New Wine in an Old Bottle: Utilizing Chemical Genetics to Dissect Apical Hook Development
Abstract
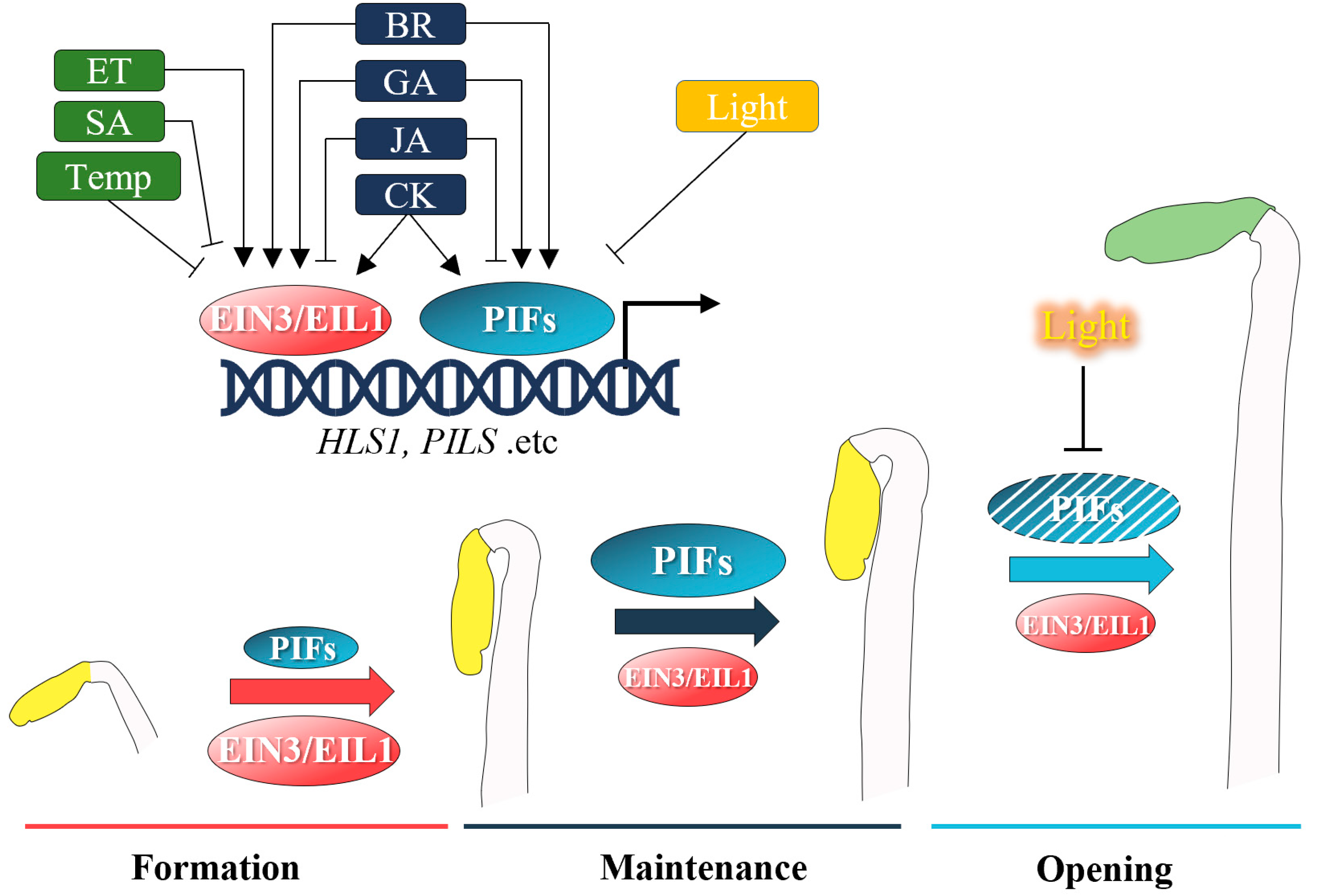
:1. Introduction
2. Recent Advances in Hormonal Regulation of the Apical Hook
3. Existing Chemical Tools That Could Help Us Understand Apical Hook Development
3.1. Chemicals Regulating Auxin Biosynthesis and Metabolism
3.2. Chemicals Regulating Auxin Transport and Signal
3.3. Other Chemicals That Regulate the Apical Hook
4. Developing Chemical Tools with Novel Targets to Further Delineate Apical Hook
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review board statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Silk, W.K.; Erickson, R.O. KINEMATICS OF HYPOCOTYL CURVATURE. Am. J. Bot. 1978, 65, 310–319. [Google Scholar] [CrossRef]
- Raz, V.; Ecker, J.R. Regulation of differential growth in the apical hook of Arabidopsis. Development 1999, 126, 3661–3668. [Google Scholar] [CrossRef] [PubMed]
- Harpham, N.V.J.; Berry, A.W.; Knee, E.M.; Roveda-Hoyos, G.; Raskin, I.; Sanders, I.O.; Smith, A.R.; Wood, C.K.; Hall, M.A. The Effect of Ethylene on the Growth and Development of Wild-type and Mutant Arabidopsis thaliana (L.) Heynh. Ann. Bot. 1991, 68, 55–61. [Google Scholar]
- Shen, X.; Li, Y.; Pan, Y.; Zhong, S. Activation of HLS1 by Mechanical Stress via Ethylene-Stabilized EIN3 Is Crucial for Seedling Soil Emergence. Front Plant Sci. 2016, 7, 1571. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guo, H. On hormonal regulation of the dynamic apical hook development. New Phytol. 2019, 222, 1230–1234. [Google Scholar] [CrossRef]
- Zhang, X.; Ji, Y.; Xue, C.; Ma, H.; Xi, Y.; Huang, P.; Wang, H.; An, F.; Li, B.; Wang, Y.; et al. Integrated Regulation of Apical Hook Development by Transcriptional Coupling of EIN3/EIL1 and PIFs in Arabidopsis. Plant Cell 2018, 30, 1971–1988. [Google Scholar] [CrossRef]
- Shi, H.; Lyu, M.; Luo, Y.; Liu, S.; Li, Y.; He, H.; Wei, N.; Deng, X.W.; Zhong, S. Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proc. Natl. Acad. Sci. USA 2018, 115, 6482–6487. [Google Scholar] [CrossRef]
- Pandey, B.K.; Huang, G.; Bhosale, R.; Hartman, S.; Sturrock, C.J.; Jose, L.; Martin, O.C.; Karady, M.; Voesenek, L.; Ljung, K.; et al. Plant roots sense soil compaction through restricted ethylene diffusion. Science 2021, 371, 276–280. [Google Scholar] [CrossRef]
- Kieber, J.J.; Rothenberg, M.; Roman, G.; Feldmann, K.A.; Ecker, J.R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 1993, 72, 427–441. [Google Scholar] [CrossRef]
- Park, E.; Kim, Y.; Choi, G. Phytochrome B Requires PIF Degradation and Sequestration to Induce Light Responses across a Wide Range of Light Conditions. Plant Cell 2018, 30, 1277–1292. [Google Scholar] [CrossRef]
- Lehman, A.; Black, R.; Ecker, J.R. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 1996, 85, 183–194. [Google Scholar] [CrossRef]
- Zádníková, P.; Petrásek, J.; Marhavy, P.; Raz, V.; Vandenbussche, F.; Ding, Z.; Schwarzerová, K.; Morita, M.T.; Tasaka, M.; Hejátko, J.; et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 2010, 137, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Abbas, M.; Alabadí, D.; Blázquez, M.A. Differential growth at the apical hook: All roads lead to auxin. Front. Plant Sci. 2013, 4, 441. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Bou Daher, F.; Liu, Y.; Steward, A.; Tillmann, M.; Zhang, X.; Wong, J.H.; Ren, H.; Cohen, J.D.; Li, C.; et al. Biphasic control of cell expansion by auxin coordinates etiolated seedling development. Sci. Adv. 2022, 8, eabj1570. [Google Scholar] [CrossRef]
- Peng, Y.; Zhang, D.; Qiu, Y.; Xiao, Z.; Ji, Y.; Li, W.; Xia, Y.; Wang, Y.; Guo, H. Growth asymmetry precedes differential auxin response during apical hook initiation in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 5–22. [Google Scholar] [CrossRef]
- Li, H.; Johnson, P.; Stepanova, A.; Alonso, J.M.; Ecker, J.R. Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev. Cell 2004, 7, 193–204. [Google Scholar] [CrossRef]
- Taniguchi, M.; Furutani, M.; Nishimura, T.; Nakamura, M.; Fushita, T.; Iijima, K.; Baba, K.; Tanaka, H.; Toyota, M.; Tasaka, M.; et al. The Arabidopsis LAZY1 Family Plays a Key Role in Gravity Signaling within Statocytes and in Branch Angle Control of Roots and Shoots. Plant Cell 2017, 29, 1984–1999. [Google Scholar] [CrossRef]
- Ge, L.; Chen, R. Negative gravitropism in plant roots. Nat. Plants 2016, 2, 16155. [Google Scholar] [CrossRef]
- Furutani, M.; Morita, M.T. LAZY1-LIKE-mediated gravity signaling pathway in root gravitropic set-point angle control. Plant Physiol. 2021, 187, 1087–1095. [Google Scholar] [CrossRef]
- Furutani, M.; Hirano, Y.; Nishimura, T.; Nakamura, M.; Taniguchi, M.; Suzuki, K.; Oshida, R.; Kondo, C.; Sun, S.; Kato, K.; et al. Polar recruitment of RLD by LAZY1-like protein during gravity signaling in root branch angle control. Nat Commun 2020, 11, 76. [Google Scholar] [CrossRef]
- Rosquete, M.R.; von Wangenheim, D.; Marhavý, P.; Barbez, E.; Stelzer, E.H.; Benková, E.; Maizel, A.; Kleine-Vehn, J. An auxin transport mechanism restricts positive orthogravitropism in lateral roots. Curr. Biol. 2013, 23, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Robert, S.; Raikhel, N.V.; Hicks, G.R. Powerful Partners: Arabidopsis and Chemical Genomics. Arab. Book 2009, 2009, e0109. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Asami, T. Chemical regulators of plant hormones and their applications in basic research and agriculture*. Biosci. Biotechnol. Biochem. 2018, 82, 1265–1300. [Google Scholar] [CrossRef] [PubMed]
- Tóth, R.; van der Hoorn, R.A.L. Emerging principles in plant chemical genetics. Trends Plant Sci. 2010, 15, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Zavaliev, R.; Mohan, R.; Chen, T.; Dong, X. Formation of NPR1 Condensates Promotes Cell Survival during the Plant Immune Response. Cell 2020, 182, 1093–1108.e1018. [Google Scholar] [CrossRef]
- Kumar, S.; Zavaliev, R.; Wu, Q.; Zhou, Y.; Cheng, J.; Dillard, L.; Powers, J.; Withers, J.; Zhao, J.; Guan, Z.; et al. Structural basis of NPR1 in activating plant immunity. Nature 2022, 605, 561–566. [Google Scholar] [CrossRef]
- Huang, P.; Dong, Z.; Guo, P.; Zhang, X.; Qiu, Y.; Li, B.; Wang, Y.; Guo, H. Salicylic Acid Suppresses Apical Hook Formation via NPR1-Mediated Repression of EIN3 and EIL1 in Arabidopsis. Plant Cell 2020, 32, 612–629. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, Z.; An, F.; Hao, D.; Li, P.; Song, J.; Yi, C.; Guo, H. Jasmonate-activated MYC2 represses ETHYLENE INSENSITIVE3 activity to antagonize ethylene-promoted apical hook formation in Arabidopsis. Plant Cell 2014, 26, 1105–1117. [Google Scholar] [CrossRef]
- Jin, H.; Pang, L.; Fang, S.; Chu, J.; Li, R.; Zhu, Z. High ambient temperature antagonizes ethylene-induced exaggerated apical hook formation in etiolated Arabidopsis seedlings. Plant Cell Environ. 2018, 41, 2858–2868. [Google Scholar] [CrossRef]
- An, F.; Zhang, X.; Zhu, Z.; Ji, Y.; He, W.; Jiang, Z.; Li, M.; Guo, H. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012, 22, 915–927. [Google Scholar] [CrossRef]
- Li, K.; Yu, R.; Fan, L.M.; Wei, N.; Chen, H.; Deng, X.W. DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat. Commun. 2016, 7, 11868. [Google Scholar] [CrossRef] [PubMed]
- Hansen, M.; Chae, H.S.; Kieber, J.J. Regulation of ACS protein stability by cytokinin and brassinosteroid. Plant J. 2009, 57, 606–614. [Google Scholar] [CrossRef] [PubMed]
- Aizezi, Y.; Shu, H.; Zhang, L.; Zhao, H.; Peng, Y.; Lan, H.; Xie, Y.; Li, J.; Wang, Y.; Guo, H.; et al. Cytokinin regulates apical hook development via the coordinated actions of EIN3/EIL1 and PIF transcription factors in Arabidopsis. J. Exp. Bot. 2021, 73, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Oh, E.; Zhu, J.Y.; Wang, Z.Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell Biol. 2012, 14, 802–809. [Google Scholar] [CrossRef]
- Zhao, N.; Zhao, M.; Tian, Y.; Wang, Y.; Han, C.; Fan, M.; Guo, H.; Bai, M.Y. Interaction between BZR1 and EIN3 mediates signalling crosstalk between brassinosteroids and ethylene. New Phytol. 2021, 232, 2308–2323. [Google Scholar] [CrossRef]
- Béziat, C.; Barbez, E.; Feraru, M.I.; Lucyshyn, D.; Kleine-Vehn, J. Light triggers PILS-dependent reduction in nuclear auxin signalling for growth transition. Nat. Plants 2017, 3, 17105. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, N.; Wang, L. Phytochrome interacting factor proteins regulate cytokinesis in Arabidopsis. Cell Reports 2021, 35, 109095. [Google Scholar] [CrossRef]
- Guzmán, P.; Ecker, J.R. Exploiting the triple response of Arabidopsis to identify ethylene-related mutants. Plant Cell 1990, 2, 513–523. [Google Scholar] [CrossRef]
- Abel, S.; Theologis, A. Odyssey of auxin. Cold Spring Harb. Perspect Biol. 2010, 2, a004572. [Google Scholar] [CrossRef]
- Hayashi, K.I. Chemical Biology in Auxin Research. Cold Spring Harb. Perspect Biol. 2021, 13, a040105. [Google Scholar] [CrossRef]
- Fukui, K.; Hayashi, K.I. Manipulation and Sensing of Auxin Metabolism, Transport and Signaling. Plant Cell Physiol. 2018, 59, 1500–1510. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y. Auxin biosynthesis. Arab. Book 2014, 12, e0173. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Brumos, J.; Li, H.; Ji, Y.; Ke, M.; Gong, X.; Zeng, Q.; Li, W.; Zhang, X.; An, F.; et al. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 2011, 23, 3944–3960. [Google Scholar] [CrossRef] [PubMed]
- Kakei, Y.; Yamazaki, C.; Suzuki, M.; Nakamura, A.; Sato, A.; Ishida, Y.; Kikuchi, R.; Higashi, S.; Kokudo, Y.; Ishii, T.; et al. Small-molecule auxin inhibitors that target YUCCA are powerful tools for studying auxin function. Plant J. 2015, 84, 827–837. [Google Scholar] [CrossRef]
- Sato, A.; Soeno, K.; Kikuchi, R.; Narukawa-Nara, M.; Yamazaki, C.; Kakei, Y.; Nakamura, A.; Shimada, Y. Indole-3-pyruvic acid regulates TAA1 activity, which plays a key role in coordinating the two steps of auxin biosynthesis. Proc. Natl. Acad. Sci. 2022, 119, e2203633119. [Google Scholar] [CrossRef]
- Narukawa-Nara, M.; Nakamura, A.; Kikuzato, K.; Kakei, Y.; Sato, A.; Mitani, Y.; Yamasaki-Kokudo, Y.; Ishii, T.; Hayashi, K.; Asami, T.; et al. Aminooxy-naphthylpropionic acid and its derivatives are inhibitors of auxin biosynthesis targeting l-tryptophan aminotransferase: Structure-activity relationships. Plant J. 2016, 87, 245–257. [Google Scholar] [CrossRef]
- Kakei, Y.; Nakamura, A.; Yamamoto, M.; Ishida, Y.; Yamazaki, C.; Sato, A.; Narukawa-Nara, M.; Soeno, K.; Shimada, Y. Biochemical and Chemical Biology Study of Rice OsTAR1 Revealed that Tryptophan Aminotransferase is Involved in Auxin Biosynthesis: Identification of a Potent OsTAR1 Inhibitor, Pyruvamine2031. Plant Cell Physiol. 2017, 58, 598–606. [Google Scholar] [CrossRef]
- Nishimura, T.; Hayashi, K.; Suzuki, H.; Gyohda, A.; Takaoka, C.; Sakaguchi, Y.; Matsumoto, S.; Kasahara, H.; Sakai, T.; Kato, J.; et al. Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J. 2014, 77, 352–366. [Google Scholar] [CrossRef]
- Tsugafune, S.; Mashiguchi, K.; Fukui, K.; Takebayashi, Y.; Nishimura, T.; Sakai, T.; Shimada, Y.; Kasahara, H.; Koshiba, T.; Hayashi, K.I. Yucasin DF, a potent and persistent inhibitor of auxin biosynthesis in plants. Sci. Rep. 2017, 7, 13992. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, H.J.; Su, Q.; Wen, J.; Wang, Y.; Song, W.; Xie, Y.; He, W.; Yang, Z.; Jiang, K.; et al. A phenotype-directed chemical screen identifies ponalrestat as an inhibitor of the plant flavin monooxygenase YUCCA in auxin biosynthesis. J. Biol. Chem. 2019, 294, 19923–19933. [Google Scholar] [CrossRef]
- Porco, S.; Pěnčík, A.; Rashed, A.; Voß, U.; Casanova-Sáez, R.; Bishopp, A.; Golebiowska, A.; Bhosale, R.; Swarup, R.; Swarup, K.; et al. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 11016–11021. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lin, J.E.; Harris, C.; Campos Mastrotti Pereira, F.; Wu, F.; Blakeslee, J.J.; Peer, W.A. DAO1 catalyzes temporal and tissue-specific oxidative inactivation of auxin in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2016, 113, 11010–11015. [Google Scholar] [CrossRef] [PubMed]
- Staswick, P.E.; Serban, B.; Rowe, M.; Tiryaki, I.; Maldonado, M.T.; Maldonado, M.C.; Suza, W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 2005, 17, 616–627. [Google Scholar] [CrossRef]
- Wojtaczka, P.; Ciarkowska, A.; Starzynska, E.; Ostrowski, M. The GH3 amidosynthetases family and their role in metabolic crosstalk modulation of plant signaling compounds. Phytochemistry 2022, 194, 113039. [Google Scholar] [CrossRef]
- Hayashi, K.-i.; Arai, K.; Aoi, Y.; Tanaka, Y.; Hira, H.; Guo, R.; Hu, Y.; Ge, C.; Zhao, Y.; Kasahara, H.; et al. The main oxidative inactivation pathway of the plant hormone auxin. Nat. Commun. 2021, 12, 6752. [Google Scholar] [CrossRef]
- Böttcher, C.; Dennis, E.G.; Booker, G.W.; Polyak, S.W.; Boss, P.K.; Davies, C. A novel tool for studying auxin-metabolism: The inhibition of grapevine indole-3-acetic acid-amido synthetases by a reaction intermediate analogue. PLoS ONE 2012, 7, e37632. [Google Scholar] [CrossRef]
- Fukui, K.; Arai, K.; Tanaka, Y.; Aoi, Y.; Kukshal, V.; Jez, J.M.; Kubes, M.F.; Napier, R.; Zhao, Y.; Kasahara, H.; et al. Chemical inhibition of auxin inactivation pathway uncovers the metabolic turnover of auxin homeostasis. bioRxiv 2021. [Google Scholar] [CrossRef]
- Xie, Y.; Zhu, Y.; Wang, N.; Luo, M.; Ohta, T.; Guo, R.; Yu, Z.; Aizezi, Y.; Zhang, L.; Yan, Y.; et al. Chemical genetic screening identifies nalacin as an inhibitor of GH3 amido synthetase for auxin conjugation. bioRxiv 2022. [Google Scholar] [CrossRef]
- Swarup, R.; Péret, B. AUX/LAX family of auxin influx carriers-an overview. Front. Plant Sci. 2012, 3, 225. [Google Scholar] [CrossRef]
- Adamowski, M.; Friml, J. PIN-dependent auxin transport: Action, regulation, and evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef]
- Barbosa, I.C.R.; Hammes, U.Z.; Schwechheimer, C. Activation and Polarity Control of PIN-FORMED Auxin Transporters by Phosphorylation. Trends Plant Sci. 2018, 23, 523–538. [Google Scholar] [CrossRef]
- Tan, S.; Luschnig, C.; Friml, J. Pho-view of Auxin: Reversible Protein Phosphorylation in Auxin Biosynthesis, Transport and Signaling. Mol. Plant 2021, 14, 151–165. [Google Scholar] [CrossRef] [PubMed]
- Lanková, M.; Smith, R.S.; Pesek, B.; Kubes, M.; Zazímalová, E.; Petrásek, J.; Hoyerová, K. Auxin influx inhibitors 1-NOA, 2-NOA, and CHPAA interfere with membrane dynamics in tobacco cells. J. Exp. Bot. 2010, 61, 3589–3598. [Google Scholar] [CrossRef] [PubMed]
- Parry, G.; Delbarre, A.; Marchant, A.; Swarup, R.; Napier, R.; Perrot-Rechenmann, C.; Bennett, M.J. Novel auxin transport inhibitors phenocopy the auxin influx carrier mutation aux1. Plant J. 2001, 25, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Teale, W.; Palme, K. Naphthylphthalamic acid and the mechanism of polar auxin transport. J. Exp. Bot. 2018, 69, 303–312. [Google Scholar] [CrossRef]
- Abas, L.; Kolb, M.; Stadlmann, J.; Janacek, D.P.; Lukic, K.; Schwechheimer, C.; Sazanov, L.A.; Mach, L.; Friml, J.; Hammes, U.Z. Naphthylphthalamic acid associates with and inhibits PIN auxin transporters. Proc. Natl. Acad. Sci. USA 2021, 118, e2020857118. [Google Scholar] [CrossRef]
- Ung, K.L.; Winkler, M.; Schulz, L.; Kolb, M.; Janacek, D.P.; Dedic, E.; Stokes, D.L.; Hammes, U.Z.; Pedersen, B.P. Structures and mechanism of the plant PIN-FORMED auxin transporter. Nature 2022. [Google Scholar] [CrossRef]
- Oochi, A.; Hajny, J.; Fukui, K.; Nakao, Y.; Gallei, M.; Quareshy, M.; Takahashi, K.; Kinoshita, T.; Harborough, S.R.; Kepinski, S.; et al. Pinstatic Acid Promotes Auxin Transport by Inhibiting PIN Internalization. Plant Physiol. 2019, 180, 1152–1165. [Google Scholar] [CrossRef]
- Hayashi, K.-i.; Neve, J.; Hirose, M.; Kuboki, A.; Shimada, Y.; Kepinski, S.; Nozaki, H. Rational Design of an Auxin Antagonist of the SCFTIR1 Auxin Receptor Complex. ACS Chem. Biol. 2012, 7, 590–598. [Google Scholar] [CrossRef]
- Hayashi, K.; Nakamura, S.; Fukunaga, S.; Nishimura, T.; Jenness, M.K.; Murphy, A.S.; Motose, H.; Nozaki, H.; Furutani, M.; Aoyama, T. Auxin transport sites are visualized in planta using fluorescent auxin analogs. Proc. Natl. Acad. Sci. USA 2014, 111, 11557–11562. [Google Scholar] [CrossRef]
- Vain, T.; Raggi, S.; Ferro, N.; Barange, D.K.; Kieffer, M.; Ma, Q.; Doyle, S.M.; Thelander, M.; Pařízková, B.; Novák, O.; et al. Selective auxin agonists induce specific AUX/IAA protein degradation to modulate plant development. Proc. Natl. Acad. Sci. USA 2019, 116, 6463–6472. [Google Scholar] [CrossRef]
- Uchida, N.; Takahashi, K.; Iwasaki, R.; Yamada, R.; Yoshimura, M.; Endo, T.A.; Kimura, S.; Zhang, H.; Nomoto, M.; Tada, Y.; et al. Chemical hijacking of auxin signaling with an engineered auxin-TIR1 pair. Nat. Chem. Biol. 2018, 14, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Baral, A.; Aryal, B.; Jonsson, K.; Morris, E.; Demes, E.; Takatani, S.; Verger, S.; Xu, T.; Bennett, M.; Hamant, O.; et al. External Mechanical Cues Reveal a Katanin-Independent Mechanism behind Auxin-Mediated Tissue Bending in Plants. Dev. Cell 2021, 56, 67–80.e63. [Google Scholar] [CrossRef] [PubMed]
- Brukhin, V.; Albertini, E. Epigenetic Modifications in Plant Development and Reproduction. Epigenomes 2021, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Callebert, P.; Vandemoortel, I.; Nguyen, L.; Audenaert, D.; Verschraegen, L.; Vandenbussche, F.; Van Der Straeten, D. TR-DB: An open-access database of compounds affecting the ethylene-induced triple response in Arabidopsis. Plant Physiol. Biochem. 2014, 75, 128–137. [Google Scholar] [CrossRef]
- Oh, K.; Hoshi, T.; Tomio, S.; Ueda, K.; Hara, K. A Chemical Genetics Strategy that Identifies Small Molecules which Induce the Triple Response in Arabidopsis. Molecules 2017, 22, 2270. [Google Scholar] [CrossRef]
- Martinez Molina, D.; Nordlund, P. The Cellular Thermal Shift Assay: A Novel Biophysical Assay for In Situ Drug Target Engagement and Mechanistic Biomarker Studies. Annu. Rev. Pharmacol. Toxicol. 2016, 56, 141–161. [Google Scholar] [CrossRef]
- Huber, K.V.; Salah, E.; Radic, B.; Gridling, M.; Elkins, J.M.; Stukalov, A.; Jemth, A.S.; Göktürk, C.; Sanjiv, K.; Strömberg, K.; et al. Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy. Nature 2014, 508, 222–227. [Google Scholar] [CrossRef]
- Gad, H.; Koolmeister, T.; Jemth, A.S.; Eshtad, S.; Jacques, S.A.; Ström, C.E.; Svensson, L.M.; Schultz, N.; Lundbäck, T.; Einarsdottir, B.O.; et al. MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool. Nature 2014, 508, 215–221. [Google Scholar] [CrossRef]
- Dejonghe, W.; Sharma, I.; Denoo, B.; De Munck, S.; Lu, Q.; Mishev, K.; Bulut, H.; Mylle, E.; De Rycke, R.; Vasileva, M.; et al. Disruption of endocytosis through chemical inhibition of clathrin heavy chain function. Nat. Chem. Biol. 2019, 15, 641–649. [Google Scholar] [CrossRef]
- Lu, Q.; Zhang, Y.; Hellner, J.; Giannini, C.; Xu, X.; Pauwels, J.; Ma, Q.; Dejonghe, W.; Han, H.; Van de Cotte, B.; et al. Proteome-wide cellular thermal shift assay reveals unexpected cross-talk between brassinosteroid and auxin signaling. Proc. Natl. Acad. Sci. USA 2022, 119, e2118220119. [Google Scholar] [CrossRef] [PubMed]
- Sousa, S.F.; Fernandes, P.A.; Ramos, M.J. Protein-ligand docking: Current status and future challenges. Proteins 2006, 65, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.N. Scoring functions for protein-ligand docking. Curr. Protein Pept. Sci. 2006, 7, 407–420. [Google Scholar] [CrossRef]
- Huang, N.; Shoichet, B.K.; Irwin, J.J. Benchmarking sets for molecular docking. J. Med. Chem. 2006, 49, 6789–6801. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Shoichet, B.K. Docking Screens for Novel Ligands Conferring New Biology. J. Med. Chem. 2016, 59, 4103–4120. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Zheng, L.; Meng, J.; Jiang, K.; Lan, H.; Wang, Z.; Lin, M.; Li, W.; Guo, H.; Wei, Y.; Mu, Y. Improving protein-ligand docking and screening accuracies by incorporating a scoring function correction term. Brief. Bioinform. 2022, 23, bbac051. [Google Scholar] [CrossRef]
- Yamamuro, C.; Zhu, J.K.; Yang, Z. Epigenetic Modifications and Plant Hormone Action. Mol. Plant 2016, 9, 57–70. [Google Scholar] [CrossRef]
- Markulin, L.; Škiljaica, A.; Tokić, M.; Jagić, M.; Vuk, T.; Bauer, N.; Leljak Levanić, D. Taking the Wheel—De novo DNA Methylation as a Driving Force of Plant Embryonic Development. Front. Plant Sci. 2021, 12, 764999. [Google Scholar] [CrossRef]
- Smolikova, G.; Leonova, T.; Vashurina, N.; Frolov, A.; Medvedev, S. Desiccation Tolerance as the Basis of Long-Term Seed Viability. Int. J. Mol. Sci. 2020, 22, 101. [Google Scholar] [CrossRef]
- Campos-Rivero, G.; Osorio-Montalvo, P.; Sánchez-Borges, R.; Us-Camas, R.; Duarte-Aké, F.; De-la-Peña, C. Plant hormone signaling in flowering: An epigenetic point of view. J. Plant Physiol. 2017, 214, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Maury, S.; Sow, M.D.; Le Gac, A.L.; Genitoni, J.; Lafon-Placette, C.; Mozgova, I. Phytohormone and Chromatin Crosstalk: The Missing Link For Developmental Plasticity? Front. Plant Sci. 2019, 10, 395. [Google Scholar] [CrossRef] [PubMed]


| Full Name | Description | Reference | CAS NO. | |
|---|---|---|---|---|
| Auxin biosynthesis | L-Kynurenine (Kyn) | TAA1/TAR2 inhibitor | [43] | 2922-83-0 |
| Pyruvamine2031 | OSTAR1 inhibitor | [47] | N.A | |
| p-Phenoxyphenyl boronic acid (PPBo) | YUCCAs inhibitor | [44] | 51067-38-0 | |
| Yucasin | YUCCAs inhibitor | [48] | 26028-65-9 | |
| Yucasin DF | YUCCAs inhibitor | [49] | 443797-96-4 | |
| Ponalrestat (PRT) | YUCCAs inhibitor | [50] | 72702-95-5 | |
| Auxin metabolism | Adenosine-5′-[2-(1H-indol-3-yl)ethyl]phosphate (AIEP) | GH3 inhibitor | [56] | 260430-02-2 |
| Kakeimide (KKI) | GH3 inhibitor | [57] | N.A | |
| Nalacin | GH3 inhibitor | [58] | 1019105-44-2 | |
| Auxin transport and signaling | 1-naphthoxyacetic acid (1-NOA) | Putative AUX1/LAXs inhibitor | [63,64] | 2976-75-2 |
| 2-naphthoxyacetic acid (2-NOA) | Putative AUX1/LAXs inhibitor | [63,64] | 120-23-0 | |
| Naphthylphthalamic acid (NPA) | PINs inhibitor | [65,66] | 132-66-1 | |
| 4-ethoxyphenylacetic acid (PISA) | Auxin transport promoter | [68] | 132-66-1 | |
| Auxinole | Auxin receptor agonist | [69] | 86445-22-9 | |
| NBD-IAA | Fluorescent auxin analog | [70] | N.A | |
| RN1-4 | Selective auxin agonists | [71] | N.A | |
| cvxIAA-ccvTIR1 pair | Engineered IAA-TIR1 pair | [72] | N.A | |
| Other regulators | Oryzalin | Apical hook suppressor | [16,73] | 19044-88-3 |
| 6825783 | Apical hook suppressor | [75] | N.A | |
| 7545271 | Apical hook promoter | [75] | N.A | |
| Apical Hook Inducer 1 (Kinetin Riboside) | Apical hook promoter | [33] | 4338-47-0 | |
| Apical Hook Inducer 2 | Apical hook promoter | unpublished | N.A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aizezi, Y.; Xie, Y.; Guo, H.; Jiang, K. New Wine in an Old Bottle: Utilizing Chemical Genetics to Dissect Apical Hook Development. Life 2022, 12, 1285. https://doi.org/10.3390/life12081285
Aizezi Y, Xie Y, Guo H, Jiang K. New Wine in an Old Bottle: Utilizing Chemical Genetics to Dissect Apical Hook Development. Life. 2022; 12(8):1285. https://doi.org/10.3390/life12081285
Chicago/Turabian StyleAizezi, Yalikunjiang, Yinpeng Xie, Hongwei Guo, and Kai Jiang. 2022. "New Wine in an Old Bottle: Utilizing Chemical Genetics to Dissect Apical Hook Development" Life 12, no. 8: 1285. https://doi.org/10.3390/life12081285
APA StyleAizezi, Y., Xie, Y., Guo, H., & Jiang, K. (2022). New Wine in an Old Bottle: Utilizing Chemical Genetics to Dissect Apical Hook Development. Life, 12(8), 1285. https://doi.org/10.3390/life12081285






