Anthocyanin Accumulation and Molecular Analysis of Correlated Genes by Metabolomics and Transcriptomics in Sister Line Apple Cultivars
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Measurement of Apple Skin Color Traits
2.2.1. Relative Total Anthocyanin Content
2.2.2. Measurement of Anthocyanin Metabolites by HPLC-ESI-MS/MS
2.3. RNA Sequencing and Analysis
2.4. WGCNA and Visualization of Gene Networks
2.5. Quantitative Real-Time PCR (qRT-PCR) Validation of RNA-Seq Results
2.6. Function Prediction of Candidate Genes
2.7. Statistical Analyses
3. Results
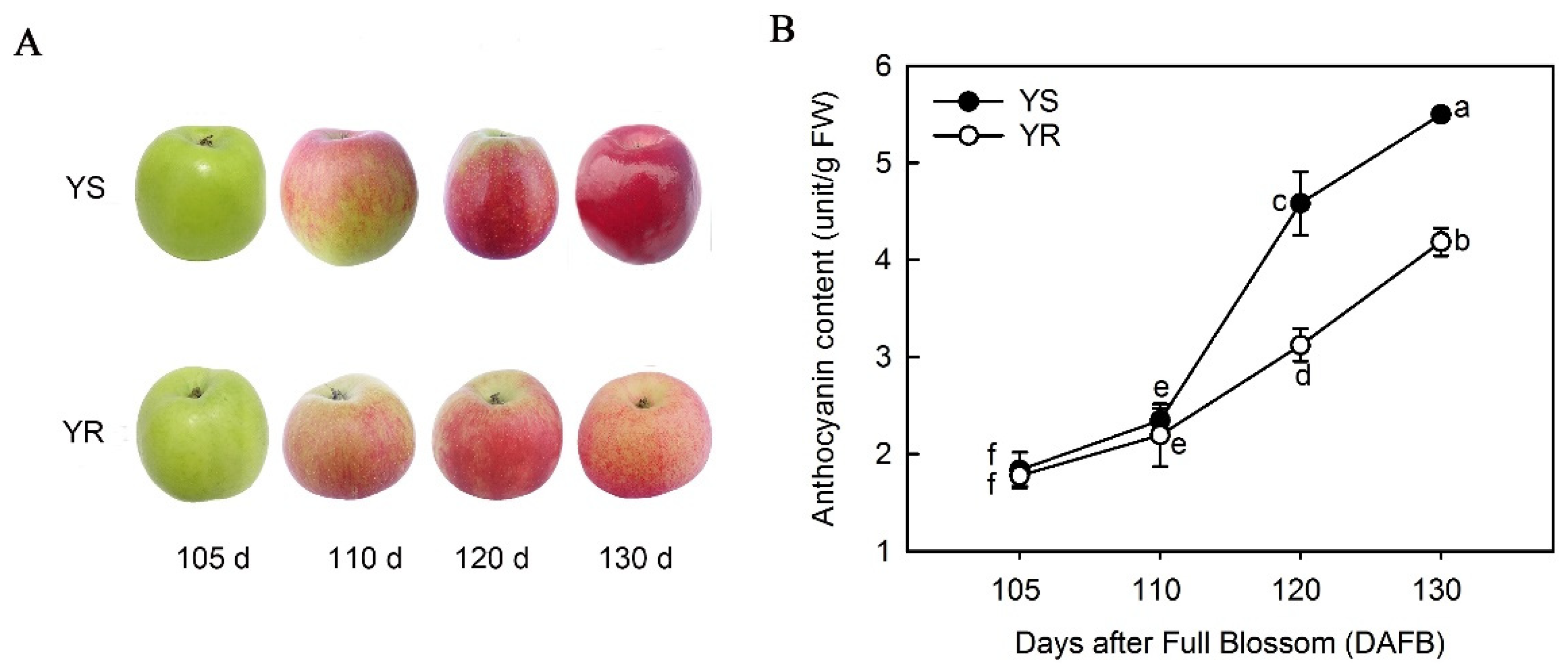
3.1. Differences in Anthocyanin Types and Total Content between Apple Cultivars
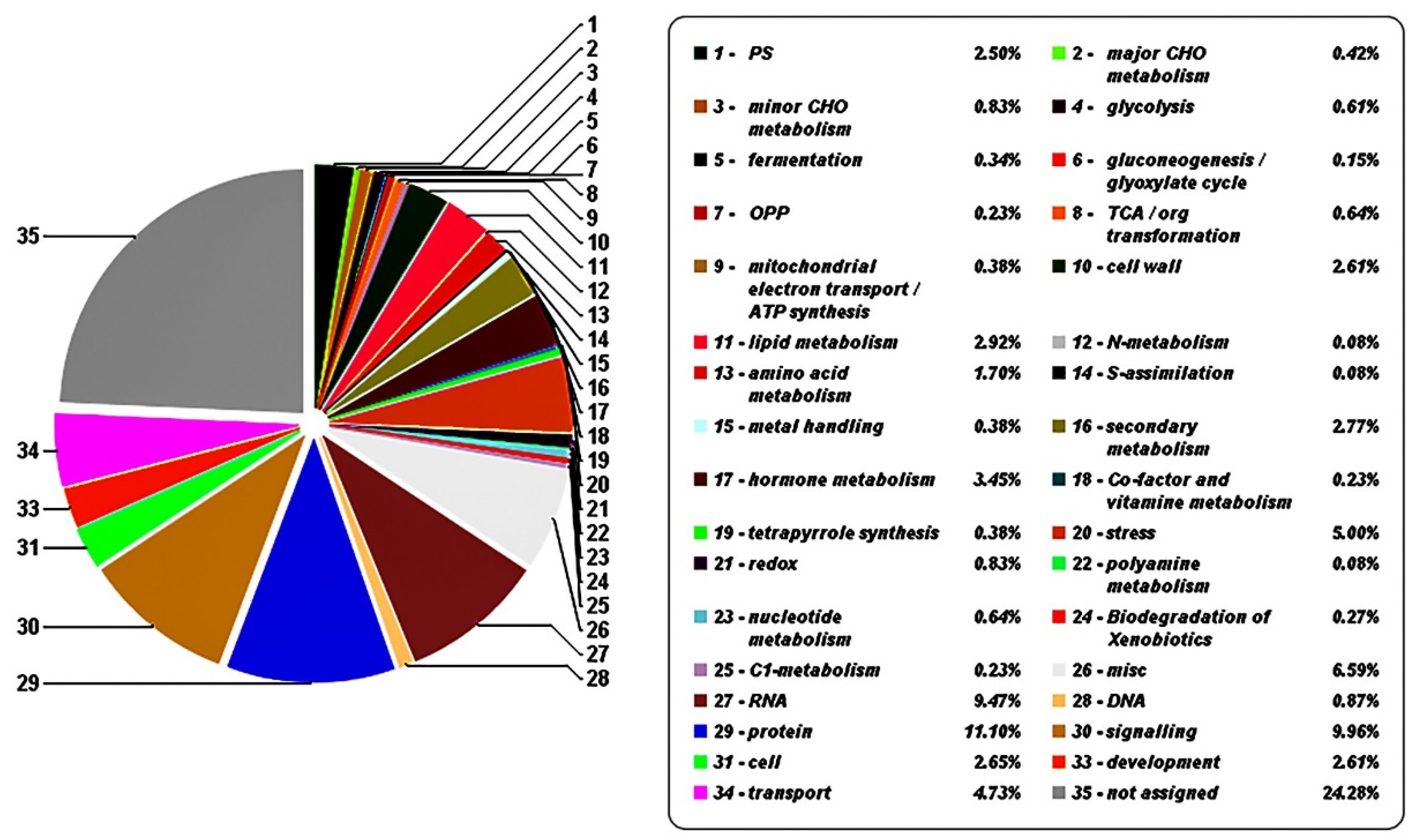
3.2. Transcriptome Sequencing and Analysis
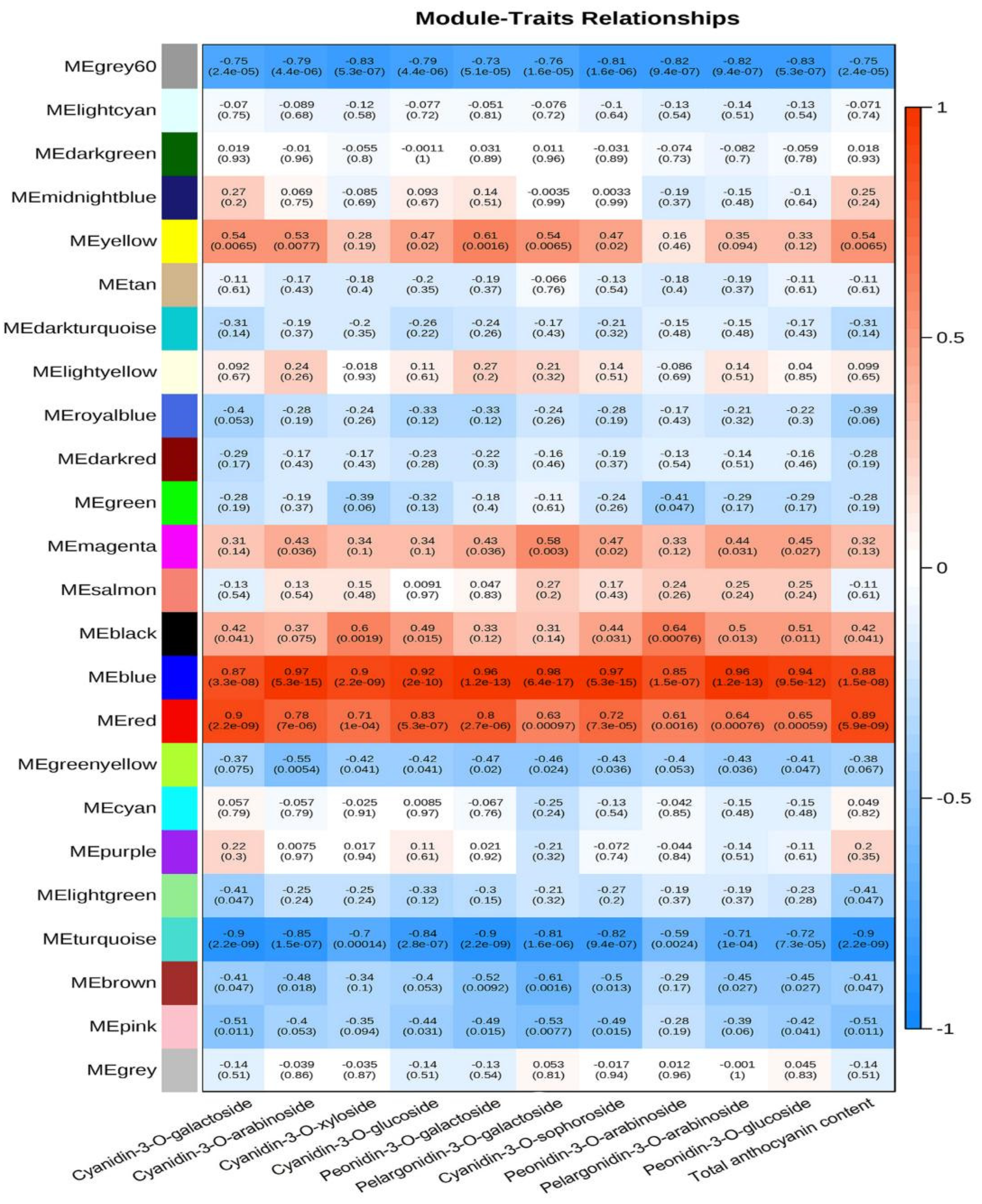
3.3. WGCNA Analysis of Anthocyanin Contents and Transcriptome Data
3.4. Expression of Genes Associated with Regulation of Anthocyanin and Phenylpropanoid Metabolism
3.5. Key Transcription Factors and Structure Genes Associated with Anthocyanin Accumulation and Their Interaction Network
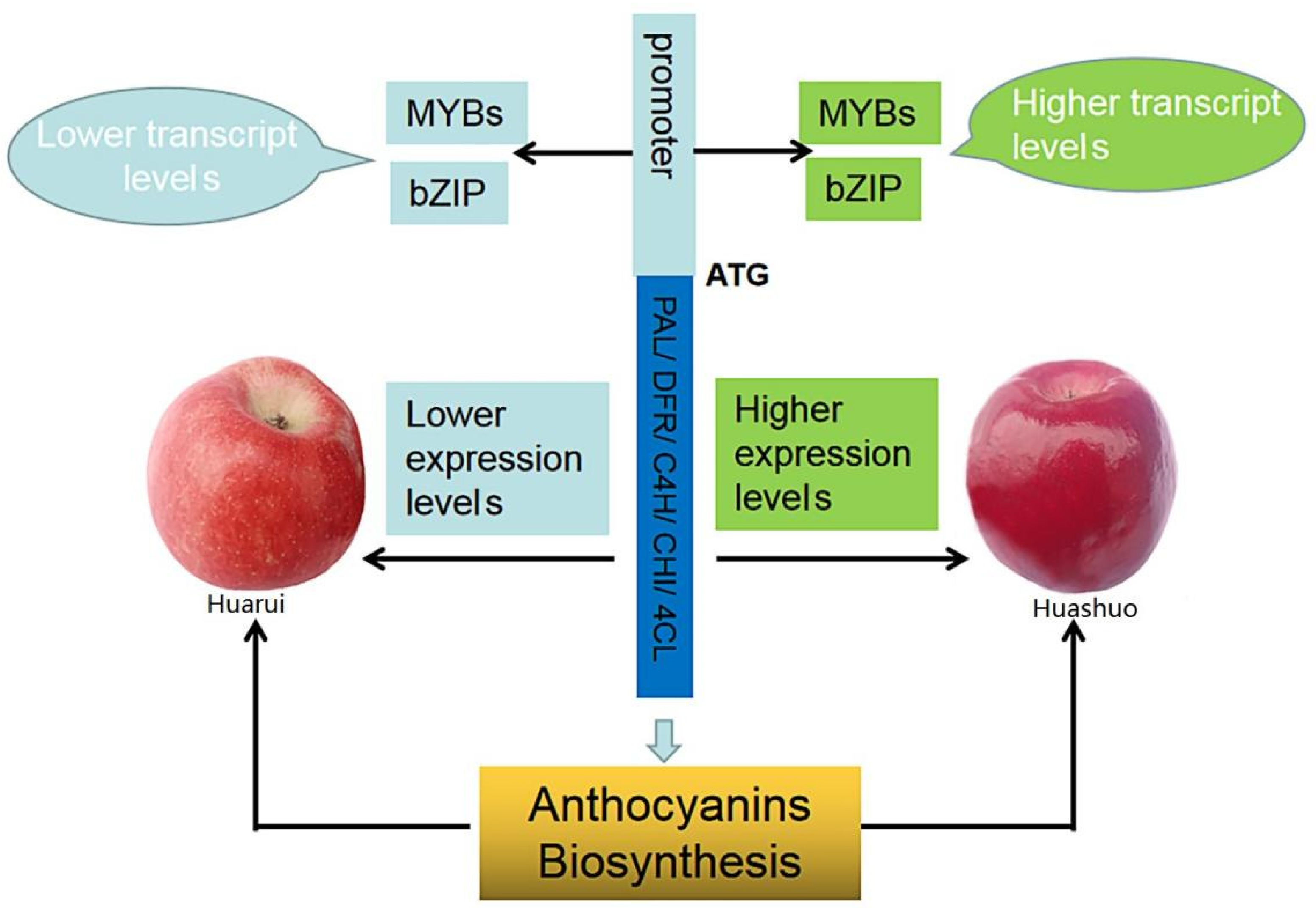
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lev-Yadun, S.; Gould, K.S. Role of anthocyanins in plant defence. In Anthocyanins; Winefifield, C., Davies, K., Gould, K.S., Eds.; Springer: New York, NY, USA, 2008; pp. 22–28. [Google Scholar]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.; Hall, R.D.; Bovy, A.G.; Luo, J. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Zhao, M.; Hu, Y.; Meng, F.; Song, X.; Tigabu, M.; Chiang, V.L.; Sederoff, R.; Ma, W.; et al. Molecular and metabolic insights into anthocyanin biosynthesis for leaf color change in chokecherry (Padus virginiana). Int. J. Mol. Sci. 2021, 22, 10697. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.; Zhao, Z. Effects of fruit bagging on anthocyanins, sugars, organic acids, and color properties of ‘granny smith’ and ‘golden delicious’ during fruit maturation. Eur. Food Res. Technol. 2013, 236, 329–339. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef]
- Qian, M.; Yu, B.; Li, X.; Sun, Y.; Zhang, D.; Teng, Y. Isolation and expression analysis of anthocyanin biosynthesis genes from the red Chinese sand pear, Pyrus pyrifolia nakai cv. Mantianhong, in response to methyl jasmonate treatment and UV-B/VIS conditions. Plant Mol. Biol. Rep. 2014, 32, 428–437. [Google Scholar] [CrossRef]
- Telias, A.; Bradeen, J.M.; Luby, J.J.; Hoover, E.E.; Allen, A.C. Regulation of anthocyanin accumulation in apple peel. Hortic. Rev. 2011, 38, 357. [Google Scholar]
- Xu, F.; Cao, S.; Shi, L.; Chen, W.; Su, X.; Yang, Z. Blue light irradiation affects anthocyanin content and enzyme activities involved in postharvest strawberry fruit. J. Agric. Food Chem. 2014, 62, 4778–4783. [Google Scholar] [CrossRef]
- Kondo, S.; Hiraoka, K.; Kobayashi, S.; Honda, C.; Terahara, N. Changes in the expression of anthocyanin biosynthetic genes during apple development. J. Am. Soc. Hortic. 2002, 127, 971–976. [Google Scholar] [CrossRef]
- Takos, A.M.; Jaffe, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef]
- Ma, C.; Liang, B.; Chang, B.; Yan, J.; Liu, L.; Wang, Y.; Yang, Y.; Zhao, Z. Transcriptome profiling of anthocyanin biosynthesis in the peel of ‘Granny Smith’ apples (Malus domestica) after bag removal. BMC Genom. 2019, 20, 353. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, N.A.; Glover, B.J. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Tao, R.; Yin, L.; Ni, J.; Yang, Q.; Yan, X.; Yang, F.; Guo, X.; Li, H.; Teng, Y. Two B-box proteins, PpBBX18 and PpBBX21, antagonistically regulate anthocyanin biosynthesis via competitive association with Pyrus pyrifolia elongated hypocotyl 5 in the peel of pear fruit. Plant J. 2019, 100, 1208–1223. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Xie, X.B.; Li, S.; Zhang, R.F.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Yao, Y.X.; You, C.X.; Zhang, X.S.; Hao, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB–bHLH–WD repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for betalain regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Ban, Y.; Honda, C.; Hatsuyama, Y.; Igarashi, M.; Bessho, H.; Moriguchi, T. Isolation and functional analysis of a MYB transcription factor gene that is a key regulator for the development of red coloration in apple skin. Plant Cell Physiol. 2007, 48, 958–970. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MDMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef]
- Chagné, D.; Lin-Wang, K.; Espley, R.V.; Volz, R.K.; How, N.M.; Rouse, S.; Brendolise, C.; Carlisle, C.M.; Kumar, S.; De Silva, N. An ancient duplication of apple MYB transcription factors is responsible for novel red fruit-flesh phenotypes. Plant Physiol. 2013, 161, 225–239. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An apple MYB transcription factor, mdMYB3, is involved in regulation of anthocyanin biosynthesis and flower development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef]
- An, X.H.; Tian, Y.; Chen, K.Q.; Liu, X.J.; Liu, D.D.; Xie, X.B.; Cheng, C.G.; Cong, P.H.; Hao, Y.J. MdMYB9 and MdMYB11 are involved in the regulation of the ja-induced biosynthesis of anthocyanin and proanthocyanidin in apples. Plant Cell Physiol. 2015, 56, 650–662. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Wang, C.; Zhang, W.; Liu, S.; Wang, W.; Yu, X.; Song, T.; Yu, M.; Yu, W.; Qu, S. The R2R3-type MYB transcription factor MdMYB90-like is responsible for the enhanced skin color of an apple bud sport mutant. Hortic. Res. 2021, 8, 156. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, N.; Liu, J.; Qu, C.; Wang, Y.; Jiang, S.; Lu, N.; Wang, D.; Zhang, Z.; Chen, X. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 2017, 94, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.X.; Xu, H.F.; Zhang, J.; Wang, N.; Fang, H.C.; Jiang, S.H.; Wang, Y.C.; Su, M.Y.; Chen, X.S. Functional identification of apple anthocyanin regulatory gene MdMYB111. Acta Hortic. Sin. 2019, 46, 832–840. [Google Scholar]
- Wang, S.; Zhang, Z.; Li, L.X.; Wang, H.B.; Zhou, H.; Chen, X.S.; Feng, S.Q. Apple MdMYB306-like inhibits anthocyanin synthesis by directly interacting with MdMYB17 and MdbHLH33. Plant J. 2022, 110, 1021–1034. [Google Scholar] [CrossRef]
- Bai, S.; Saito, T.; Honda, C.; Hatsuyama, Y.; Ito, A.; Moriguchi, T. An apple B-box protein, MdCOL11, is involved in UV-B- and temperature-induced anthocyanin biosynthesis. Planta 2014, 240, 1051–1062. [Google Scholar] [CrossRef]
- Fang, H.; Dong, Y.; Yue, X.; Hu, J.; Jiang, S.; Xu, H.; Wang, Y.; Su, M.; Zhang, J.; Zhang, Z.; et al. The B-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 2019, 42, 2090–2104. [Google Scholar] [CrossRef]
- Plunkett, B.J.; Henry-Kirk, R.; Friend, A.; Diack, R.; Helbig, S.; Mouhu, K.; Tomes, S.; Dare, A.P.; Espley, R.V.; Putterill, J.; et al. Apple B-box factors regulate light-responsive anthocyanin biosynthesis genes. Sci. Rep. 2019, 9, 17762. [Google Scholar] [CrossRef]
- An, J.P.; Wang, X.F.; Espley, R.V.; Lin-Wang, K.; Bi, S.Q.; You, C.X.; Hao, Y.J. An apple B-box protein MdBBX37 modulates anthocyanin biosynthesis and hypocotyl elongation synergistically with MdMYBs and MdHY5. Plant Cell Physiol. 2020, 61, 130–143. [Google Scholar] [CrossRef]
- Sun, Q.; Jiang, S.; Zhang, T.; Xu, H.; Fang, H.; Zhang, J.; Su, M.; Wang, Y.; Zhang, Z.; Wang, N.; et al. Apple NAC transcription factor MdNAC52 regulates biosynthesis of anthocyanin and proanthocyanidin through MdMYB9 and MdMYB11. Plant Sci. 2019, 289, 110286. [Google Scholar] [CrossRef]
- Mao, Z.; Jiang, H.; Wang, S.; Wang, Y.; Yu, L.; Zou, Q.; Liu, W.; Jiang, S.; Wang, N.; Zhang, Z.; et al. The MdHY5-MdWRKY41-MdMYB transcription factor cascade regulates the anthocyanin and proanthocyanidin biosynthesis in red-fleshed apple. Plant Sci. 2021, 306, 110848. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Fang, H.; Wang, J.; Yue, X.; Su, M.; Mao, Z.; Zou, Q.; Jiang, H.; Guo, Z.; Yu, L.; et al. Ultraviolet B-induced MdWRKY72 expression promotes anthocyanin synthesis in apple. Plant Sci. 2020, 292, 110377. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Qu, F.J.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17023. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Su, J.; Zhu, Y.; Yao, G.; Allan, A.C.; Ampomah-Dwamena, C.; Shu, Q.; Lin-Wang, K.; Zhang, S.; Wu, J. The involvement of PyBZIPa in light-induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G-boxes in its promoter. Hortic. Res. 2019, 6, 134. [Google Scholar] [CrossRef]
- Karagiannis, E.; Michailidis, M.; Tanou, G.; Scossa, F.; Sarrou, E.; Stamatakis, G.; Samiotaki, M.; Martens, S.; Fernie, A.R.; Molassiotis, A. Decoding altitude-activated regulatory mechanisms occurring during apple peel ripening. Hortic. Res. 2020, 7, 120. [Google Scholar] [CrossRef]
- Jia, D.; Li, Z.; Dang, Q.; Shang, L.; Shen, J.; Leng, X.; Wang, Y.; Yuan, Y. Anthocyanin biosynthesis and methylation of the MdMYB10 promoter are associated with the red blushed-skin mutant in the red striped-skin “changfu 2” apple. J. Agric. Food Chem. 2020, 68, 4292–4304. [Google Scholar] [CrossRef]
- Zhang, L.; Hu, J.; Han, X.; Li, J.; Gao, Y.; Richards, C.M.; Zhang, C.; Tian, Y.; Liu, G.; Gul, H.; et al. A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 2019, 10, 1494. [Google Scholar] [CrossRef]
- Yan, Z.L.; Zhang, H.T.; Zhang, R.P.; Liu, Z.Z.; Guo, G.N. Performance of new apple variety ‘huashuo’ in 14 producing areas in China. China Fruits 2016, 90–94. [Google Scholar]
- Xu, H.; Zou, Q.; Yang, G.; Jiang, S.; Fang, H.; Wang, Y.; Zhang, J.; Zhang, Z.; Wang, N.; Chen, X. MdMYB6 regulates anthocyanin formation in apple both through direct inhibition of the biosynthesis pathway and through substrate removal. Hortic. Res. 2020, 7, 72. [Google Scholar] [CrossRef]
- Pirie, A.; Mullins, M.G. Changes in anthocyanin and phenolics content of grapevine leaf and fruit tissues treated with sucrose, nitrate, and abscisic acid. Plant Physiol. 1976, 58, 468–472. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. Htseq—A python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Simko, V. R Package “Corrplot”: Visualization of a Correlation Matrix. Version 0.84. 2017. Available online: https://github.com/taiyun/corrplot (accessed on 27 November 2021).
- Langfelder, P.; Mehrabian, M.; Schadt, E.E.; Lusis, A.J.; Horvath, S. Weighted gene co-expression network analysis of adipose and liver reveals gene modules related to plasma HDL levels and containing candidate genes at loci identified in genome wide association studies. J. Am. Heart Assoc. 2008, 118, S_327. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, 808–815. [Google Scholar] [CrossRef]
- Shumayla; Mendu, V.; Singh, K.; Upadhyay, S.K. Insight into the roles of proline-rich extensin-like receptor protein kinases of bread wheat (Triticum aestivum L.). Life 2022, 12, 941. [Google Scholar] [CrossRef]
- Rathour, M.; Alok, A.; Upadhyay, S.K. Investigation of roles of TaTALE genes during development and stress response in bread wheat. Plants 2022, 11, 587. [Google Scholar] [CrossRef]
- Ji, X.H.; Zhang, R.; Wang, N.; Yang, L.; Chen, X.S. Transcriptome profiling reveals auxin suppressed anthocyanin biosynthesis in red-fleshed apple callus (Malus sieversii f. niedzwetzkyana). Plant Cell Tissue Organ Cult. (PCTOC) 2015, 123, 389–404. [Google Scholar] [CrossRef]
- Gao, H.N.; Jiang, H.; Cui, J.Y.; You, C.X.; Li, Y.Y. Review: The effects of hormones and environmental factors on anthocyanin biosynthesis in apple. Plant Sci. 2021, 312, 111024. [Google Scholar] [CrossRef]
- Honda, C.; Moriya, S. Anthocyanin biosynthesis in apple fruit. Hortic. J. 2018, 87, 305–314. [Google Scholar] [CrossRef]
- Sun, B.H.; Francis, F. Apple anthocyanins: Identification of cyanidin-7-arabinoside. J. Food Sci. 1967, 32, 647–649. [Google Scholar] [CrossRef]
- Tsao, R.; Yang, R.; Young, J.C.; Zhu, H. Polyphenolic profiles in eight apple cultivars using high-performance liquid chromatography (HPLC). J. Agric. Food Chem. 2003, 51, 6347–6353. [Google Scholar] [CrossRef] [PubMed]
- Ubi, B.E.; Honda, C.; Bessho, H.; Kondo, S.; Wada, M.; Kobayashi, S.; Moriguchi, T. Expression analysis of anthocyanin biosynthetic genes in apple skin: Effect of UV-B and temperature. Plant Sci. 2006, 170, 571–578. [Google Scholar] [CrossRef]
- Wu, B.H.; Cao, Y.G.; Guan, L.; Xin, H.P.; Li, J.H.; Li, S.H. Genome-wide transcriptional profiles of the berry skin of two red grape cultivars (Vitis vinifera) in which anthocyanin synthesis is sunlight-dependent or-independent. PLoS ONE 2014, 9, e105959. [Google Scholar] [CrossRef]
- Zhang, L.; Fan, S.; An, N.; Zuo, X.; Gao, C.; Zhang, D.; Han, M. Identification and expression analysis of pal gene family in apple. Acta Agric. Zhejiangensis 2018, 30, 2031–2043. [Google Scholar]
- Gleitz, J.; Seitz, H.U. Induction of chalcone synthase in cell suspension cultures of carrot (Daucus carota L. spp. sativus) by ultraviolet light: Evidence for two different forms of chalcone synthase. Planta 1989, 179, 323–330. [Google Scholar] [CrossRef]
- Lancaster, J.; Dougall, D.K. Regulation of skin color in apples. Crit. Rev. Plant Sci. 1992, 10, 487–502. [Google Scholar] [CrossRef]
- Kim, S.H.; Lee, J.R.; Hong, S.T.; Yoo, Y.K.; An, G.; Kim, S.R. Molecular cloning and analysis of anthocyanin biosynthesis genes preferentially expressed in apple skin. Plant Sci. 2003, 165, 403–413. [Google Scholar] [CrossRef]
- Huang, L.N.; Wu, G.B.; Zhang, S.; Kuang, F.Y.; Chen, F.H. The identification and functional verification of the cinnamate 4-hydroxylase gene from wax apple fruit and its role in lignin biosynthesis during nitric oxide-delayed postharvest cottony softening. Postharvest Biol. Technol. 2019, 158, 110964. [Google Scholar] [CrossRef]
- Ring, L.; Yeh, S.-Y.; Hücherig, S.; Hoffmann, T.; Blanco-Portales, R.; Fouche, M.; Villatoro, C.; Denoyes, B.; Monfort, A.; Caballero, J.L.; et al. Metabolic interaction between anthocyanin and lignin biosynthesis is associated with peroxidase FaPRX27 in strawberry fruit. Plant Physiol. 2013, 163, 43–60. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Nie, P.; Zhang, H.; Wang, L.; Wang, H.; Zhang, L. Dynamic changes of anthocyanin accumulation and endogenous hormone contents in blueberry. J. Beijing For. Univ. 2017, 39, 64–71. [Google Scholar]
- Li, W.F.; Mao, J.; Yang, S.J.; Guo, Z.G.; Ma, Z.H.; Dawuda, M.M.; Zuo, C.W.; Chu, M.Y.; Chen, B.H. Anthocyanin accumulation correlates with hormones in the fruit skin of ‘red delicious’ and its four generation bud sport mutants. BMC Plant Biol. 2018, 18, 363. [Google Scholar] [CrossRef] [PubMed]
- Figueroa, N.E.; Gatica-Meléndez, C.; Figueroa, C.R. Ethylene application at the immature stage of Fragaria chiloensis fruit represses the anthocyanin biosynthesis with a concomitant accumulation of lignin. Food Chem. 2021, 358, 129913. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Yu, K.; Shi, Y.; Wang, J.; Duan, C. Transcription factor VviMYB86 oppositely regulates proanthocyanidin and anthocyanin biosynthesis in grape berries. Front. Plant Sci. 2021, 11, 613677. [Google Scholar] [CrossRef]
- Wei, X.; Ju, Y.; Ma, T.; Zhang, J.; Fang, Y.; Sun, X. New perspectives on the biosynthesis, transportation, astringency perception and detection methods of grape proanthocyanidins. Crit. Rev. Food Sci. 2021, 61, 2372–2398. [Google Scholar] [CrossRef]
- Appelhagen, I.; Lu, G.-H.; Huep, G.; Schmelzer, E.; Weisshaar, B.; Sagasser, M. Transparent testa1 interacts with R2R3-MYB factors and affects early and late steps of flavonoid biosynthesis in the endothelium of Arabidopsis thaliana seeds. Plant J. 2011, 67, 406–419. [Google Scholar] [CrossRef]
- An, X.-H.; Tian, Y.; Chen, K.-Q.; Wang, X.-F.; Hao, Y.-J. The apple WD40 protein mdttg1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. J. Plant Physiol. 2012, 169, 710–717. [Google Scholar] [CrossRef]
- Feyissa, D.N.; Løvdal, T.; Olsen, K.M.; Slimestad, R.; Lillo, C. The endogenous GL3, but not EGL3, gene is necessary for anthocyanin accumulation as induced by nitrogen depletion in arabidopsis rosette stage leaves. Planta 2009, 230, 747–754. [Google Scholar] [CrossRef]
- Baxter, I.R.; Young, J.C.; Armstrong, G.; Foster, N.; Bogenschutz, N.; Cordova, T.; Peer, W.A.; Hazen, S.P.; Murphy, A.S.; Harper, J.F. A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2005, 102, 2649–2654. [Google Scholar] [CrossRef]
- White-Cooper, H.; Leroy, D.; MacQueen, A.; Fuller, M.T. Transcription of meiotic cell cycle and terminal differentiation genes depends on a conserved chromatin associated protein, whose nuclear localisation is regulated. Development 2000, 127, 5463–5473. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.A.; Shalit-Kaneh, A.; Chu, D.N.; Hsu, P.Y.; Harmer, S.L. The reveille clock genes inhibit growth of juvenile and adult plants by control of cell size. Plant Physiol. 2017, 173, 2308–2322. [Google Scholar] [CrossRef] [PubMed]
- An, J.P.; Yao, J.F.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ. 2018, 41, 2678–2692. [Google Scholar] [CrossRef]
- Ang, L.-H.; Chattopadhyay, S.; Wei, N.; Oyama, T.; Okada, K.; Batschauer, A.; Deng, X.-W. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1998, 1, 213–222. [Google Scholar] [CrossRef]
- Li, Y.Y.; Mao, K.; Zhao, C.; Zhao, X.Y.; Zhang, H.L.; Shu, H.R.; Hao, Y.J. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 2012, 160, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Hudson, M.E.; Lisch, D.R.; Quail, P.H. The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome a-signaling pathway. Plant J. 2003, 34, 453–471. [Google Scholar] [CrossRef]
- Oyama, T.; Shimura, Y.; Okada, K. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 1997, 11, 2983–2995. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, C.; Liu, L.; Wei, Z.; Liu, J.; Li, M.; Yan, Z.; Gao, D. Anthocyanin Accumulation and Molecular Analysis of Correlated Genes by Metabolomics and Transcriptomics in Sister Line Apple Cultivars. Life 2022, 12, 1246. https://doi.org/10.3390/life12081246
Shi C, Liu L, Wei Z, Liu J, Li M, Yan Z, Gao D. Anthocyanin Accumulation and Molecular Analysis of Correlated Genes by Metabolomics and Transcriptomics in Sister Line Apple Cultivars. Life. 2022; 12(8):1246. https://doi.org/10.3390/life12081246
Chicago/Turabian StyleShi, Caiyun, Li Liu, Zhifeng Wei, Junwei Liu, Ming Li, Zhenli Yan, and Dengtao Gao. 2022. "Anthocyanin Accumulation and Molecular Analysis of Correlated Genes by Metabolomics and Transcriptomics in Sister Line Apple Cultivars" Life 12, no. 8: 1246. https://doi.org/10.3390/life12081246
APA StyleShi, C., Liu, L., Wei, Z., Liu, J., Li, M., Yan, Z., & Gao, D. (2022). Anthocyanin Accumulation and Molecular Analysis of Correlated Genes by Metabolomics and Transcriptomics in Sister Line Apple Cultivars. Life, 12(8), 1246. https://doi.org/10.3390/life12081246





