Transcriptome Analysis of Sugarcane Young Leaves and Protoplasts after Enzymatic Digestion
Abstract
:1. Introduction
2. Result
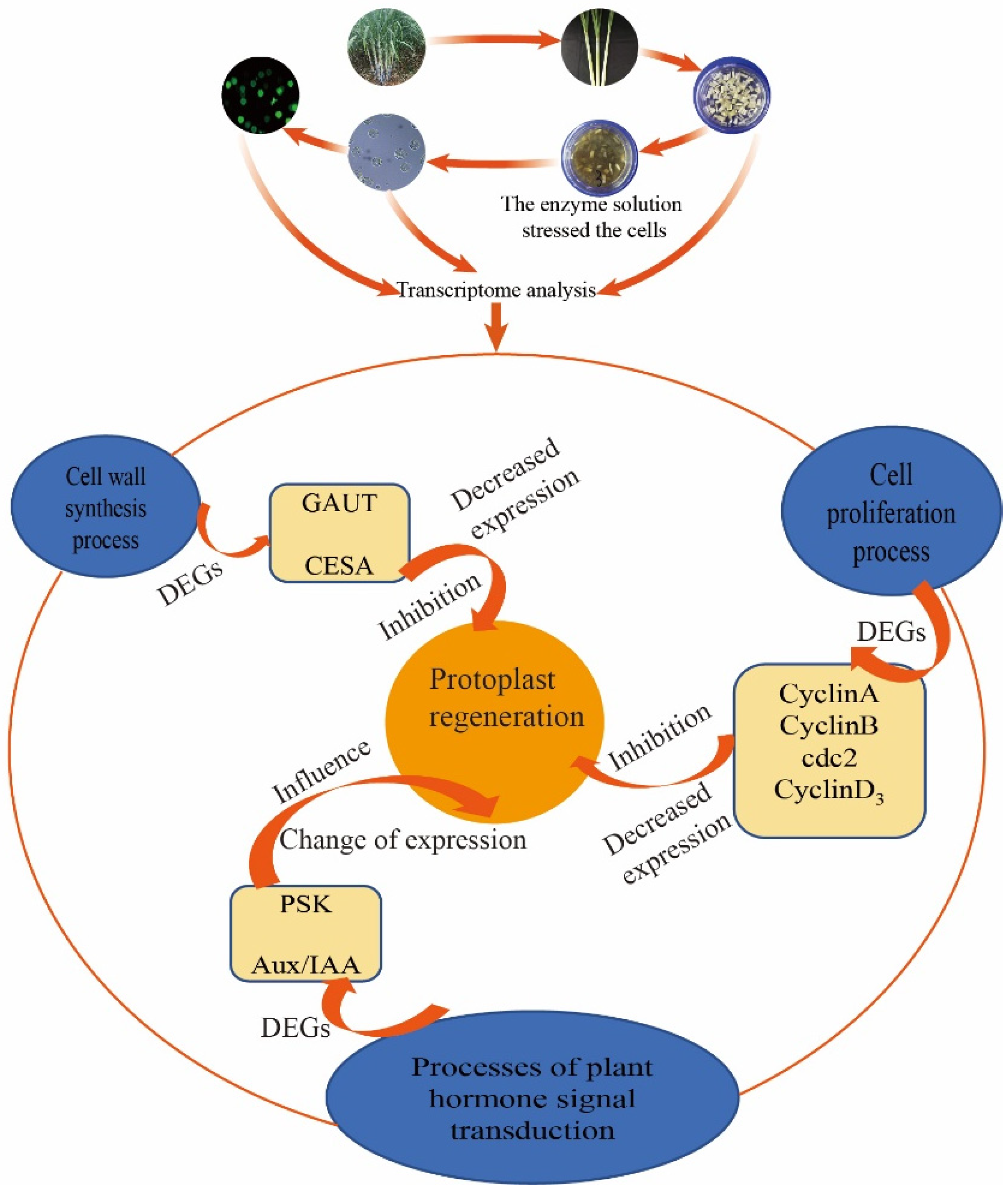
2.1. Detection of Protoplast Activity in Sugarcane
2.2. Quantitative Analysis of Differentially Expressed Genes by RNA-Seq
2.3. Validation of RNA-Seq Data by Quantitative RT-PCR
2.4. Cluster Analysis of Differential Gene Expression Levels
2.5. GO Function Analysis of DEGs
2.6. KEGG Enrichment Analysis of DEGs
2.7. Analysis of Related DEGs in Young Sugarcane Leaves before and after Enzymatic Digestion
2.8. Differential Expression of Regeneration Key Genes in Young Leaves before and after Enzymolysis
3. Discussion
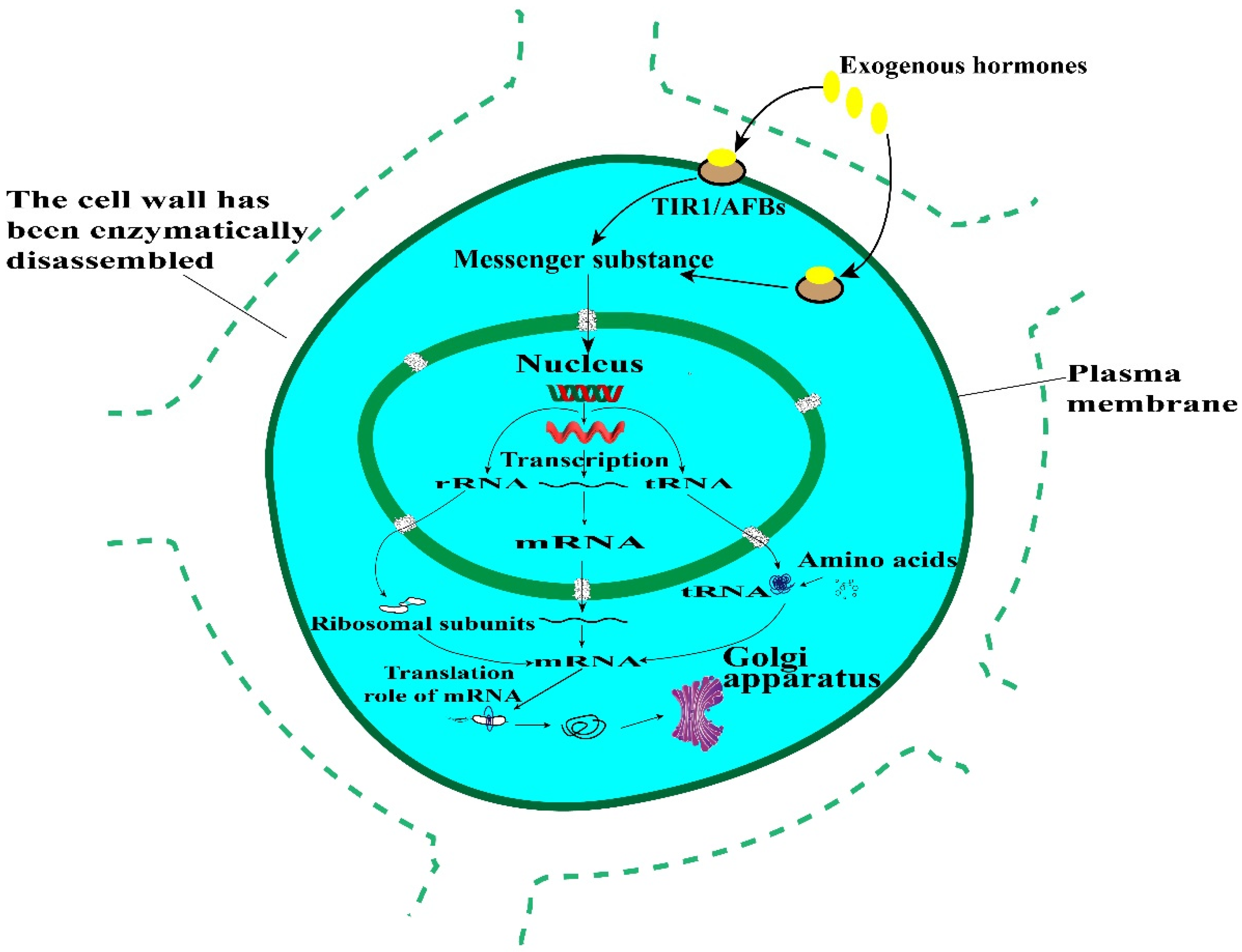
3.1. Enzymatic Hydrolysis Had a Significant Effect on Gene Expression in Sugarcane Protoplast
3.2. The Effect of Enzymatic Hydrolysis on Regeneration Related Genes May Lead to Difficulties in Plant Regeneration from Protoplasts
4. Material and Method
4.1. Plant Material Preparation
4.2. Preparation of RNA-Seq Libraries
4.3. Quality Control of Sequencing Data and Identification of Differentially Expressed Genes
4.4. Functional Annotation for DEGs
4.5. Real-Time Quantitative PCR Analysis of Genes
4.6. Data Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tiwari, J.K.; Devi, S.; Ali, N.; Luthra, S.K.; Kumar, V.; Bhardwaj, V.; Singh, R.K.; Rawat, S.; Chakrabarti, S.K. Progress in somatic hybridization research in potato during the past 40 years. Plant Cell Tissue Organ Cult. 2018, 132, 225–238. [Google Scholar] [CrossRef]
- Collonnier, C.; Mulya, K.; Fock, I.; Mariska, I.; Servaes, A.; Vedel, F.; Siljak-Yakovlev, S.; Souvannavong, V.; Ducreux, G.; Sihachakr, D. Source of resistance against Ralstonia solanacearum in fertile somatic hybrids of eggplant (Solanum melongena L.) with Solanum aethiopicum L. Plant Sci. 2001, 160, 301–313. [Google Scholar] [CrossRef]
- Luthra, S.K.; Tiwari, J.K.; Kumar, V.; Lal, M. Evaluation of Interspecific Somatic Hybrids of Potato (Solanum tuberosum) and Wild S. cardiophyllum for Adaptability, Tuber Dry Matter, Keeping Quality and Late Blight Resistance. Agric. Res. 2019, 8, 158–164. [Google Scholar] [CrossRef]
- Aleza, P.; Garcia-Lor, A.; Juárez, J.; Navarro, L. Recovery of citrus cybrid plants with diverse mitochondrial and chloroplastic genome combinations by protoplast fusion followed by in vitro shoot, root, or embryo micrografting. Plant Cell Tissue Organ Cult. 2016, 126, 205–217. [Google Scholar] [CrossRef]
- Cheng, A.; Xia, G. Somatic hybridisation between common wheat and Italian ryegrass. Plant Sci. 2004, 166, 1219–1226. [Google Scholar] [CrossRef]
- Xiang, F.; Xia, G.; Chen, H. Effect of UV dosage on somatic hybridization between common wheat (Triticum aestivum L.) and Avena sativa L. Plant Sci. 2003, 164, 697–707. [Google Scholar] [CrossRef]
- Xu, C.; Xia, G.; Zhi, D.; Xiang, F.; Chen, H. Integration of maize nuclear and mitochondrial DNA into the wheat genome through somatic hybridization. Plant Sci. 2003, 165, 1001–1008. [Google Scholar] [CrossRef]
- Kiran Kumar, M.; Sandeep, B.V.; Sudhakar Rao, P. Development of salt tolerant callus cultures by somatic hybridization between Oryza sativa and mangrove grass Myriostachya wightiana. Ann. Agrar. Sci. 2018, 16, 396–404. [Google Scholar] [CrossRef]
- Feng, Y.Y.; Takahashi, H.; Akagi, H.; Mori, K. New interspecific rice genome constructions obtained by symmetric cell fusion. Plant Breed. 2006, 125, 403–404. [Google Scholar] [CrossRef]
- Kang, H.H.; Naing, A.H.; Kim, C.K. Protoplast Isolation and Shoot Regeneration from Protoplast-Derived Callus of Petunia hybrida Cv. Mirage Rose. Biology 2020, 9, 228. [Google Scholar] [CrossRef]
- Su, W.Y.; Yang, H.C.; Tan, Y.L.; Zhou, Z.L.; Qin, Y.Y.; Li, X. Xiangying mianfa youhua Yumuer yuanshengzhiti zhibei tiaojian [Response surface methodology to optimize the preparation conditions for the protoplasmic system of Tamarindus indica]. Nangfang Nongye Xuebao [J. South. Agric.] 2020, 51, 169–175. [Google Scholar]
- Xu, M.; Du, Q.; Tian, C.; Wang, Y.; Jiao, Y. Stochastic gene expression drives mesophyll protoplast regeneration. Sci. Adv. 2021, 7, eabg8466. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Meng, D.; McGrouther, K.; Zhang, J.; Cheng, L. Efficient isolation of Magnolia protoplasts and the application to subcellular localization of MdeHSF1. Plant Methods 2017, 13, 44. [Google Scholar] [CrossRef]
- He, R.T.; Qin, W. Ganzhe he yancao youye yuansheng zhiti de hesuan hanliang yu hesuan mei huoxing jiqi yingxiang yinsu [Nucleic acid content and nuclease activity of young leaf protoplasts of sugarcane and tobacco and the factors affecting them]. Guangxi zhiwu [GUIHAIA] 1997, 1, 90–94. [Google Scholar]
- He, R.T.; Qin, W. Ganzhe he yancao youye yuansheng zhiti fenli qianhou de motou xing yu fuan suan han-liang de bianhua [Changes in membrane permeability and proline content of sugarcane and tobacco young leaf pro-toplasts before and after separation]. Guangxi nongye kexue [J. Guangxi Agric. Biol. Sci.] 2002, 4, 255–257. [Google Scholar]
- Taylor, P.W.J.; Ko, H.-L.; Adkins, S.W.; Rathus, C.; Birch, R.G. Establishment of embryogenic callus and high protoplast yielding suspension cultures of sugarcane (Saccharum spp. hybrids). Plant Cell Tissue Organ Cult. 1992, 28, 69–78. [Google Scholar] [CrossRef]
- He, M.; Zhang, Z.J.; Zhong, H.M.; Zhang, M.; Zhu, B.J. Yingxiang ganzhe peixing yushang zuzhi yuansheng zhiti peiyang de zhuyao yinsu [Major factors affecting the protoplast culture of sugarcane embryonic healing tissue]. Xinan Nongye Xuebao [Southwest China J. Agric. Sci.] 1994, 4, 71–76. [Google Scholar] [CrossRef]
- Li, J.; Liao, X.; Zhou, S.; Liu, S.; Jiang, L.; Wang, G. Efficient protoplast isolation and transient gene expression system for Phalaenopsis hybrid cultivar ‘Ruili Beauty’. Vitro Cell. Dev. Biol.-Plant 2018, 54, 87–93. [Google Scholar] [CrossRef]
- Bai, L.; Cheng, Y.; She, J.; He, Z.; Liu, H.; Zhang, G.; Cao, R.; Chen, Y. Development of an efficient protoplast isolation and transfection system for castor bean (Ricinus communis L.). Plant Cell Tissue Organ Cult. 2020, 143, 457–464. [Google Scholar] [CrossRef]
- Ma, W.; Yi, F.; Xiao, Y.; Yang, G.; Chen, F.; Wang, J. Isolation of leaf mesophyll protoplasts optimized by orthogonal design for transient gene expression in Catalpa bungei. Sci. Hortic. 2020, 274, 109684. [Google Scholar] [CrossRef]
- Jiang, Y.S.; Song, L.; Zhao, D.G. Baimai gen yuansheng zhiti zhibei meijie ye youhua ji zaisheng bi xianwei su hanliang fenxi[Optimization of enzymatic digestion solution for protoplasm preparation and analysis of cellulose content in regenerated walls of Passion root]. Fenzi zhiwu yuzhong [Mol. Plant Breed.] 2017, 15, 2274–2277. [Google Scholar] [CrossRef]
- Zhao, H.J.; Zhang, B.; Chen, A.P.; Li, P.Y.; Xiao, Y. Meijie dui muxu ziye yuansheng zhiti fenli xiaoguo de yingxiang[Effect of enzymatic digestion on the separation effect of protoplasts from alfalfa cotyledons]. Caodi Xuebao [Acta Agrestia Sin.] 2008, 1, 50–53. [Google Scholar]
- Chen, R.; Fan, Y.; Zhou, H.; Mo, S.; Zhou, Z.; Yan, H.; Luo, T.; Huang, X.; Weng, M.; Lakshmanan, P.; et al. Global transcriptome changes of elongating internode of sugarcane in response to mepiquat chloride. BMC Genom. 2021, 22, 79. [Google Scholar] [CrossRef] [PubMed]
- Li, S.L.; Liu, F.J.; Li, Z.G.; Lai, P.H.; Long, A.S. Ganzhe yuansheng zhiti RNA tiqu tiaojian he fangfa de youhua [Optimization of conditions and methods for RNA extraction from sugarcane protoplasts]. Redai Zuowu Xuebao [Chin. J. Trop. Crops] 2019, 40, 1537–1545. [Google Scholar]
- Wu, S.J.; Ding, Y.H.; Wang, X.Q. Xiaoli wanxian yuansheng zhiti xibao bi zaisheng guocheng guan-cha [Observations on the regeneration process of protoplast cell wall of small standing bowl moss]. Beijing Nongxue Xuebao [J. Beijing Univ. Agric.] 2016, 31, 1–4. [Google Scholar] [CrossRef]
- Sterling, J.D.; Atmodjo, M.A.; Inwood, S.E.; Kumar Kolli, V.S.; Quigley, H.F.; Hahn, M.G.; Mohnen, D. Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc. Natl. Acad. Sci. USA 2006, 103, 5236–5241. [Google Scholar] [CrossRef]
- Daras, G.; Templalexis, D.; Avgeri, F.; Tsitsekian, D.; Karamanou, K.; Rigas, S. Updating Insights into the Catalytic Domain Properties of Plant Cellulose synthase (CesA) and Cellulose synthase-like (Csl) Proteins. Molecules 2021, 26, 4335. [Google Scholar] [CrossRef]
- Caffall, K.H.; Pattathil, S.; Phillips, S.E.; Hahn, M.G.; Mohnen, D. Arabidopsis thaliana T-DNA Mutants Implicate GAUT Genes in the Biosynthesis of Pectin and Xylan in Cell Walls and Seed Testa. Mol. Plant 2009, 2, 1000–1014. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Liu, Z.W.; Zhang, Y.; Yang, J.; Ma, Z.Y.; Wang, X.F. Haidao mian GAUT jiyin jiazu de jianding jiqi zai xianwei fayu zhongde biaoda fenxi [Identification of GAUT gene family in Sea Island cotton and its expression analysis in cotton fiber development]. Zhiwu Yichuan Ziyuan Xuebao [J. Plant Genet. Resour.] 2018, 19, 722–730. [Google Scholar] [CrossRef]
- Nagar, P.; Kumar, A.; Jain, M.; Kumari, S.; Mustafiz, A. Genome-wide analysis and transcript profiling of PSKR gene family members in Oryza sativa. PLoS ONE 2020, 15, e0236349. [Google Scholar] [CrossRef]
- Matsubayashi, Y.; Takagi, L.; Sakagami, Y. Phytosulfokine-α, a sulfated pentapeptide, stimulates the proliferation of rice cells by means of specific high- and low-affinity binding sites. Proc. Natl. Acad. Sci. USA 1997, 94, 13357–13362. [Google Scholar] [CrossRef] [PubMed]
- Eun, C.-H.; Ko, S.-M.; Matsubayashi, Y.; Sakagami, Y.; Kamada, H. Phytosulfokine-α requires auxin to stimulate carrot non-embryogenic cell proliferation. Plant Physiol. Biochem. 2003, 41, 447–452. [Google Scholar] [CrossRef]
- Zhang, A.; Yang, X.; Lu, J.; Song, F.; Sun, J.; Wang, C.; Lian, J.; Zhao, L.; Zhao, B. OsIAA20, an Aux/IAA protein, mediates abiotic stress tolerance in rice through an ABA pathway. Plant Sci. 2021, 308, 110903. [Google Scholar] [CrossRef] [PubMed]
- Basunia, M.A.; Nonhebel, H.M.; Backhouse, D.; McMillan, M. Localised expression of OsIAA29 suggests a key role for auxin in regulating development of the dorsal aleurone of early rice grains. Planta 2021, 254, 40. [Google Scholar] [CrossRef]
- Nukina, K.; Hayashi, A.; Shiomi, Y.; Sugasawa, K.; Ohtsubo, M.; Nishitani, H. Mutations at multiple CDK phosphorylation consensus sites on Cdt2 increase the affinity of CRL4Cdt2 for PCNA and its ubiquitination activity in S phase. Genes Cells 2018, 23, 200–213. [Google Scholar] [CrossRef]
- Negis, Y.; Unal, A.Y.; Korulu, S.; Karabay, A. Cell cycle markers have different expression and localization patterns in neuron-like PC12 cells and primary hippocampal neurons. Neurosci. Lett. 2011, 496, 135–140. [Google Scholar] [CrossRef]
- Moritani, M.; Ishimi, Y. Inhibition of DNA binding of MCM2-7 complex by phosphorylation with cyclin-dependent kinases. J. Biochem. 2013, 154, 363–372. [Google Scholar] [CrossRef]
- Gibbs, E.; Pan, Z.-Q.; Niu, H.; Hurwitz, J. Studies on the Phosphorylation of HSSB-p34 and -p107 by Cyclin-dependent Kinases. J. Biol. Chem. 1996, 271, 22847–22854. [Google Scholar] [CrossRef]
- Chupeau, M.-C.; Granier, F.; Pichon, O.; Renou, J.-P.; Gaudin, V.; Chupeau, Y. Characterization of the Early Events Leading to Totipotency in an Arabidopsis Protoplast Liquid Culture by Temporal Transcript Profiling. Plant Cell 2013, 25, 2444–2463. [Google Scholar] [CrossRef]
- Xu, X.Y.; Xu, X.L.; Zhang, Y.H.; Liu, Y.L.; Chen, X. Ganju yuansheng zhiti zongRNA tiqu fangfa yanjiu [Study on the extraction method of total RNA from citrus protoplasts]. Yangzhou Daxue Xuebao [J. Yangzhou Univ.] 2016, 37, 111–114. [Google Scholar] [CrossRef]
- Mironov, V.; De Veylder, L.; Van Montagu, M.; Inzé, D. Cyclin-Dependent Kinases and Cell Division in Plants—The Nexus. Plant Cell 1999, 11, 509–521. [Google Scholar] [CrossRef] [PubMed]
- Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa–DPa transcription factor. EMBO J. 2002, 21, 1360–1368. [CrossRef] [PubMed]
- Wang, R.; Li, S.H.; Sun, S.W.; Wang, F.R. NDPK1 zai xibao fenlie zhongde gongneng fenxi [Functional analysis of NDPK1 in cell division]. Fenzi Zhiwu Yuzhong [Mol. Plant Breed.] 2018, 16, 3369–3373. [Google Scholar] [CrossRef]
- Moser, B.A.; Russell, P. Cell cycle regulation in Schizosaccharomyces pombe. Curr. Opin. Microbiol. 2000, 3, 631–636. [Google Scholar] [CrossRef]
- Song, Y.N. Ganzhe Yuansheng Zhiti de Ronghe Ji Zahe Xibao Zaisheng Tiaojian de Youhua [Optimization of Fusion of Sugarcane Protoplasts and Regeneration Conditions of Heteronuclear Cells]. Guangxi daxue [Guangxi University]. [Online]. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?dbcode=CMFD&dbname=CMFD201802&filename=1018132375.nh&uniplatform=NZKPT&v=WuUXeyaZs4ajY2jWKd-iEmJMSW5TpLxlc1tte_k2V-9A_Ni-aLMDiUbk8cSk321Y (accessed on 7 July 2022).
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
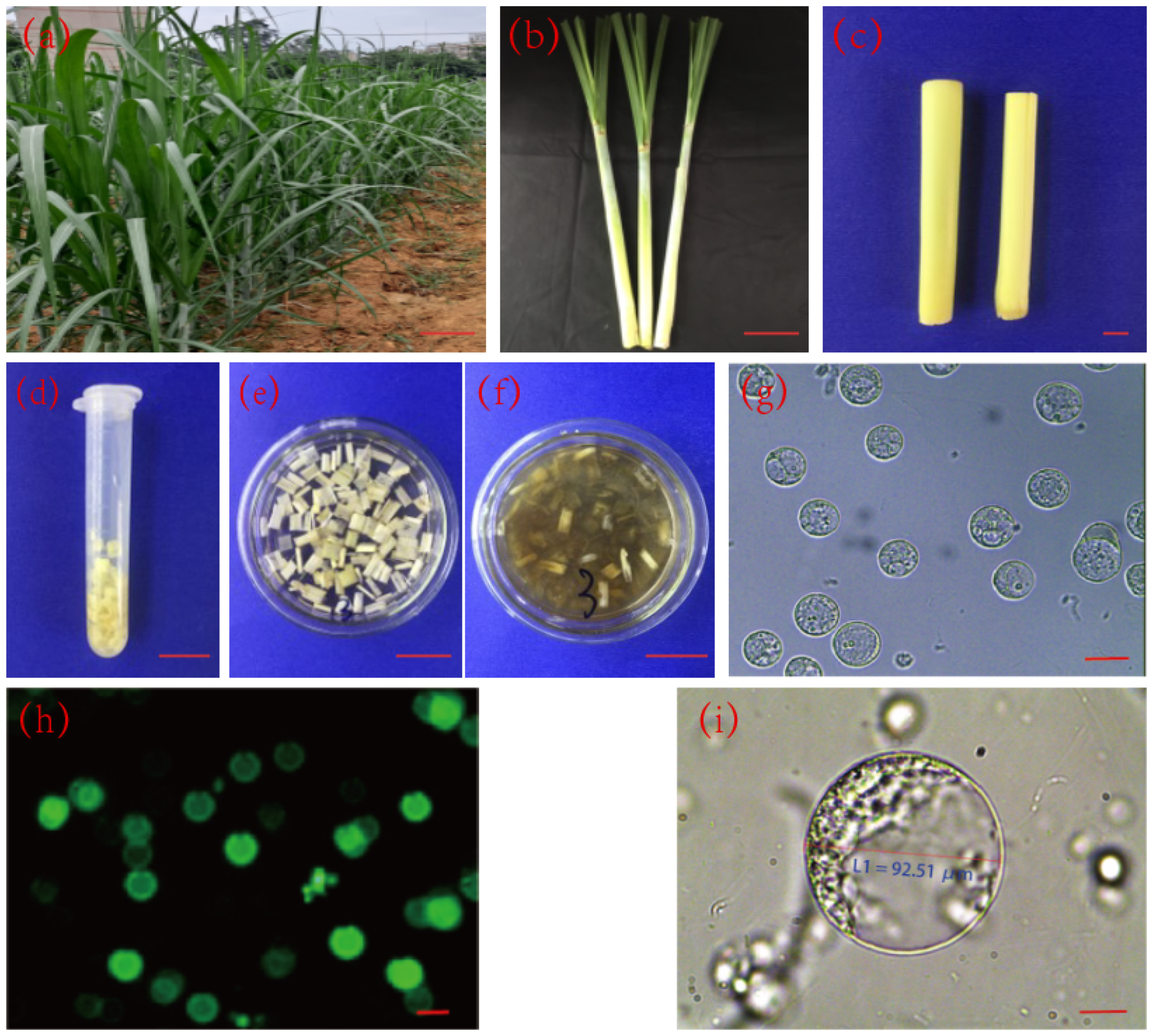

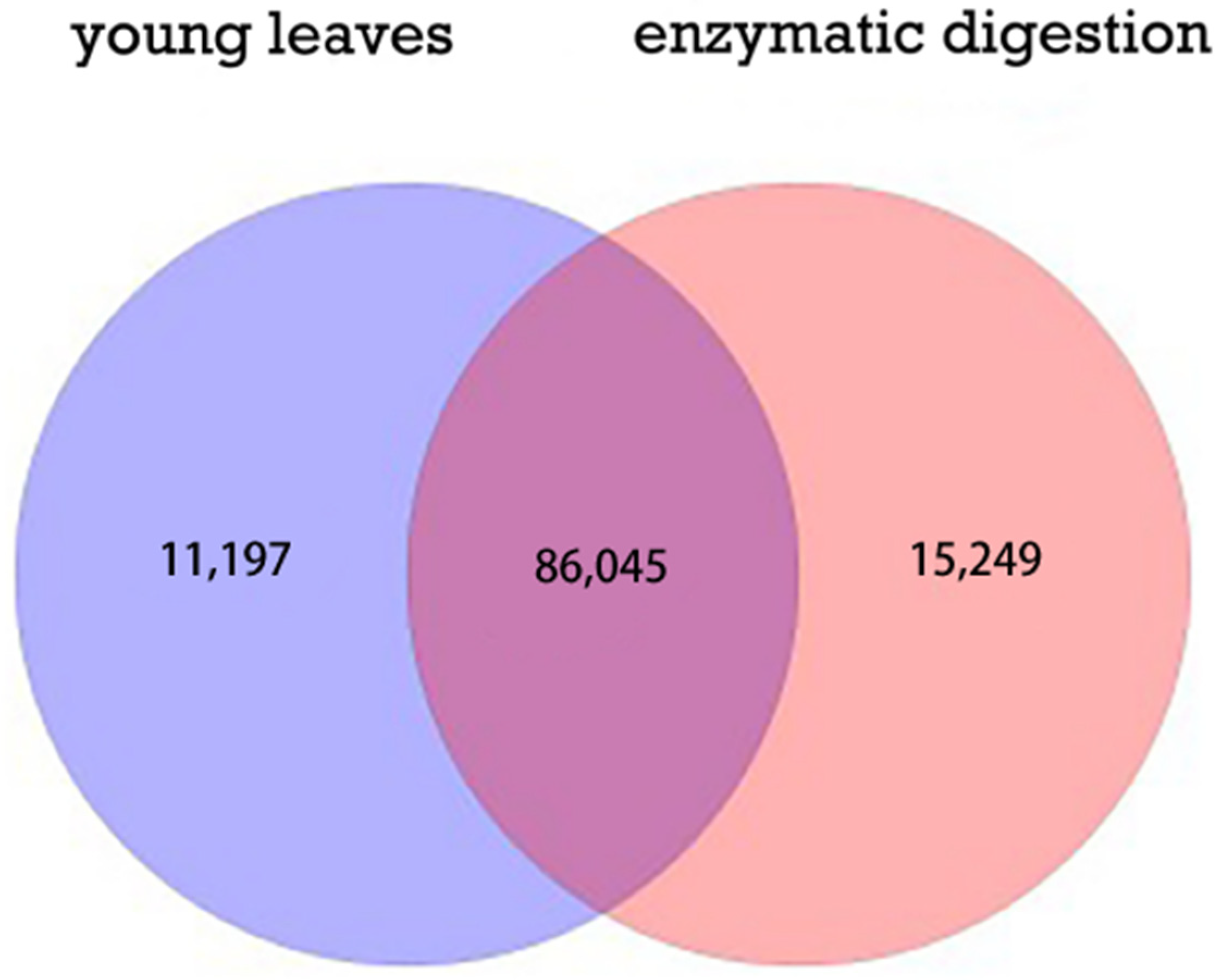


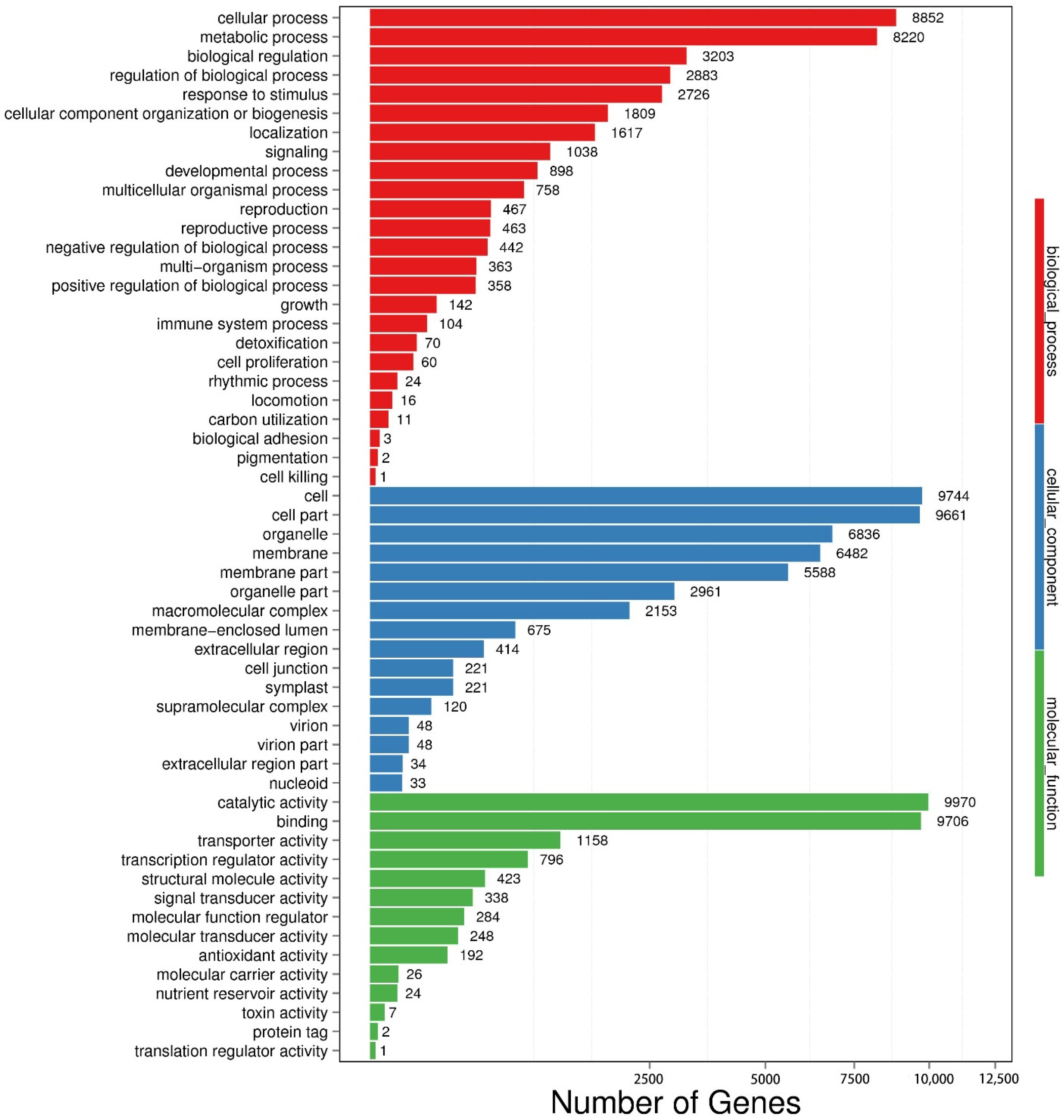


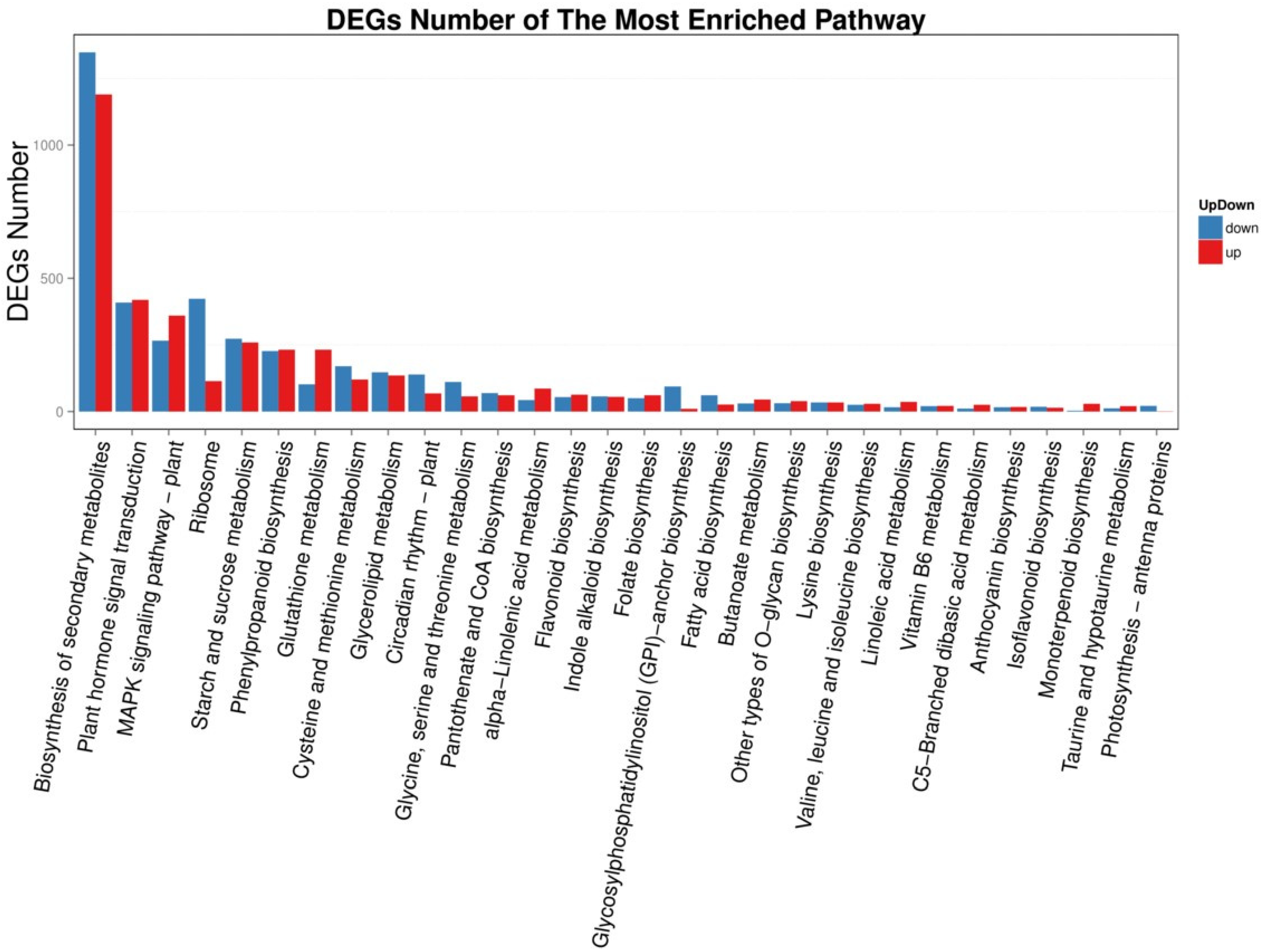

| Sample | Total Number | Total Length | Mean Length | N50 | N70 | N90 | GC (%) |
|---|---|---|---|---|---|---|---|
| enzymatic digestion 1 | 78,382 | 73,051,813 | 931 | 1560 | 949 | 369 | 49.07 |
| enzymatic digestion 2 | 77,229 | 72,426,174 | 937 | 1569 | 960 | 372 | 49.22 |
| young leaves 1 | 67,147 | 65,903,089 | 981 | 1611 | 1028 | 397 | 50.23 |
| young leaves 2 | 68,813 | 66,219,582 | 962 | 1598 | 1002 | 388 | 50.14 |
| Total unigenes | 117,411 | 1.29 × 108 | 1102 | 1800 | 1186 | 474 | 49.35 |
| Pathway | Gene Number | Background Gene Number | Rich Factor | Q-Value |
|---|---|---|---|---|
| Glutathione metabolism | 334 | 540 | 0.618518519 | 6.42 × 10−6 |
| Pantothenate and CoA biosynthesis | 130 | 197 | 0.659898477 | 4.60 × 10−4 |
| Photosynthesis—antenna proteins | 22 | 24 | 0.916666667 | 9.50 × 10−4 |
| Circadian rhythm—plant | 207 | 340 | 0.608823529 | 2.08 × 10−3 |
| C5-Branched dibasic acid metabolism | 36 | 46 | 0.782608696 | 2.61 × 10−3 |
| Starch and sucrose metabolism | 532 | 945 | 0.562962963 | 3.13 × 10−3 |
| Glycosyl phosphatidylinositol (GPI)-anchor biosynthesis | 104 | 161 | 0.645962733 | 3.68 × 10−3 |
| Ribosome | 537 | 958 | 0.560542797 | 3.80 × 10−3 |
| Glycerolipid metabolism | 282 | 490 | 0.575510204 | 1.26 × 10−2 |
| Folate biosynthesis | 111 | 179 | 0.620111732 | 1.54 × 10−2 |
| Vitamin B6 metabolism | 41 | 59 | 0.694915254 | 2.82 × 10−2 |
| Phenylpropanoid biosynthesis | 459 | 833 | 0.551020408 | 3.84 × 10−2 |
| Butanoate metabolism | 75 | 119 | 0.630252101 | 3.85 × 10−2 |
| Glycine, serine and threonine metabolism | 168 | 290 | 0.579310345 | 5.70 × 10−2 |
| Monoterpenoid biosynthesis | 32 | 46 | 0.695652174 | 5.97 × 10−2 |
| Lysine biosynthesis | 68 | 109 | 0.623853211 | 6.59 × 10−2 |
| Plant hormone signal transduction | 828 | 1551 | 0.53384913 | 7.14 × 10−2 |
| Cysteine and methionine metabolism | 290 | 522 | 0.555555556 | 7.41 × 10−2 |
| Alpha-linolenic acid metabolism | 129 | 221 | 0.583710407 | 7.41 × 10−2 |
| Indole alkaloid biosynthesis | 112 | 192 | 0.583333333 | 1.09 × 10−1 |
| GO Term | Number of Up-Regulated Genes | Number of Down-Regulated Genes |
|---|---|---|
| Cell cycle | 36 | 80 |
| Development | 132 | 426 |
| Cell differentiation | 40 | 104 |
| Cell wall | 154 | 258 |
| Cell proliferation | 10 | 50 |
| Oxidative stress | 84 | 105 |
| MAPK signaling pathway | 2 | 0 |
| Cell death | 17 | 17 |
| Growth | 42 | 102 |
| Cell killing | 1 | 0 |
| Transcription regulator activity | 42 | 71 |
| Antioxidant activity | 1 | 5 |
| Plant hormone signal transduction | 0 | 3 |
| Protein kinase activity | 465 | 337 |
| Signal transduction | 215 | 308 |
| response to external stimulus | 72 | 108 |
| Morphogenesis | 21 | 49 |
| Post-embryonic development | 2 | 7 |
| Defense response | 177 | 169 |
| Response to stress | 60 | 36 |
| Cell death in response to oxidative stress | 0 | 2 |
| Regulation of response to reactive oxygen species | 8 | 20 |
| Osmotic regulation | 9 | 9 |
| Gene ID | Primer 5′—3′ | |
|---|---|---|
| Unigene17671_All | F | CCGAGACCAAAGACATCTTGC |
| R | GGGATCAGCTTCGTCATCAC | |
| CL11183.Contig2_All | F | GGGCTACTCGAAGCTGATTG |
| R | CGAATCGGACTCTAGGGTTGA | |
| CL4403.Contig2_All | F | CCTACGCCGATTTCTACCAG |
| R | GCTTGCCAAAGACTTGCCTC | |
| CL12119.Contig2_All | F | GCACTACCAGCATGGGTTTAG |
| R | GCTCCCGTGGCATACTACAA | |
| CL5995.Contig1_All | F | GGCACAGGGCTAGTTTTAGAC |
| R | CCACCAGAGTACATTCCACG | |
| CL15852.Contig2_All | F | CAAGAAGGCTGGCAGGTGGAAG |
| R | CACGAGCAAGTCCTCTGACAGTTC | |
| CL1500.Contig7_All | F | AAGGATGTGAATGCCGCTGTGG |
| R | CGCTGGTGGAGTTGGAGATCATG | |
| CL7155.Contig2_All | F | AGGCGGAATGATAGGTCGAGGTC |
| R | CGTCGTAGCGGTCGGAGGAG | |
| CL5141.Contig1_All | F | CCAGTTCTGCCTCAACCACTTCTC |
| R | GTGCCTGCCGTCTGCTTCTC | |
| CL3148.Contig3_All | F | CCAGTCGCCATCACCATCATCATC |
| R | CTCCTCCTCGCCGCTGTCAG | |
| GADPH | F | AAGGGTGGTGCCAAGAAGG |
| R | CAAGGGGAGCAAGGCAGTT | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Wang, R.; Han, S.; Li, Z.; Xiao, J.; Li, Y.; Wang, L.; Li, S. Transcriptome Analysis of Sugarcane Young Leaves and Protoplasts after Enzymatic Digestion. Life 2022, 12, 1210. https://doi.org/10.3390/life12081210
Zhang D, Wang R, Han S, Li Z, Xiao J, Li Y, Wang L, Li S. Transcriptome Analysis of Sugarcane Young Leaves and Protoplasts after Enzymatic Digestion. Life. 2022; 12(8):1210. https://doi.org/10.3390/life12081210
Chicago/Turabian StyleZhang, Demei, Rui Wang, Shijian Han, Zhigang Li, Jiming Xiao, Yangrui Li, Lingqiang Wang, and Suli Li. 2022. "Transcriptome Analysis of Sugarcane Young Leaves and Protoplasts after Enzymatic Digestion" Life 12, no. 8: 1210. https://doi.org/10.3390/life12081210






