgal lipid bodies. Biotechnol. Biofuels 2015, 8, 164. [Google Scholar] [CrossRef]
- Pereira, H.; Schulze, P.S.; Schüler, L.M.; Santos, T.; Barreira, L.; Varela, J. Fluorescence activated cell-sorting principles and applications in microalgal biotechnology. Algal Res. 2018, 30, 113–120. [Google Scholar] [CrossRef]
- Yu, X.-J.; Huang, C.-Y.; Chen, H.; Wang, D.-S.; Chen, J.-L.; Li, H.-J.; Liu, X.-Y.; Wang, Z.; Sun, J.; Wang, Z.-P. High-Throughput Biochemical Fingerprinting of Oleaginous Aurantiochytrium sp. Strains by Fourier Transform Infrared Spectroscopy (FT-IR) for Lipid and Carbohydrate Productions. Molecules 2019, 24, 1593. [Google Scholar] [CrossRef]
- Hong, M.-E.; Choi, S.P.; Park, Y.-I.; Kim, Y.-K.; Chang, W.S.; Kim, B.W.; Sim, S.J. Astaxanthin production by a highly photosensitive Haematococcus mutant. Process Biochem. 2012, 47, 1972–1979. [Google Scholar] [CrossRef]
- Bajhaiya, A.K.; Dean, A.P.; Driver, T.; Trivedi, D.K.; Rattray, N.J.W.; Allwood, J.W.; Goodacre, R.; Pittman, J.K. High-throughput metabolic screening of microalgae genetic variation in response to nutrient limitation. Metabolomics 2015, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Radzun, K.A.; Wolf, J.; Jakob, G.; Zhang, E.; Stephens, E.; Ross, I.; Hankamer, B. Automated nutrient screening system enables high-throughput optimisation of microalgae production conditions. Biotechnol. Biofuels 2015, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Unthan, S.; Radek, A.; Wiechert, W.; Oldiges, M.; Noack, S. Bioprocess automation on a Mini Pilot Plant enables fast quantitative microbial phenotyping. Microb. Cell Fact. 2015, 14, 32. [Google Scholar] [CrossRef] [PubMed]
- Morschett, H.; Wiechert, W.; Oldiges, M. Automation of a Nile red staining assay enables high throughput quantification of microalgal lipid production. Microb. Cell Fact. 2016, 15, 1–11. [Google Scholar] [CrossRef]
- Kosa, G.; Shapaval, V.; Kohler, A.; Zimmermann, B. FTIR spectroscopy as a unified method for simultaneous analysis of intra- and extracellular metabolites in high-throughput screening of microbial bioprocesses. Microb. Cell Fact. 2017, 16, 195. [Google Scholar] [CrossRef]
- Morschett, H.; Freier, L.; Rohde, J.; Wiechert, W.; Von Lieres, E.; Oldiges, M. A framework for accelerated phototrophic bioprocess development: Integration of parallelized microscale cultivation, laboratory automation and Kriging-assisted experimental design. Biotechnol. Biofuels 2017, 10, 1–13. [Google Scholar] [CrossRef]
- Sivakaminathan, S.; Hankamer, B.; Wolf, J.; Yarnold, J. High-throughput optimisation of light-driven microalgae biotechnologies. Sci. Rep. 2018, 8, 11687. [Google Scholar] [CrossRef]
- Zeng, W.; Guo, L.; Xu, S.; Chen, J.; Zhou, J. High-Throughput Screening Technology in Industrial Biotechnology. Trends Biotechnol. 2020, 38, 888–906. [Google Scholar] [CrossRef]
- Rohe, P.; Venkanna, D.; Kleine, B.; Freudl, R.; Oldiges, M. An automated workflow for enhancing microbial bioprocess optimization on a novel microbioreactor platform. Microb. Cell Fact. 2012, 11, 144. [Google Scholar] [CrossRef]
- Tillich, U.M.; Wolter, N.; Schulze, K.; Kramer, D.; Brödel, O.; Frohme, M. High-throughput cultivation and screening platform for unicellular phototrophs. BMC Microbiol. 2014, 14, 239. [Google Scholar] [CrossRef] [PubMed]
- Sesen, M.; Alan, T.; Neild, A. Droplet control technologies for microfluidic high throughput screening (μHTS). Lab Chip 2017, 17, 2372–2394. [Google Scholar] [CrossRef]
- Wang, B.L.; Ghaderi, A.; Zhou, H.; Agresti, J.J.; Weitz, D.A.; Fink, G.R.; Stephanopoulos, G. Microfluidic high-throughput culturing of single cells for selection based on extracellular metabolite production or consumption. Nat. Biotechnol. 2014, 32, 473–478. [Google Scholar] [CrossRef] [PubMed]
- Lippi, L.; Bähr, L.; Wüstenberg, A.; Wilde, A.; Steuer, R. Exploring the potential of high-density cultivation of cyanobacteria for the production of cyanophycin. Algal Res. 2018, 31, 363–366. [Google Scholar] [CrossRef]
- Dienst, D.; Wichmann, J.; Mantovani, O.; Rodrigues, J.S.; Lindberg, P. High density cultivation for efficient sesquiterpenoid biosynthesis in Synechocystis sp. PCC 6803. Sci. Rep. 2020, 10, 5932. [Google Scholar] [CrossRef]
- Hlavova, M.; Turoczy, Z.; Bisova, K. Improving microalgae for biotechnology—From genetics to synthetic biology. Biotechnol. Adv. 2015, 33, 1194–1203. [Google Scholar] [CrossRef]
- Qin, Y.; Wu, L.; Wang, J.; Han, R.; Shen, J.; Wang, J.; Xu, S.; Paguirigan, A.L.; Smith, J.L.; Radich, J.P.; et al. A Fluorescence-Activated Single-Droplet Dispenser for High Accuracy Single-Droplet and Single-Cell Sorting and Dispensing. Anal. Chem. 2019, 91, 6815–6819. [Google Scholar] [CrossRef]
- Hyka, P.; Lickova, S.; Přibyl, P.; Melzoch, K.; Kovar, K. Flow cytometry for the development of biotechnological processes with microalgae. Biotechnol. Adv. 2013, 31, 2–16. [Google Scholar] [CrossRef]
- Doan, T.-T.Y.; Obbard, J.P. Enhanced lipid production in Nannochloropsis sp. using fluorescence-activated cell sorting. GCB Bioenergy 2010, 3, 264–270. [Google Scholar] [CrossRef]
- Satpati, G.G.; Mallick, S.K.; Pal, R. An Alternative High-Throughput Staining Method for Detection of Neutral Lipids in Green Microalgae for Biodiesel Applications. Biotechnol. Bioproc. E 2015, 20, 1044–1055. [Google Scholar] [CrossRef]
- Satpati, G.G.; Pal, R. Rapid detection of neutral lipid in green microalgae by flow cytometry in combination with Nile red staining—an improved technique. Ann. Microbiol. 2015, 65, 937–949. [Google Scholar] [CrossRef]
- Rumin, J.; Bonnefond, H.; Saint-Jean, B.; Rouxel, C.; Sciandra, A.; Bernard, O.; Cadoret, J.-P.; Bougaran, G. The use of fluorescent Nile red and BODIPY for lipid measurement in microalgae. Biotechnol. Biofuels 2015, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Kishi, M.; Takahashi, K.; Furuya, K.; Wahid, M.E.A.; Khatoon, H.; Kasan, N.A. Isolation of lipid-rich marine microalgae by flow cytometric screening with Nile Red staining. Aquac. Int. 2019, 27, 509–518. [Google Scholar] [CrossRef]
- Govender, T.; Ramanna, L.; Rawat, I.; Bux, F. BODIPY staining, an alternative to the Nile Red fluorescence method for the evaluation of intracellular lipids in microalgae. Bioresour. Technol. 2012, 114, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Abatemarco, J.; Sarhan, M.F.; Wagner, J.M.; Lin, J.-L.; Liu, L.; Hassouneh, W.; Yuan, S.-F.; Alper, H.S.; Abate, A.R. RNA-aptamers-in-droplets (RAPID) high-throughput screening for secretory phenotypes. Nat. Commun. 2017, 8, 332. [Google Scholar] [CrossRef] [PubMed]
- Saad, M.G.; Dosoky, N.S.; Khan, M.S.; Zoromba, M.S.; Mekki, L.; El-Bana, M.; Nobles, D.; Shafik, H.M. High-Throughput Screening of Chlorella Vulgaris Growth Kinetics inside a Droplet-Based Microfluidic Device under Irradiance and Nitrate Stress Conditions. Biomolecules 2019, 9, 276. [Google Scholar] [CrossRef]
- Kim, H.S.; Hsu, S.-C.; Han, S.-I.; Thapa, H.R.; Guzman, A.R.; Browne, D.R.; Tatli, M.; Devarenne, T.P.; Stern, D.B.; Han, A. High-throughput droplet microfluidics screening platform for selecting fast-growing and high lipid-producing microalgae from a mutant library. Plant Direct 2017, 1, e00011. [Google Scholar] [CrossRef]
- Ren, L.; Yang, S.; Zhang, P.; Qu, Z.; Mao, Z.; Huang, P.-H.; Chen, Y.; Wu, M.; Wang, L.; Li, P.; et al. Standing Surface Acoustic Wave (SSAW)-Based Fluorescence-Activated Cell Sorter. Small 2018, 14, e1801996. [Google Scholar] [CrossRef]
- Kim, H.S.; Devarenne, T.P.; Han, A. A high-throughput microfluidic single-cell screening platform capable of selective cell extraction. Lab Chip 2015, 15, 2467–2475. [Google Scholar] [CrossRef]
- Kim, H.S.; Guzman, A.R.; Thapa, H.R.; Devarenne, T.P.; Han, A. A droplet microfluidics platform for rapid microalgal growth and oil production analysis. Biotechnol. Bioeng. 2016, 113, 1691–1701. [Google Scholar] [CrossRef]
- Bardin, D.; Kendall, M.R.; Dayton, P.A.; Lee, A.P. Parallel generation of uniform fine droplets at hundreds of kilohertz in a flow-focusing module. Biomicrofluidics 2013, 7, 34112. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-T.; Wang, C.Y. Review of Microfluidic Photobioreactor Technology for Metabolic Engineering and Synthetic Biology of Cyanobacteria and Microalgae. Micromachines 2016, 7, 185. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bleisch, R.; Freitag, L.; Ihadjadene, Y.; Sprenger, U.; Steingröwer, J.; Walther, T.; Krujatz, F. Strain Development in Microalgal Biotechnology—Random Mutagenesis Techniques. Life 2022, 12, 961. https://doi.org/10.3390/life12070961
Bleisch R, Freitag L, Ihadjadene Y, Sprenger U, Steingröwer J, Walther T, Krujatz F. Strain Development in Microalgal Biotechnology—Random Mutagenesis Techniques. Life. 2022; 12(7):961. https://doi.org/10.3390/life12070961
Chicago/Turabian StyleBleisch, Richard, Leander Freitag, Yob Ihadjadene, Una Sprenger, Juliane Steingröwer, Thomas Walther, and Felix Krujatz. 2022. "Strain Development in Microalgal Biotechnology—Random Mutagenesis Techniques" Life 12, no. 7: 961. https://doi.org/10.3390/life12070961
APA StyleBleisch, R., Freitag, L., Ihadjadene, Y., Sprenger, U., Steingröwer, J., Walther, T., & Krujatz, F. (2022). Strain Development in Microalgal Biotechnology—Random Mutagenesis Techniques. Life, 12(7), 961. https://doi.org/10.3390/life12070961








