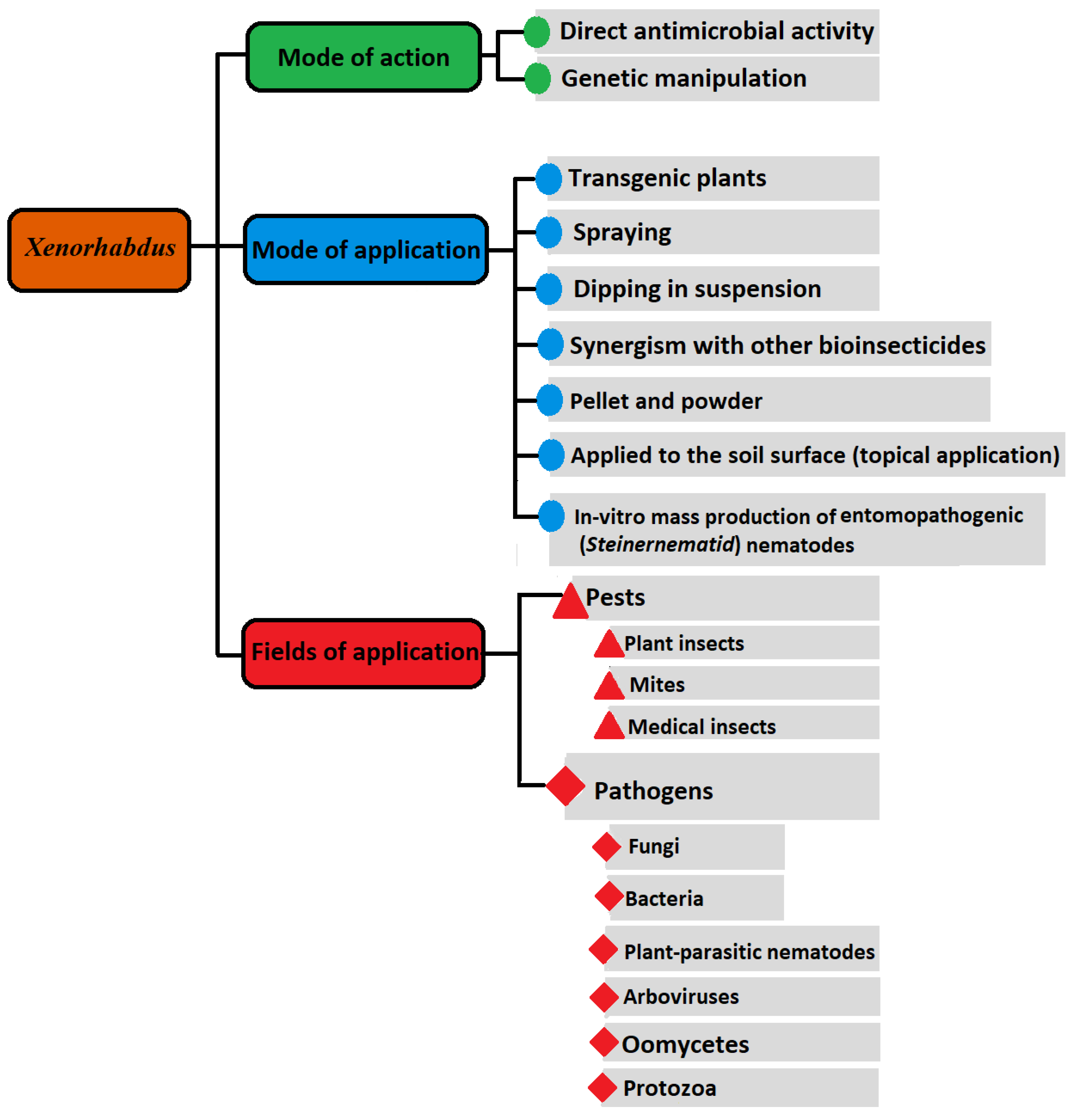
Xenorhabdus spp.: An Overview of the Useful Facets of Mutualistic Bacteria of Entomopathogenic Nematodes
Abstract
:1. Introduction
2. Identification, Taxonomy, Lifestyle, and Diversity of Xenorhabdus spp.
2.1. Their Identification/Taxonomy
2.2. The Lifestyle and Diversity of Xenorhabdus spp.
3. Pathogenicity of Xenorhabdus spp.
3.1. Magnitude and Profile of Pathogenicity
3.2. Xenorhabdus Bacterial Mechanism via Their Secreted Materials
4. The Positive and Negative Aspects of Xenorhabdus spp.
4.1. Major and Direct Favorable Aspects
4.2. Favorable Aspects That Need Further Exploration
4.3. Cost-Effective Xenorhabdus Mass Culture, Formulation, and Application
4.4. Other Aspects of Xenorhabdus spp.
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiao, Y.; Wu, K. Recent progress on the interaction between insects and Bacillus thuringiensis crops. Philos. Trans. R. Soc. 2019, 374, 20180316. [Google Scholar] [CrossRef] [PubMed]
- da Silva, W.J.; Pilz-Júnior, H.L.; Heermann, R.; da Silva, O.S. The great potential of entomopathogenic bacteria Xenorhabdus and Photorhabdus for mosquito control: A review. Parasites Vectors 2020, 13, 376. [Google Scholar] [CrossRef] [PubMed]
- Abd-Elgawad, M.M.M. Status of entomopathogenic nematodes in integrated pest management strategies in Egypt. In Biocontrol Agents: Entomopathogenic and Slug Parasitic Nematodes; Abd-Elgawad, M.M.M., Askary, T.H., Coupland, J., Eds.; CAB International: Wallingford, UK, 2017; pp. 473–501. [Google Scholar]
- Migunova, V.D.; Sasanelli, N. Bacteria as biocontrol tool against phytoparasitic nematodes. Plants 2021, 10, 389. [Google Scholar] [CrossRef]
- Javed, N.; Kamran, M.; Abbas, H. Toxic secretions of Xenorhabdus and their efficacy against crop insect pests. In Biocontrol Agents: Entomopathogenic and Slug Parasitic Nematodes; Abd-Elgawad, M.M.M., Askary, T.H., Coupland, J., Eds.; CAB International: Wallingford, UK, 2017; pp. 223–230. [Google Scholar]
- Eroglu, C.; Cimenb, H.; Ulug, D.; Karagoz, M.; Hazir, S.; Cakmaka, I. Acaricidal effect of cell-free supernatants from Xenorhabdus and Photorhabdus bacteria against Tetranychus urticae (Acari: Tetranychidae). J. Invertebr. Pathol. 2019, 106, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Ffrench-Constant, R.; Waterfield, N.; Daborn, P. Insecticidal toxins from Photorhabdus and Xenorhabdus. In Encyclopedia of Microbiology, 4th ed.; Schmid, T.M., Ed.; Academic Press: New York, NY, USA, 2019; pp. 704–715. [Google Scholar] [CrossRef]
- Muangpat, P.; Suwannaroj, M.; Yimthin, T.; Fukruksa, C.; Sitthisak, S.; Chantratita, N.; Vitta, A.; Thanwisai, A. Antibacterial activity of Xenorhabdus and Photorhabdus isolated from entomopathogenic nematodes against antibiotic-resistant bacteria. PLoS ONE 2020, 15, e0234129. [Google Scholar] [CrossRef]
- Dong, Y.; Li, X.; Duan, J.; Qin, Y.; Yang, X.; Ren, J.; Li, G. Improving the yield of xenocoumacin 1 enabled by in situ product removal. ACS Omega 2020, 5, 20391–20398. [Google Scholar] [CrossRef]
- Hug, J.J.; Krug, D.; Müller, R. Bacteria as genetically programmable producers of bioactive natural products. Nat. Rev. Chem. 2020, 4, 172–193. [Google Scholar] [CrossRef]
- Zhang, H.; Mao, J.; Liu, F.; Zeng, F. Expression of a nematode symbiotic bacterium-derived protease inhibitor protein in tobacco enhanced tolerance against Myzus persicae. Plant Cell Rep. 2012, 31, 1981–1989. [Google Scholar] [CrossRef]
- Thomas, G.M.; Poinar, G.O. Xenorhabdus gen. nov., a genus of entomopathogenic nematophilic bacteria of the family Enterobacteriaceae. Int. J. Syst. Bacteriol. 1979, 29, 352–360. [Google Scholar] [CrossRef] [Green Version]
- Boemare, N.E.; Akhurst, R.J.; Mourant, R.G. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int. J. Syst. Bacteriol. 1993, 43, 249–255. [Google Scholar] [CrossRef]
- Fischer-Le Saux, M.; Viallard, V.; Brunel, B.; Normand, P.; Boemare, N.E. Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subsp. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperate subsp. temperatas ubsp. nov. and P. asymbiotica sp. nov. Int. J. Syst. Bacteriol. 1999, 49, 1645–1656. [Google Scholar] [PubMed]
- Boemare, N. Biology, taxonomy and systematics of Photorhabdus and Xenorhabdus. In Entomopathogenic Nematology; Gaugler, R., Ed.; CAB International: Wallingford, UK, 2002; pp. 35–56. [Google Scholar]
- Sajnaga, E.; Kazimierczak, W. Evolution and taxonomy of nematode-associated entomopathogenic bacteria of the genera Xenorhabdus and Photorhabdus: An overview. Symbiosis 2020, 80, 1–13. [Google Scholar] [CrossRef]
- Shapiro-Ilan, D.; Hazir, S.; Glazer, I. Advances in use of entomopathogenic nematodes. In Integrated Management of Insect Pests: Current and Future Developments; Kogan, M., Heinrichs, E.A., Eds.; Burleigh Dodds Science Publication: Cambridge, UK, 2020; pp. 1–30. [Google Scholar]
- Abd-Elgawad, M.M.M. Photorhabdus spp.: An overview of the beneficial aspects of mutualistic bacteria of insecticidal nematodes. Plants 2021, 10, 1660. [Google Scholar] [CrossRef] [PubMed]
- Akhurst, R.J. Taxonomic study of Xenorhabdus, a genus of bacteria symbiotically associated with insect pathogenic nematodes. Int. J. Syst. Bacteriol. 1983, 33, 38–45. [Google Scholar] [CrossRef]
- Akhurst, R.J.; Boemare, N.E. A numerical taxonomic study of the genus Xenorhabdus (Enterobacteriaceae) and proposed elevation of the subspecies of X. nematophilus to species. J. Gen. Microbiol. 1988, 134, 1835–1845. [Google Scholar] [CrossRef]
- Koppenhöfer, H.S. Bacterial symbionts of Steinernema and Heterorhabditis. In Entomopathogenic Nematodes: Systematics, Phylogeny and Bacterial Symbionts; Nguyen, K.B., Hunt, D.J., Eds.; Brill Academic Publishers: Leiden, The Netherlands; Boston, MA, USA, 2007; pp. 735–808. [Google Scholar]
- Tomar, P.; Thakur, N.; Yadav, A.N. Endosymbiotic microbes from entomopathogenic nematode (EPNs) and their applications as biocontrol agents for agro-environmental sustainability. Egypt. J. Biol. Pest. Control 2022, 32, 80. [Google Scholar] [CrossRef]
- Castaneda-Alvarez, C.; Prodan, S.; Zamorano, A.; San-Blas, E.; Aballay, E. Xenorhabdus lircayensis sp. nov., the symbiotic bacterium associated with the entomopathogenic nematode Steinernema unicornum. Int. J. Syst. Evol. Microbiol. 2021, 71, 005151. [Google Scholar] [CrossRef]
- Donmez Ozkan, H.; Cimen, H.; Ulug, D.; Wenski, S.; Yigit Ozer, S.; Telli, M.; Aydin, N.; Bode, H.B.; Hazir, S. Nematode-associated bacteria: Production of antimicrobial agent as a presumptive nominee for curing endodontic infections caused by Enterococcus faecalis. Front. Microbiol. 2019, 10, 2672. [Google Scholar] [CrossRef]
- Goodrich-Blair, H.; Clarke, D.J. Mutualism and pathogenesis in Xenorhabdus and Photorhabdus: Two roads to the same destination. Mol. Microbiol. 2007, 64, 260–268. [Google Scholar] [CrossRef]
- Ciche, T.A.; Kim, K.S.; Kaufmann-Daszczuk, B.; Nguyen, K.C.Q.; Hall, D.H. Cell invasion and matricide during Photorhabdus luminescens transmission by Heterorhabditis bacteriophora nematodes. Appl. Environ. Microbiol. 2008, 74, 2275–2287. [Google Scholar] [CrossRef]
- Chaston, J.M.; Suen, G.; Tucker, S.L.; Andersen, A.W.; Bhasin, A.; Bode, E. The entomopathogenic bacterial endosymbionts Xenorhabdus and Photorhabdus: Convergent lifestyles from divergent genomes. PLoS ONE 2011, 6, e27909. [Google Scholar] [CrossRef] [PubMed]
- An, R.; Grewal, P.S. Molecular mechanisms of persistence of mutualistic bacteria Photorhabdus in the entomopathogenic nematode host. PLoS ONE 2010, 5, e13154. [Google Scholar] [CrossRef] [PubMed]
- Lefoulon, E.; McMullen, J.G.; Stock, S.P. Transcriptomic analysis of Steinernema nematodes highlights metabolic costs associated to Xenorhabdus endosymbiont association and rearing conditions. Front. Physiol. 2022, 13, 821845. [Google Scholar] [CrossRef] [PubMed]
- Koppenhöfer, H.S.; Gaugler, R. Entomopathogenic nematode and bacteria mutualism. In Defensive Mutualism in Microbial Symbiosis; White, J., Torres, M., Eds.; CRC Press: Boca Raton, FL, USA, 2009; pp. 99–116. [Google Scholar]
- Hillman, K.; Goodrich-Blair, H. Are you my symbiont? Microbial polymorphic toxins and antimicrobial compounds as honest signals of beneficial symbiotic defensive traits. Curr. Opin. Microbiol. 2016, 31, 184–190. [Google Scholar] [CrossRef]
- Henry, L.M.; Peccoud, J.; Simon, J.C.; Hadfield, J.D.; Maiden, M.J.; Ferrari, J.; Godfray, H.C. Horizontally transmitted symbionts and host colonization of ecological niches. Curr. Biol. 2013, 9, 1713–1717. [Google Scholar] [CrossRef]
- Maher, A.M.D.; Asaiyah, M.A.M.; Brophy, C.; Griffin, C.T. An entomopathogenic nematode extends its niche by associating with different symbionts. Microb. Ecol. 2017, 73, 211–223. [Google Scholar] [CrossRef]
- Chang, D.Z.; Serra, L.; Lu, D.; Mortazavi, A.; Dillman, A.R.A. Core set of venom proteins is released by entomopathogenic nematodes in the genus Steinernema. PLoS Pathog. 2019, 15, e1007626. [Google Scholar] [CrossRef]
- Murfin, K.E.; Whooley, A.C.; Klassen, J.L.; Goodrich-Blair, H. Comparison of Xenorhabdus bovienii bacterial strain genomes reveals diversity in symbiotic functions. BMC Genom. 2015, 16, 889. [Google Scholar] [CrossRef]
- McMullen, J.G.; Peterson, B.F.; Forst, S.; Goodrich-Blair, H.; Stock, S.P. Fitness costs of symbiont switching using entomopathogenic nematodes as a model. BMC Evol. Biol. 2017, 17, 100. [Google Scholar] [CrossRef] [Green Version]
- Tailliez, P.; Laroui, C.; Ginibre, N.; Paule, A.; Pages, S.; Boemare, N. Phylogeny of Photorhabdus and Xenorhabdus based on universally conserved protein-coding sequences and implications for the taxonomy of these two genera. Proposal of new taxa. X. vietnamensis sp. nov., P. luminescens subsp. caribbeanensis subsp. nov., P. luminescens subsp. hainanensis subsp. nov., P. temperata subsp. khanii subsp. nov., P. temperata subsp. tasmaniensis subsp. nov., and the reclassification of P. luminescens subsp. thracensis as P. temperata subsp. thracensis comb. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 1921–1937. [Google Scholar]
- Askary, T.H.; Abd-Elgawad, M.M.M. Opportunities and challenges of entomopathogenic nematodes as biocontrol agents in their tripartite interactions. Egypt. J. Biol. Pest Cont. 2021, 31, 42. [Google Scholar] [CrossRef]
- Baiocchi, T.; Abd-Elgawad, M.M.M.; Dillman, A.R. Genetic improvement of entomopathogenic nematodes for enhanced biological control. In Biocontrol Agents: Entomopathogenic and Slug Parasitic Nematodes; Abd-Elgawad, M.M.M., Askary, T.H., Coupland, J., Eds.; CAB International: Wallingford, UK, 2017; pp. 505–517. [Google Scholar]
- Hazir, S.; Shapiro-Ilan, D.I.; Bock, C.H.; Hazir, C.; Leite, L.G.; Hotchkiss, M.W. Relative potency of culture supernatants of Xenorhabdus and Photorhabdus spp. on growth of some fungal phytopathogens. Eur. J. Plant Pathol. 2016, 146, 369–381. [Google Scholar] [CrossRef]
- Dreyer, J.; Malan, A.P.; Dicks, L.M.T. Bacteria of the genus Xenorhabdus, a novel source of bioactive compounds. Front. Microbiol. 2018, 9, 3177. [Google Scholar] [CrossRef]
- Cimen, H.; Touray, M.; Gulsen, S.H.; Hazir, S. Natural products from Photorhabdus and Xenorhabdus: Mechanisms and impacts. Appl. Microbiol. Biotechnol. 2022, 106, 4387–4399. [Google Scholar] [CrossRef]
- Booysen, E.; Rautenbach, M.; Stander, M.A.; Dicks, L.M. Profiling the production of antimicrobial secondary metabolites by Xenorhabdus khoisanae J194 under different culturing conditions. Front. Chem. 2021, 9, 626653. [Google Scholar] [CrossRef]
- Nunez-Valdez, M.E.; Lanois, A.; Pages, S.; Duvic, B.; Gaudriault, S. Inhibition of Spodoptera frugiperda phenoloxidase activity by the products of the Xenorhabdus rhabduscin gene cluster. PLoS ONE 2019, 22, e0212809. [Google Scholar]
- Plichta, K.L.; Joyce, S.A.; Clarke, D.; Waterfield, N.; Stock, S.P. Heterorhabditis gerrardi n. sp. (Nematoda: Heterorhabditidae): The hidden host of Photorhabdus asymbiotica (Enterobacteriaceae:g-Proteobacteria). J. Helminthol. 2009, 83, 309–320. [Google Scholar] [CrossRef]
- Abd-Elgawad, M.M.M. Towards optimization of entomopathogenic nematodes for more service in the biological control of insect pests. Egypt. J. Biol. Pest Cont. 2019, 29, 77. [Google Scholar] [CrossRef]
- Koppenhöfer, A.M.; Shapiro-Ilan, D.I.; Hiltpold, I. Entomopathogenic nematodes in sustainable food production. Front. Sustain. Food Syst. 2020, 4, 125. [Google Scholar] [CrossRef]
- Morgan, J.A.W.; Kuntzelmann, V.; Tavernor, S.; Ousley, M.A.; Winstanley, C. Survival of Xenorhabdus nematophilus and Photorhabdus luminescens in water and soil. J. Appl. Microbiol. 1997, 83, 665–670. [Google Scholar] [CrossRef]
- Antonello, A.M.; Sartori, T.; Silva, M.B.; Prophiro, J.S.; Pinge-Filho, P.; Heermann, R. Anti-Trypanosoma activity of bioactive metabolites from Photorhabdus luminescens and Xenorhabdus nematophila. Exp. Parasitol. 2019, 204, 107724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Nangong, Z.; Kong, F.; Song, P.; Wang, Q. Biological activity of Xenorhabdus nematophila HB310 against Locusta migratoria manilensis. Chin. J. Pest Sci. 2013, 15, 516–522. [Google Scholar]
- Gulsen, S.H.; Tileklioglu, E.; Bode, E.; Cimen, H.; Ertabaklar, H.; Ulug, D.; Ertug, S.; Wenski, S.L.; Touray, M.; Hazir, C.; et al. Antiprotozoal activity of different Xenorhabdus and Photorhabdus bacterial secondary metabolites and identification of bioactive compounds using the easyPACId approach. Sci. Rep. 2022, 12, 10779. [Google Scholar] [CrossRef]
- Sergeant, M.; Baxter, L.; Jarrett, P.; Shaw, E.; Ousley, M.; Winstanley, C.; Alun, J.; Morgan, W. Identification, typing and insecticidal activity of Xenorhabdus isolates from entomopathogenic nematodes in United Kingdom soil and characterization of the xpt toxin loci. Appl. Environ. Microbiol. 2006, 72, 5895–5907. [Google Scholar] [CrossRef]
- Pidot, S.J.; Coyne, S.; Kloss, F.; Hertweck, C. Antibiotics from neglected bacterial sources. Int. J. Med. Microbiol. 2014, 304, 14–22. [Google Scholar] [CrossRef]
- Abebew, D.; Sayedain, F.S.; Bode, E.; Bode, H.B. Uncovering nematicidal natural products from Xenorhabdus bacteria. J. Agric. Food Chem. 2022, 70, 498–506. [Google Scholar] [CrossRef]
- Dreyer, J.; Rautenbach, M.; Booysen, E.; Van Staden, A.; Deane, S.; Dicks, L. Xenorhabdus khoisanae SB10 produces Lys-rich PAX lipopeptides and a Xenocoumacin in its antimicrobial complex. BMC Microbiol. 2019, 19, 1–11. [Google Scholar] [CrossRef]
- Abdel-Razek, A.S. Pathogenic effects of Xenorhabdus nematophilus and Photorhabdus luminescens (Enterobacteriaceae) against pupae of the Diamondback Moth, Plutella xylostella (L.). J. Pest Sci. 2003, 76, 108–111. [Google Scholar] [CrossRef]
- Alotaibi, S.S.; Darwish, H.; Alharthi, S.; Alghamdi, A.; Noureldeen, A.; Fallatah, A.M.; Fodor, A.; Al-Barty, A.; Albogami, B.; Baazeem, A. Control potentials of three entomopathogenic bacterial isolates for the carob moth, Ectomyelois ceratoniae (Lepidoptera: Pyralidae) in pomegranates. Agriculture 2021, 11, 1256. [Google Scholar] [CrossRef]
- Sayedain, F.S.; Ahmadzadeh, M.; Talaei-Hassanloui, R.; Olia, M.; Bode, H.B. Nematicidal effect of cell-free culture filtrates of EPN-symbiotic bacteria on Meloidogyne javanica. Biol. Cont. Pests Pl. Dis. 2019, 8, 17–26. [Google Scholar]
- Sicard, M.; Le Brun, N.; Pages, S.; Godelle, B.; Boemare, N.; Moulia, C. Effect of native Xenorhabdus on the fitness of their Steinernema hosts: Contrasting types of interaction. Parasitol. Res. 2003, 91, 520–524. [Google Scholar] [CrossRef]
- Kim, I.H.; Aryal, S.K.; Aghai, D.T.; Casanova-Torres, Á.M.; Hillman, K.; Kozuch, M.P.; Mans, E.J.; Mauer, T.J.; Ogier, J.C.; Ensign, J.C.; et al. The insect pathogenic bacterium Xenorhabdus innexi has attenuated virulence in multiple insect model hosts yet encodes a potent mosquitocidal toxin. BMC Genom. 2017, 18, 927. [Google Scholar] [CrossRef]
- Askary, T.H.; Abd-Elgawad, M.M.M. Beneficial nematodes in agroecosystems: A global perspective. In Biocontrol Agents: Entomopathogenic and Slug Parasitic Nematodes; Abd-Elgawad, M.M.M., Askary, T.H., Coupland, J., Eds.; CAB International: Wallingford, UK, 2017; pp. 3–25. [Google Scholar]
- Eom, S.; Park, Y.; Kim, H.; Kim, Y. Development of a high efficient“dual Bt-plus” insecticide using a primary form of an entomopathogenic bacterium, Xenorhabdus nematophila. J. Microbiol. Biotechnol. 2014, 24, 507–521. [Google Scholar] [CrossRef]
- Seongchae, J.; Yonggyun, K. Synergistic effect of entomopathogenic bacteria (Xenorhabdus sp. and Photorhabdus temperata ssp. temperata) on the pathogenicity of Bacillus thuringiensis ssp. Aizawai against Spodoptera exigua (Lepidoptera: Noctuidae). Environ. Entomol. 2006, 35, 1584–1589. [Google Scholar]
- Seo, S.; Lee, S.; Hong, Y.; Kim, Y. Phospholipase A2 inhibitors synthesized by two entomopathogenic bacteria, Xenorhabdus nematophila and Photorhabdus temperata subsp. temperata. Appl. Environ. Microbiol. 2012, 78, 3816–3823. [Google Scholar] [CrossRef]
- Mollah, M.I.; Kim, Y. Virulent secondary metabolites of entomopathogenic bacteria genera, Xenorhabdus and Photorhabdus, inhibit phospholipase A2 to suppress host insect immunity. BMC Microbiol. 2020, 20, 359. [Google Scholar] [CrossRef]
- Mahar, A.N.; Munir, M.; Gowen, S.R.; Hague, N.G.M. Role of entomopathogenic bacteria, Photorhabdus luminescens and its toxic secretions against Galleria mellonella larvae. J. Entomol. 2005, 2, 69–76. [Google Scholar] [CrossRef]
- Wang, H.; Dong, H.; Qian, H.; Xia, R.; Cong, B. Isolation, bioassay and characterisation of Xenorhabdus sp. SY5, a highly virulent symbiotic bacterium of an entomopathogenic nematode isolated from China. Nematology 2013, 15, 153–163. [Google Scholar] [CrossRef]
- Hajihassani, A.; Gitonga, D.; Timper, P.; Shapiro-Ilan, D. Effects of application timing on the efficacy of Xenorhabdus and Photorhabdus metabolites for control of Meloidogyne incognita. In Proceedings of the 7th International Congress of Nematology, Antibes Juan-les-Pins, France, 1–5 May 2022; p. 598. [Google Scholar]
- Castaneda-Alvarez, C.; Prodan, S.; San-Blas, E.; Aballay, E. Symbiotic bacteria of entomopathogenic nematodes for the biocontrol of dagger nematode Xiphinema index. In Proceedings of the 7th International Congress of Nematology, Antibes Juan-les-Pins, France, 1–5 May 2022; p. 587. [Google Scholar]
- Vicente-Díez, I.; Blanco-Pérez, R.; Chelkha, M.; Pou1, A.; Campos-Herrera, R. Plasticity in the use of Xenorhabdus nematophila and Photorhabdus laumondii against Botrytis cinerea. In Proceedings of the 7th International Congress of Nematology, Antibes Juan-les-Pins, France, 1–5 May 2022; p. 660. [Google Scholar]
- Shapiro-Ilan, D.I.; Bock, C.H.; Hotchkiss, M.W. Suppression of pecan and peach pathogens on different substrates using Xenorhabdus bovienii and Photorhabdus luminescens. Biol. Cont. 2014, 77, 1–6. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, Q.; Han, Y.; Han, J.; Yan, Z.; Wang, Y.; Zhang, X. Nematophin, an antimicrobial dipeptide compound from Xenorhabdus nematophila YL001 as a potent biopesticide for Rhizoctonia solani control. Front. Microbiol. 2019, 10, 1765. [Google Scholar] [CrossRef]
- Fang, X.; Zhang, M.; Tang, Q.; Wang, Y.; Zhang, X. Inhibitory effect of Xenorhabdus nematophila TB on plant pathogens Phytophthora capsici and Botrytis cinerea in vitro and in planta. Sci. Rep. 2014, 4, 4300. [Google Scholar] [CrossRef]
- Qin, Y.; Jia, F.; Li, X.; Li, B.; Ren, J.; Yang, X.; Li, G. Improving the yield of xenocoumacin 1 by PBAD promoter replacement in Xenorhabdus nematophila CB6. Agriculture 2021, 11, 1251. [Google Scholar] [CrossRef]
- Yang, X.; Qiu, D.; Yang, H.; Liu, Z.; Zeng, H.; Yuan, J. Antifungal activity of xenocoumacin 1 from Xenorhabdus nematophilus var. pekingensis against Phytophthora infestans. World J. Microb. Biotechnol. 2011, 27, 523–528. [Google Scholar] [CrossRef]
- Zhou, T.; Yang, X.; Qiu, D.; Zeng, H. Inhibitory effects of xenocoumacin 1 on the different stages of Phytophthora capsici and its control effect on Phytophthora blight of pepper. BioControl 2017, 62, 151–160. [Google Scholar] [CrossRef]
- Li, B.; Kong, L.; Qiu, D.; Francis, F.; Wang, S. Biocontrol potential and mode of action of entomopathogenic bacteria Xenorhabdus budapestensis C72 against Bipolaris maydis. Biol. Cont. 2021, 158, 104605. [Google Scholar] [CrossRef]
- Chacón-Orozco, J.G.; Bueno, C., Jr.; Shapiro Ilan, D.I.; Hazir, S.; Leite, L.G.; Harakava, R. Antifungal activity of Xenorhabdus spp. and Photorhabdus spp. against the soybean pathogenic Sclerotinia sclerotiorum. Sci. Rep. 2020, 10, 20649. [Google Scholar] [CrossRef]
- Ciezki, K.J. New Insights into the Role of Antimicrobials of Xenorhabdus in Interspecies Competition. Ph.D. Dissertation, University of Wisconsin-Milwaukee, Milwaukee, WI, USA, 2017; 103p. [Google Scholar]
- Oliva, B.; O’Neill, A.; Wilson, J.M.; O’Hanlon, P.J.; Chopra, I. Antimicrobial properties and mode of action of the pyrrothine holomycin. Antimicrob. Agents Chemother. 2001, 45, 532–539. [Google Scholar] [CrossRef]
- Shrestha, S.; Kim, Y. An entomopathogenic bacterium, Xenorhabdus nematophila, inhibits hemocyte phagocytosis of Spodoptera exigua by inhibiting phospholipase A2. J. Invertebr. Pathol. 2007, 96, 64–70. [Google Scholar] [CrossRef]
- Xiao, Y.; Meng, F.; Qiu, D.; Yang, X. Two novel antimicrobial peptides purified from the symbiotic bacteria Xenorhabdus budapestensis NMC-10. Peptides 2012, 35, 253–260. [Google Scholar] [CrossRef]
- Li, B.; Qiu, D.; Wang, S. Complete genome sequence data of Xenorhabdus budapestensis strain C72, a candidate biological control agent from China. Plant Dis. 2021, 105, 3276–3278. [Google Scholar] [CrossRef]
- Ji, D.; Yi, Y.; Kang, G.-H.; Choi, Y.-H.; Kim, P.; Baek, N.-I.; Kim, Y. Identification of an antibacterial compound, benzylideneacetone, from Xenorhabdus nematophila against major plant-pathogenic bacteria. FEMS Microbiol. Lett. 2004, 239, 241–248. [Google Scholar] [CrossRef]
- Böszörményi, E.; Ersek, T.; Fodor, A.; Fodor, A.M.; Földes, L.S.; Hevesi, M.; Hogan, J.S.; Katona, Z.; Klein, M.G.; Kormany, A.; et al. Isolation and activity of Xenorhabdus antimicrobial compounds against the plant pathogens Erwinia amylovora and Phytophthora nicotianae. J. Appl. Microbiol. 2009, 107, 746–759. [Google Scholar] [CrossRef]
- Rathore, J.S. Expression, purification, and functional characterization of atypical xenocin, its immunity protein, and their domains from Xenorhabdus nematophila. Int. J. Bacteriol. 2013, 2013, 746862. [Google Scholar] [CrossRef]
- Xi, X.; Lu, X.; Zhang, X.; Bi, Y.; Li, X.; Yu, Z. Two novel cyclic depsipeptides xenematides F and G from the entomopathogenic bacterium Xenorhabdus budapestensis. J. Antibiot. 2019, 72, 736–743. [Google Scholar] [CrossRef]
- Fuchs, S.W.; Grundmann, F.; Kurz, M.; Kaiser, M.; Bode, H.B. Fabclavines: Bioactive peptide-polyketide-polyamino hybrids from Xenorhabdus. ChemBioChem 2014, 15, 512–516. [Google Scholar] [CrossRef]
- Park, D.; Ciezki, K.; van der Hoeven, R.; Singh, S.; Reimer, D.; Bode, H.B.; Forst, S. Genetic analysis of xenocoumacin antibiotic production in the mutualistic bacterium Xenorhabdus nematophila. Mol. Microbiol. 2009, 73, 938–949. [Google Scholar] [CrossRef]
- Li, J.; Chen, G.; Webster, J.M. Nematophin, a novel antimicrobial substance produced by Xenorhabdus nematophilus (Enterobactereaceae). Can. J. Microbiol. 1997, 43, 770–773. [Google Scholar] [CrossRef]
- Zhao, L.; Kaiser, M.; Bode, H.B. Rhabdopeptide/xenortide-like peptides from Xenorhabdus innexi with terminal amines showing potent antiprotozoal activity. Org. Lett. 2018, 20, 5116–5120. [Google Scholar] [CrossRef]
- Fuchs, S.W.; Proschak, A.; Jaskolla, T.W.; Karas, M.; Bode, H.B. Structure elucidation and biosynthesis of lysine-rich cyclic peptides in Xenorhabdus nematophila. Org. Biomol. Chem. 2011, 9, 3130–3132. [Google Scholar] [CrossRef]
- Zhou, Q.; Grundmann, F.; Kaiser, M.; Schiell, M.; Gaudriault, S.; Batzer, A. Structure and biosynthesis of xenoamicins from entomopathogenic Xenorhabdus. Chem. Eur. J. 2013, 19, 16772–16779. [Google Scholar] [CrossRef]
- Kronenwerth, M.; Brachmann, A.O.; Kaiser, M.; Bode, H.B. Bioactive derivatives of isopropylstilbene from mutasynthesis and chemical synthesis. ChemBioChem 2014, 15, 2689–2691. [Google Scholar] [CrossRef]
- Lang, G.; Kalvelage, T.; Peters, A.; Wiese, J.; Imhoff, J.F. Linear and cyclic peptides from the entomopathogenic bacterium Xenorhabdus nematophilus. J. Nat. Prod. 2008, 71, 1074–1077. [Google Scholar] [CrossRef]
- Nollmann, F.I.; Dowling, A.; Kaiser, M.; Deckmann, K.; Grösch, S.; French-Constant, R.; Bode, H.B. Synthesis of szentiamide, a depsipeptide from entomopathogenic Xenorhabdus szentirmaii with activity against Plasmodium falciparum. Beilstein J. Org. Chem. 2012, 8, 528–533. [Google Scholar] [CrossRef]
- Grundmann, F.; Kaiser, M.; Kurz, M.; Schiell, M.; Batzer, A.; Bode, H.B. Structure determination of the bioactive depsipeptide xenobactin from Xenorhabdus sp. PB30. 3. RSC Adv. 2013, 3, 22072–22077. [Google Scholar] [CrossRef]
- Tobias, N.J.; Wolff, H.; Djahanschiri, B.; Grundmann, F.; Kronenwerth, M.; Shi, Y.M. Natural product diversity associated with the nematode symbionts Photorhabdus and Xenorhabdus. Nat. Microbiol. 2017, 2, 1676–1685. [Google Scholar] [CrossRef]
- Kepenekci, I.; Hazir, S.; Oksal, E.; Lewis, E.E. Application methods of Steinernema feltiae, Xenorhabdus bovienii and Purpureocillium lilacinum to control root-knot nematodes in greenhouse tomato systems. Crop Prot. 2018, 108, 31–38. [Google Scholar] [CrossRef]
- Erler, S.; Popp, M.; Lattorff, M. Dynamics of immune system gene expression upon bacterial challenge and wounding in a social insect (Bombus terrestris). PLoS ONE 2011, 6, e18126. [Google Scholar] [CrossRef]
- Abd El-Zaher, F.H.; Abd-Elgawad, M.M.M.; Abd El-Maksoud, H.K. Use of the entomopathogenic nematode symbiont Photorhabdus luminescens as a biocontrol agent. B- Factors affecting the cell-free filtrates from the bacterium. J. Appl. Sci. Res. 2012, 8, 4600–4614. [Google Scholar]
- Boemare, N.; Boyer-Giglio, M.; Thaler, J.; Akhurst, R.; Brehelin, M. Lysogeny and bacteriocinogeny in Xenorhabdus nematophilus and other Xenorhabdus spp. Appl. Environ. Microbiol. 1992, 58, 3032–3037. [Google Scholar] [CrossRef]
- Proschak, A.; Zhou, Q.; Schöner, T.; Thanwisai, A.; Kresovic, D.; Dowling, A.; Ffrench-Constant, R.; Proshack, E.; Bode, H.B. Biosynthesis of the insecticidal xenocyloins in Xenorhabdus bovienii. Chem. Biol. Chem. 2014, 15, 369–372. [Google Scholar] [CrossRef]
- Park, H.B.; Perez, C.E.; Perry, E.K.; Crawford, J.M. Activating and attenuating the amicoumacin antibiotics. Molecules 2016, 21, 824. [Google Scholar] [CrossRef]
- Li, J.; Chen, G.; Webster, J.M.; Czyzewska, E. Antimicrobial metabolites from a bacterial symbiont. J. Nat. Prod. 1995, 58, 1081–1086. [Google Scholar] [CrossRef]
- Wenski, S.L.; Cimen, H.; Berghaus, N.; Fuchs, S.W.; Hazir, S.; Bode, H.B. Fabclavine diversity in Xenorhabdus bacteria. Beilstein J. Org. Chem. 2020, 16, 956–965. [Google Scholar] [CrossRef]
- Gualtieri, M.; Villain-Guillot, P.; Givaudan, A.; Pages, S. Nemaucin, an Antibiotic Produced by Entomopathogenic Xenorhabdus cabanillasii. Google Patents. WO2012085177 A1, 28 June 2012. pp. 1–39. [Google Scholar]
- Reimer, D.; Cowles, K.N.; Proschak, A.; Nollmann, F.I.; Dowling, A.J.; Kaiser, M.; Constant, R.F.; Goodrich-Blair, H.; Bode, H.B. Rhabdopeptides as insect-specific virulence factors from entomopathogenic bacteria. Chem. Biol. Chem. 2013, 14, 1991–1997. [Google Scholar] [CrossRef]
- Houard, J.; Aumelas, A.; Noel, T.; Givaudan, A.; Fitton-Ouhabi, V.; Villain-Guillot, P.; Gualtieri, M. Cabanillasin, a new antifungal metabolite, produced by entomopathogenic Xenorhabdus cabanillasii JM26. J. Antibiot. 2013, 66, 617–620. [Google Scholar] [CrossRef]
- Bode, E.; He, Y.; Vo, T.D.; Schultz, R.; Kaiser, M.; Bode, H.B. Biosynthesis and function of simple amides in Xenorhabdus doucetiae. Environ. Microbiol. 2017, 19, 4564–4575. [Google Scholar] [CrossRef]
- Hacker, C.; Cai, X.; Kegler, C.; Zhao, L.; Weickhmann, A.K.; Wurm, J.P.; Bode, H.B.; Wöhnert, J. Structure-based redesign of docking domain interactions modulates the product spectrum of a rhabdopeptide-synthesizing NRPS. Nat. Commun. 2018, 9, 4366. [Google Scholar] [CrossRef]
- Brachmann, A.O.; Reimer, D.; Lorenzen, W.; Augusto, A.E.; Kopp, Y.; Piel, J.; Bode, H.B. Reciprocal cross talk between fatty acid and antibiotic biosynthesis in a nematode symbiont. Angew. Chem. 2012, 124, 12252–12255. [Google Scholar] [CrossRef]
- Qin, Z.; Huang, S.; Yu, Y.; Deng, H. Dithiolopyrrolone natural products: Isolation, synthesis and biosynthesis. Mar. Drugs 2013, 11, 3970–3997. [Google Scholar] [CrossRef]
- Cai, X.; Nowak, S.; Wesche, F.; Bischoff, I.; Kaiser, M.; Fürst, R.; Bode, H.B. Entomopathogenic bacteria use multiple mechanisms for bioactive peptide library design. Nat. Chem. 2017, 9, 379–386. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, S.; Fang, X.; Liu, Q.; Gao, J.; Bilal, M.; Wang, Y.; Zahng, X. Regulation of antimicrobial activity and xenocoumacins biosynthesis by pH in Xenorhabdus nematophila. Microb. Cell. Fact. 2017, 16, 203. [Google Scholar] [CrossRef]
- Esmati, N.; Maddirala, A.; Hussein, N.; Amawi, H.; Tiwari, A.; Andreana, P. Efficient syntheses and anti-cancer activity of xenortides A-D including ent/epi-stereoisomers. Org. Biomol. Chem. 2018, 16, 5332–5342. [Google Scholar] [CrossRef]
- Crawford, J.M.; Portmann, C.; Zhang, X.; Roeffaers, M.B.; Clardy, J. Small molecule perimeter defense in entomopathogenic bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 10821–10826. [Google Scholar] [CrossRef]
- Nollmann, F.I.; Dauth, C.; Mulley, G.; Kegler, C.; Kaiser, M.; Waterfield, N.R.; Bode, H.B. Insect-specific production of new GameXPeptides in Photorhabdus luminescens TTO1, widespread natural products in entomopathogenic bacteria. Chem. Biol. Chem. 2015, 16, 205–208. [Google Scholar] [CrossRef]
- Dongare, R.K.; Inamdar, S.N.; Tigote, R.M. Dft based investigations of antibiotic and antifungal activity of allantofuranone and related γ-lactone compounds. J. Adv. Sci. Res. 2021, 12, 336–339. [Google Scholar]
- Fodor, A.; Gualtieri, M.; Zeller, M.; Tarasco, E.; Klein, M.G.; Fodor, A.M.; Haynes, L.; Lengyel, K.; Forst, S.A.; Furgani, G.M.; et al. Type strains of entomopathogenic nematode-symbiotic bacterium species, Xenorhabdus szentirmaii (EMC) and X. budapestensis (EMA), are exceptional sources of non-ribosomal templated, large-target-spectral, thermotolerant-antimicrobial peptides (by both), and iodinin (by EMC). Pathogens 2022, 11, 342. [Google Scholar] [CrossRef]
- Kepenekci, I.; Hazir, S.; Lewis, E.E. Evaluation of entomopathogenic nematodes and the supernatants of the in vitro culture medium of their mutualistic bacteria for the control of the root-knot nematodes Meloidogyne incognita and M. arenaria. Pest Manag. Sci. 2016, 72, 327–334. [Google Scholar] [CrossRef]
- Grundmann, F.; Kaiser, M.; Schiell, M.; Batzer, A.; Kurz, M.; Thanwisai, A.; Chantratita, N.; Bode, H.B. Antiparasitic chaiyaphumines from entomopathogenic Xenorhabdus sp. PB61.4. J. Nat. Prod. 2014, 77, 779–783. [Google Scholar] [CrossRef]
- Ruiu, L.; Satta, A.; Floris, I. Emerging entomopathogenic bacteria for insect pest management. Bull. Insectol. 2013, 66, 181–186. [Google Scholar]
- Hemalatha, D.; Prabhu, S.; Rani, W.B.; Anandham, R. Isolation and characterization of toxins from Xenorhabdus nematophilus against Ferrisia virgata (Ckll.) on tuberose, Polianthes tuberosa. Toxicon 2018, 146, 42–49. [Google Scholar] [CrossRef]
- Vicente-Díez, I.; Blanco-Pérez, R.; Chelkha, M.; Puelles, M.; Pou, A.; Campos-Herrera, R. Exploring the use of entomopathogenic nematodes and the natural products derived from their symbiotic bacteria to control the grapevine moth, Lobesia botrana (Lepidoptera: Tortricidae). Insects 2021, 12, 1033. [Google Scholar] [CrossRef]
- Shi, H.; Zeng, H.; Yang, X.; Zhao, J.; Chen, M.; Qiu, D. An insecticidal protein from Xenorhabdus ehlersii triggers prophenoloxidase activation and hemocyte decrease in Galleria mellonella. Curr. Microbiol. 2012, 64, 604–610. [Google Scholar] [CrossRef]
- Shi, H.X.; Zengm, H.M.; Yang, X.F.; Liu, Z.; Qiu, D. An insecticidal protein from Xenorhabdus ehlersii stimulates the innate immune response in Galleria mellonella. World J. Microbiol. Biotechnol. 2013, 29, 1705–1711. [Google Scholar] [CrossRef]
- Bussaman, P.; Sa-Uth, C.; Rattanasena, P.; Chandrapatya, A. Acaricidal activities of whole cell suspension, cell-free supernatant, and crude cell extract of Xenorhabdus stokiae against mushroom mite (Luciaphorus sp.). J. Zhejiang Univ. Sci. B 2012, 13, 261–266. [Google Scholar] [CrossRef]
- Bussaman, P.; Rattanasena, P. Additional property of Xenorhabdus stockiae for inhibiting cow mastitis-causing bacteria. Biosci. Biotechnol. Res. Asia 2016, 13, 1871–1878. [Google Scholar] [CrossRef]
- Namsena, P.; Bussaman, P.; Rattanasena, P. Bioformulation of Xenorhabdus stockiae PB09 for controlling mushroom mite, Luciaphorus perniciosus Rack. Bioresour. Bioprocess. 2016, 3, 19. [Google Scholar] [CrossRef]
- Zhou, X.; Kaya, H.K.; Heungens, K.; Goodrich-Blair, H. Response of ants to a deterrent factor(s) produced by the symbiotic bacteria of entomopathogenic nematodes. Appl. Environ. Microbiol. 2002, 68, 6202–6209. [Google Scholar] [CrossRef] [Green Version]
- Gulcu, B.; Hazir, S.; Kaya, H.K. Scavenger deterrent factor (SDF) from symbiotic bacteria of entomopathogenic nematodes. J. Invertebr. Pathol. 2012, 110, 326–333. [Google Scholar] [CrossRef]
- Park, Y.; Kyo Jung, J.; Kimm, Y.A. Mixture of Bacillus thuringiensis subsp. israelensis with Xenorhabdus nematophila-cultured broth enhances toxicity against mosquitoes Aedes albopictus and Culex pipiens pallens (Diptera: Culicidae). J. Econ. Entomol. 2016, 109, 1086–1093. [Google Scholar] [CrossRef]
- Gaugler, R. (Ed.) Entomopathogenic Nematology; CAB International: New York, NY, USA, 2002; 365p. [Google Scholar]
- Grewal, P.S.; Ehlers, R.-U.; Shapiro-Ilan, D.I. (Eds.) Nematodes as Biocontrol Agents; CAB International: Wallingford, UK, 2005; 523p. [Google Scholar]
- Abd-Elgawad, M.M.M.; Askary, T.H.; Coupland, J. (Eds.) Biocontrol Agents: Entomopathogenic and Slug Parasitic Nematodes; CAB International: Wallingford, UK, 2005; 662p. [Google Scholar]
- Grewal, P.S.; Martin, W.R.; Miller, R.; Lewis, E.E. Suppression of plant-parasitic nematode populations in turfgrass by application of entomopathogenic nematodes. Biocontrol Sci. Technol. 1997, 7, 393–399. [Google Scholar] [CrossRef]
- Kumari, P.; Mahapatro, G.K.; Banerjee, N.; Sarin, N.B. Ectopic expression of GroEL from Xenorhabdus nematophila in tomato enhances resistance against Helicoverpa armigera and salt and thermal stress. Transgenic Res. 2015, 24, 859–873. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Bilgrami, A.L.; Shapiro-Ilan, D.; Gaugler, R. Stability of entomopathogenic bacteria, Xenorhabdus nematophila and Photorhabdus luminescens, during in vitro culture. J. Int. Microbiol. Biotechnol. 2007, 34, 73–81. [Google Scholar] [CrossRef]
- Han, Y.; Gao, J.; Zhang, S.; Han, J.; Yan, Z.; Ta, Y.; Wang, Y. Increasing the production of xenocoumacin 1 by optimizing the fermentation process of Xenorhabdus nematophila. Res. Square 2021, 1–23. [Google Scholar] [CrossRef]
- Gulcu, B. Comparison of powder and liquid forms of antifungal metabolites produced by Xenorhabdus szentirmaii, the symbionts of entomopathogenic nematodes, against gray mold Botrytis cinerea. J. Agric. Sci. Technol. 2022, 24, 457–464. [Google Scholar]
- Sergeant, M.; Jarrett, P.; Ousely, M.; Morgan, A.W. Interactions of insecticidal toxin gene products from Xenorhabdus nematophila PMFI296. Appl. Environ. Microbiol. 2003, 69, 3344–3349. [Google Scholar] [CrossRef] [PubMed]
- Abd-Elgawad, M.M.M.; Askary, T.H. Factors affecting success of biological agents used in controlling plant-parasitic nematodes. Egypt. J. Biol. Pest Cont. 2020, 30, 17. [Google Scholar] [CrossRef]
- Coupland, J.; Abd-Elgawad, M.M.M.; Askary, T.H. Beneficial nematodes and the changing scope of crop protection. In Biocontrol Agents: Entomopathogenic and Slug Parasitic Nematodes; Abd-Elgawad, M.M.M., Askary, T.H., Coupland, J., Eds.; CAB International: Wallingford, UK, 2017; pp. 26–42. [Google Scholar]
| Bacterial spp. | Bioactive Complexes/ Secondary Metabolite | Biological Asset | References |
|---|---|---|---|
| X. beddingii | R-type bacteriocins | Bactericidal | [102] |
| X. bovienii | Xenocyloins | Insecticidal | [103] |
| Amicoumacin | Antibacterial, insecticidal, antifungal, anticancer, and anti-inflammatory | [104] | |
| Indoles | Antibiotic | [105] | |
| Dithiolopyrrolones | Antibiotic | [105] | |
| X. budapestensis | Bicornitun | Antibacterial and antifungal | [98] |
| GP-19 | Antibacterial and antifungal | [83] | |
| EP-20 | Antifungal | ||
| Fabclavine | Antibacterial, antifungal, antiprotozoal, and cytotoxic | [106] | |
| X. cabanillasii | Nemaucin | Antibacterial and antifungal | [107] |
| Rhabdopeptide | Antiprotozoal, insecticidal, and cytotoxic | [108] | |
| Cabanillasin | Antifungal | [109] | |
| X. doucetiae | Xenoamicin | Antiprotozoal | [110] |
| Xenorhabdin | Antibacterial | ||
| Xenocoumacin | Antibacterial, antifungal, and antiulcer | ||
| X. ehlersii | XeGroEL protein | Insecticidal | [42] |
| X. indica | Taxlllaids | Antiprotozoal and cytotoxic | [95] |
| X. kozodoii | Xenocoumacin | Antibacterial, antifungal, and antiulcer | [98] |
| Xenorhabdusspp. | Xenobactin | Antibacterial and antiprotozoal | [97] |
| X. khoisanaestrain SB10 | PAX lipopeptides | Antimicrobial | [43] |
| Xenocoumacin | Antimicrobial | [43] | |
| X. innexi | Rhabdopeptides | Antiprotozoal, insecticidal, and cytotoxic | [111] |
| X. mauleonii | Xenoamicin | Antiprotozoal | [98] |
| xenocoumacin | Antibacterial, antifungal, and antiulcer | ||
| xenorhabdin | Antibacterial | ||
| X. nematophila | Pristinamycin | Antibacterial | [112] |
| Xenorhabdins | Antibacterial | [113] | |
| Xenorxides | Antibacterial and antifungal | ||
| PAX peptides | Antibacterial and antifungal | [40] | |
| Nematophin | Antibacterial and antifungal | [114] | |
| Xenocin | Antibacterial | [87] | |
| Xenorhabdicin (R-type bacteriocins) | Antibacterial | [44] | |
| Xenocoumacins | Antibacterial, antifungal, and antiulcer | [115] | |
| Xenortides | Antiprotozoal and cytotoxic | [116] | |
| Rhabdopeptides | Antiprotozoal, insecticidal, and cytotoxic | [92] | |
| Xenematides | Antibacterial and insecticidal | [117] | |
| Rhabducin | Insecticidal | [117] | |
| Oxindole and Benzylidene-acetone | Antibacterial, immuno-suppressant, and insecticidal | [41,85] | |
| X. szentirmaii | Fabclavines | Antibacterial, antifungal, antiprotozoal, and cytotoxic | [106] |
| Szentiamide and Xenocin (peptide xcinA sequenced) | Antibacterial, antifungal, and/or cytotoxicity | [41,118] | |
| Xenofuranones | A and B Insecticidal | [119] | |
| Xenocoumacin | Antimicrobial | [55] | |
| PAX lipopeptides | Antimicrobial | [55] | |
| Iodinin (1,6-dihydroxyphenazine 5,10-dioxide) | Antimicrobial multidrug-resistant pathogens | [120] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abd-Elgawad, M.M.M. Xenorhabdus spp.: An Overview of the Useful Facets of Mutualistic Bacteria of Entomopathogenic Nematodes. Life 2022, 12, 1360. https://doi.org/10.3390/life12091360
Abd-Elgawad MMM. Xenorhabdus spp.: An Overview of the Useful Facets of Mutualistic Bacteria of Entomopathogenic Nematodes. Life. 2022; 12(9):1360. https://doi.org/10.3390/life12091360
Chicago/Turabian StyleAbd-Elgawad, Mahfouz M. M. 2022. "Xenorhabdus spp.: An Overview of the Useful Facets of Mutualistic Bacteria of Entomopathogenic Nematodes" Life 12, no. 9: 1360. https://doi.org/10.3390/life12091360
APA StyleAbd-Elgawad, M. M. M. (2022). Xenorhabdus spp.: An Overview of the Useful Facets of Mutualistic Bacteria of Entomopathogenic Nematodes. Life, 12(9), 1360. https://doi.org/10.3390/life12091360