3.1. Basic Model—Length Distributions and Error Threshold Behaviour
The model was run with parameters shown in
Table 2. We used values reported in the literature to approximate the orders of magnitude between our parameters. The rate of non-enzymatic addition of a 25-base character with strand displacement,
, was defined as our point of reference with a value of 1 h
−1, and we assumed that the
rate (which does not require strand displacement) is 100 times faster. When probing for an error threshold, as we do later in this section, the rate of cell death
is defined as 1
10
−3 h
−1. This is much less than the rate of cell division when the mutation rate is low, but when
approaches the error threshold, cell division becomes very slow and eventually falls below
. When
u is small and we are not close to an error threshold, it makes little difference whether
is zero or has a small positive value. We expect folding of a strand into its secondary structure to be a relatively fast event, so we set
to 100 h
−1. We do not know exact values for these rate parameters but we aim to correctly identify fast and slow steps. Though we use hours as our time unit for simplicity, it is only the relative size of the rates that effects the outcome of our model.
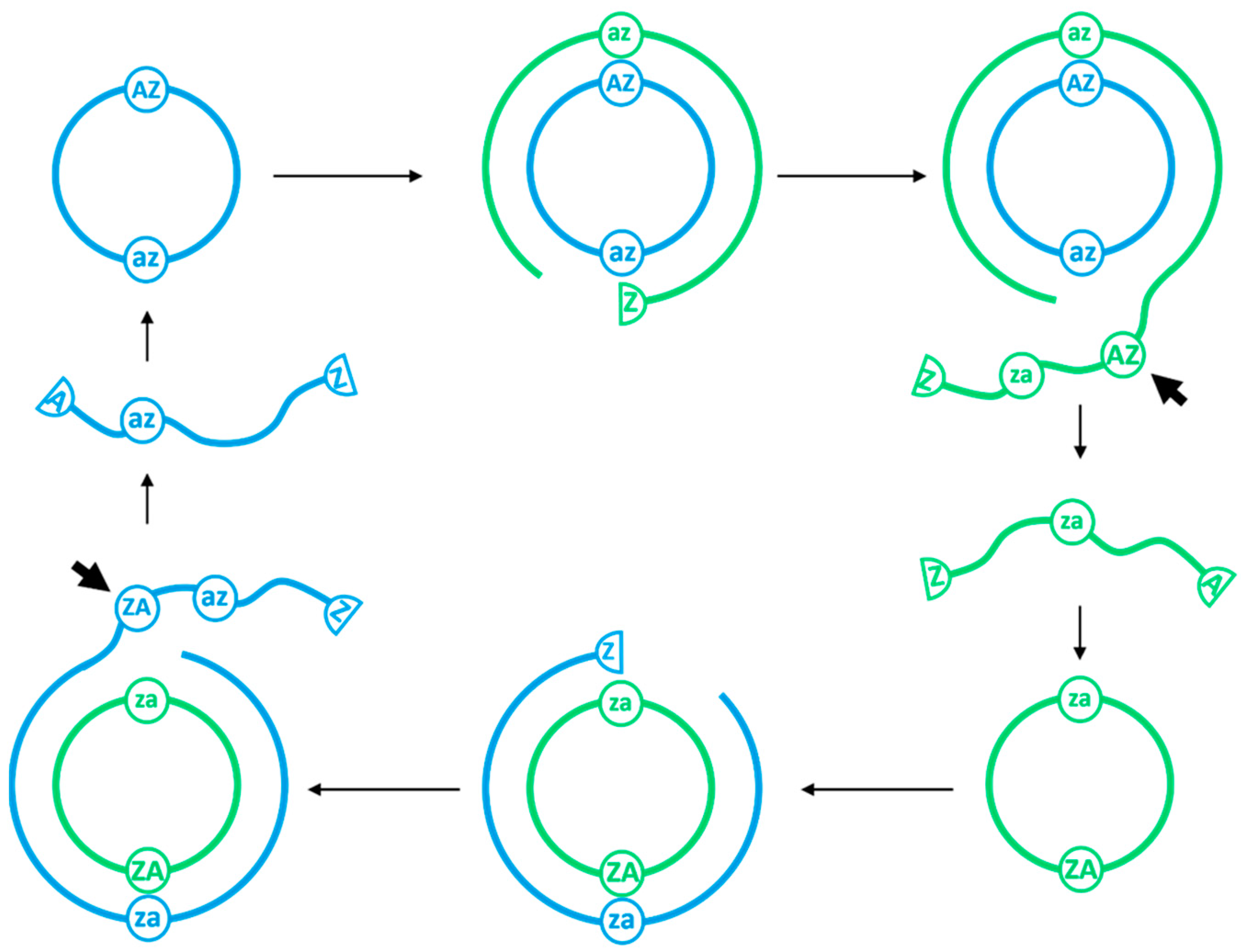
We begin with a single circular
ZzaA sequence per cell. After allowing the system to reach a steady state, the mean number of strands per cell of each length and type was counted and averaged over time. Strands are classified as one of four types, indicated by colours in
Figure 3. Reproducing strands are those which have one complete
AZ motif and one complete
za. Non-Cleaving strands have one complete
AZ motif and no
za. Halving strands are those which have one complete
AZ and more than one complete
za. Fragments are those which do not have
Z and
A at the ends, and which therefore cannot circularize.
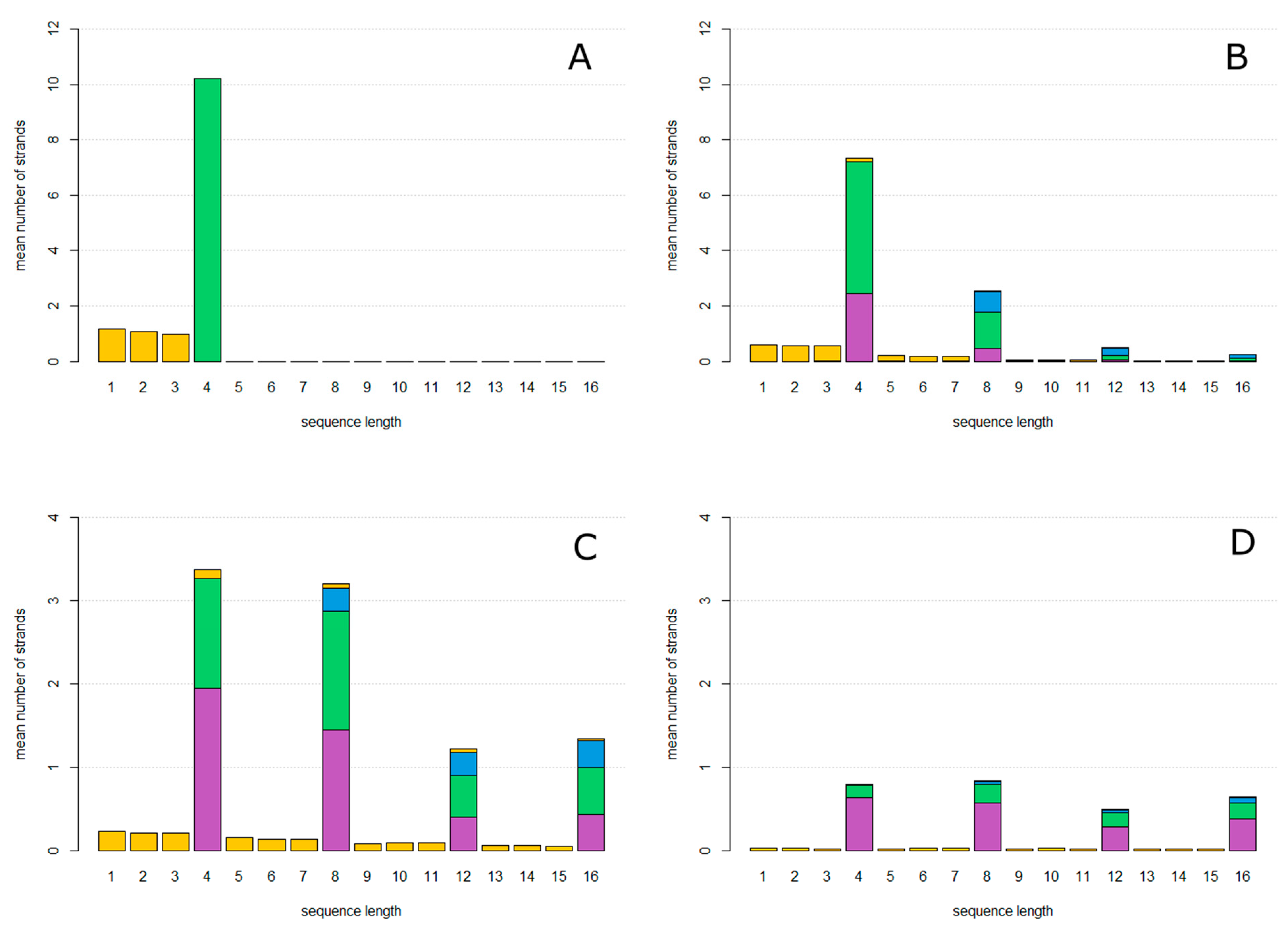
In
Figure 3A, the mutation probability is 0. The only sequences that arise are accurate copies of
ZzaA, which are reproducing strands of length 4, and fragments of lengths less than 4 which are created as the first product strand of each new rolling circle. In
Figure 3B, the mutation probability is
u = 0.15. Circles of lengths equal to multiples of 4 are now seen, as well as fragments of all the other lengths. Non-Cleaving and Halving circles are found as well as Reproductive circles.
As the mutation probability increases as shown in
Figure 3C (
u = 0.3) and
Figure 3D (
u = 0.5), the number of reproductive circles decreases and the relative proportion halving and non-cleaving strands increases until the system meets an error threshold where replication can no longer keep up with the loss rate, as shown in
Figure 4. At this point, all types of strands disappear, because they cannot be maintained without continued replication of the reproductive circles. The observed error threshold of
u = 0.6 is the probability of a deleterious mutation in a single character (
A,
Z etc.). As each character represents a length roughly 25 bases,
, where
is the point mutation rate per base, and
is the fraction of mutations that are deleterious, i.e., the fraction of mutations within the structural region of the hammerhead that destroy the function of the hammerhead. This region contains loops whose sequence may not be important and paired regions in which compensatory changes may occur. Thus,
may be considerably less than 1. If we assume
, then an error threshold of 0.6 in
u corresponds to a threshold of 0.048 in
, which is not unreasonably small. The threshold value of 0.6 is dependent on the loss rate of cells, and would be higher if the loss rate were lower.
3.2. Insertions and Deletions
The 4-mer
ZzaA is the shortest sequence having both the hammerhead and its complement. It is therefore the most rapidly replicating type of circle in this model. There is no reason why the self-cleaving ribozymes should initially appear on a minimal length sequence. However, we expect there to be strong selection for speed of replication; therefore if the hammerhead motifs initially appear on a longer circle, we expect relatively quick evolution toward the minimal-length circle due to deletions of the non-functional parts of the circle. Insertions and deletions (indels) can occur via the slipped-strand mispairing mechanism [
41]. This tends to produce short tandem repeats which are often seen in viral genes [
41,
42].
Indels are introduced into the model in the following way. Each time a character is copied, an insertion occurs with probability , a deletion occurs with probability , or neither with probability . In the case of an insertion, a * character is inserted in the growing sequence after the character that was just copied. In the case of a deletion, the character that was just copied is deleted, but the position of growing end on the template (indicated by last) still advances by one.
Figure 5 considers a case where the first replicating circle that arises happens to be an 8-mer
Zza****A. Each cell in the population begins with one copy of this 8-mer. The mutation rate is
u = 0.15 and the indel rate is
uindel = 0.015. After a relatively number of time steps in the Gillespie algorithm T = 2 × 10
4, shown in
Figure 5A, 7-mers have evolved from this by deletion, and double-length circles have arisen from both the 8-mer and the 7-mer by the doubling mechanism described above. After a much longer number of steps T = 2 × 10
6, shown in
Figure 5B, reproducing circles of lengths 4, 5 and 6 have also evolved by deletion. The length distribution converges to a state in which the 4-mers are most frequent, together with multiples of the 4-mer created by the doubling mechanism, and small numbers of 5, 6, and 7-mers which are replenished by the insertions occurring in the 4-mer. In the case where
<< u, the 5, 6 and 7-mers will be much less frequent than the 4-mers because selection favours rapid replication. In this case, the length distribution will consist of the minimal-length 4-mer and its multiples that arise via the doubling mechanism, as was already shown in
Figure 3B.
Although we do not know with any certainty the rates of the mutation and selection processes that would have applied in the RNA World, we expect that point mutation rates associated with non-enzymatic replication would be quite large, as measure experimentally [
7,
8]. On the other hand, indel rates might be much smaller than this. We expect selection for increased replication rate to be strong, and therefore to operate on a short time scale of a few cell divisions. Under the assumption that insertions and deletions were rare and occurred with roughly equal frequency, then rare deletions would spread rapidly due to selection, whereas rare insertions would be eliminated by selection as fast as they could originate. This would result in a population dominated by minimal-length circles, which would have no additional non-coding regions that might evolve to encode beneficial genes. In order for a substantial numbers of longer sequences to be maintained in the population (with space available for beneficial genes), there must be a mechanism of lengthening circles that operates sufficiently rapidly to counter selection for rapidly replicating, minimal-length circles. The doubling mechanism that we have seen here, does indeed work rapidly, because it occurs on the time scale of single point mutations that render the HHRz non-functional. Thus, we argue that the doubling mechanism is important for creating a population containing circles substantially longer than the minimum length.
3.3. Beneficial Genes
Up to this point, we have considered circles that replicate but encode no additional function. We now introduce a ribozyme with beneficial function for the cell, represented by character B. Copying B gives its complement b, and vice versa. Mutations may occur with probability u to give a non-functional character *. We assume the B ribozyme is functional only when it is in a folded linear strand. It is not functional when it is part of a circular strand being used as a template, or part of the complementary strand that is being synthesized on a rolling circle. The b character is also assumed to be non-functional. The simulation keeps track of the number of functional ribozymes in each cell (i.e., the number of occurrences of the B character in folded linear sequences). Each functional ribozyme gives an increase in the rates of polymerization. For a cell with ribozymes, the rate of strand-displacement is increased to . We are assuming that beneficial ribozyme speeds up polymerization by contributing in some way to the metabolic reactions in the cell. Replication of all strands in the cell is benefitted equally, not just the strand on which the B gene is present. The B ribozyme does not represent a polymerase that binds to one specific template at a time, and it does not require to bind to a template in order to give the beneficial effect.
With the rules of the model as specified above, when a new linear strand is cleaved from a rolling circle, it folds at a rate . Cleavage can only occur between the A and Z characters. Therefore, every sequence ends with A. If the strand begins with a Z character (arising from the previous cleavage or from starting synthesis with this character), then the strand can circularize. We have assumed that folding of the sequence represents formation of the structure that brings to two ends together and allows circularization. A strand beginning with Z always forms a circle. A fragmentary strand not beginning with Z cannot form a circle and always forms a folded linear strand. When B genes are not present in the model, a folded linear strand plays no role, but when B genes are included, a folded linear strand becomes a beneficial ribozyme whenever it contains a B character. Thus with these rules of the model, the B gene can only have a useful function if it arises in a fragmentary sequence produced from the first incomplete cycle of a rolling circle. B genes in complete copies of the circle, always end up as new circular templates without contributing to the function of the cell.
This suggests that we need a new kind of hammerhead ribozyme whose rate of circularization is tunable. We represent the tunable ribozyme by the characters AY, and their complement ya. Whenever AY appears in the tail of a rolling circle, cleavage of the strand is assumed to be immediate (as with AZ). This produces an unfolded linear strand. Folding of all linear strands occurs at the same rate . If the strand begins with Y, then when folding occurs, a circle is produced with probability and a folded linear strand with probability . Strands beginning with Z always form a circle ( = 1). Fragmentary strands that begin with neither Y nor Z always form a folded linear strand (.
Adding the
parameter introduces a trade-off between circularization, which produces a new template, and folding to a linear strand, which allows the expression of a beneficial ribozyme. A wide variety of hammerhead ribozymes are known with various distinct configurations, sequences, and rates [
26]. Metallic ion concentration has also been suggested to change catalytic activity in the HHRz [
25]. Thus it is likely that such tunability is evolutionarily possible. As a minimal-length 4-mer without
B genes will always benefit from forming a circle, there should be strong selection for rapidly circularizing ribozymes on minimal circles. Therefore, we began with
Z-type ribozymes (with
) on the minimal circles.
Y-type ribozymes (with
) can only be beneficial when
B genes also exist.
We now wish to determine the range of parameters for which the presence of a B gene increases the reproduction rate of the cells which contain it. The most common type of sequence arising, other than the minimal 4-mer is the double-length 8-mer. Therefore, we suppose that the B gene arises initially on an 8-mer with sequence YzaB***A, which we call a plus strand because it encodes a functional B ribozyme. The complementary minus strand (accounting for circularity) is Zya***bA. The minus strand always benefits from circularization; therefore, we assume a Z-type ribozyme on the minus strand. The plus strand has a trade-off between circularization and expression of the B genes; therefore, we assume a Y-type ribozyme on the plus strand.
We ran simulations in which each cell began with one copy of the 4-mer plus strand. After reaching a steady state, the simulation was run for a period of 10
6 simulation steps and the average number of divisions per cell per unit time was measured. This is shown in
Figure 6. For comparison, we also measure the division rate of cells that begin with the minimal 4-mer
ZzzA (shown as a dashed line in
Figure 6). The mutation and indel probabilities
u and
uindel were set to zero in these simulations, so the only replicating sequences that arise are the 8-mers or 4-mers that we begin with, and there is only one type of replicating sequence in each cell.
When there is no benefit of the
B gene (
in
Figure 6A), cells containing the 8-mers always reproduce more slowly than cells containing the 4-mers. In this case, the division rate of the 8-mer cells is maximum when
is 1. There is no advantage to
not forming a circle if the
B gene does not produce a benefit to the cell. When
, there is an optimal value of
in the range 0.3 to 0.5 for the parameters shown. The trade-off favors increasing the expression of the
B gene at the expense of reducing the number of templates. For
, the division rate of the 8-mer cells remains lower than the 4-mer cells across the whole range of
. For
, the division rate of the 8-cells just exceeds the 4-mer cells when
is close to its optimal value. For
, the division rate of the 8-cells exceeds the 4-mer cells across most of the range of
. Higher values of
β are shown in
Figure 6B. For
β > 1, the 8-mer cells reproduce much faster than the 4-mer cells. It can also be seen that even when
= 1 (when a
Y gene always circularizes), the division rate of the 8-mer cells still increases with
. So there is some benefit given by the
B genes even when the only functional
B ribozymes are on the incomplete fragments.
3.4. Spread of a Beneficial Gene
So far, we have shown that cells containing double-length circles with a beneficial genes can sometimes out-compete cells containing minimal-length circles. However, we assumed above that the longer circle with the beneficial gene was already established in a separate cell from the cells containing minimal 4-mers. More realistically, the beneficial gene is likely to first arise as a single copy inside a cell that also contains minimal 4-mers. The longer circle is at a disadvantage relative to minimal 4-mers in the same cell because it replicates more slowly. Spread of the beneficial 8-mer requires the selection at the cell level to exceed the disadvantage at the molecular level.
In this section, we begin with a population of cells containing minimal 4-mers and allow it to come to an equilibrium under mutation
u = 0.15, so that the cells also contain 8-mers, 12-mers, etc. We then change one
* character on a single 8-mer to a
B. We choose the 8-mer because it is the most abundant strand greater than 4. We also change the
Z character on this same strand to a
Y with
= 0.3, because this is close to the value at which the 8-mer cells reproduce most rapidly in
Figure 6.
After introduction of the single B gene, we track how many cells contain circles with the B gene or its complement b. Cells can be divided into four types: those with the beneficial genes, B or b, and no 4-mers, those with 4-mers and no beneficial gene, those with both, and those with neither. 8-mer circles without B or b will arise from duplication of the 4-mers or mutation of B and b characters to *. These circles are counted as having neither a 4-mer or a beneficial 8-mer. Multiples of the 8-mer which have a B gene will arise by duplication of the 8-mers, and strands shorter than 8 with the benefit can arise from deletions. These are counted as having a benefit but no 4-mers.
The results of such a simulation are shown in
Figure 7. The first arrow (at time close to 40 h) shows the point at which the single
B gene was added with
β = 5. Cells which contain both 4-mers and beneficial 8-mers remain rare for a long time after this. The second arrow (at time close to 50 h) shows the point at which the first cell containing beneficial 8-mers and no 4-mers appears. The appearance of the first cell of this type requires a cell-division event in which all beneficial 8-mers from a mixed cell segregate to one daughter cell while all 4-mers segregate to the other daughter. This is relatively rare, but once cells of this type are created, they rapidly multiply. In
Figure 7, cells containing only beneficial 8-mers become the dominant type by about time 80 h. Mixed cells disappear at around time 80 h because they are out-competed by the cells with only beneficial 8-mers. However, this simulation also includes indels; therefore 4-mers can also be created by deletions occurring in 8-mers. This recreates mixed cells later in the simulation (around time 100 h), however these mixed cells do not take over the population, because selection against them is quite strong.
The scenario seen in
Figure 7 shows how a beneficial gene arising on a longer sequence can eventually spread to a high frequency in the population. This requires the creation of a cell that contains only beneficial 8-mer circles without any minimal circles, which is relatively rare. It is more likely that the beneficial 8-mers will disappear whilst they are still rare in the population, either due to deleterious mutations in the
B gene or due to death of the mixed cells containing the
B genes. If the beneficial 8-mers disappear before the creation of a cell containing only beneficial 8-mers, then they will not spread through the population.
We measured how often this occurs by running our simulation multiple times. In each run, a single
B gene was introduced on an 8-mer. The simulation was continued until one of two stop criteria was reached: either (i) the beneficial gene disappeared completely; or (ii) the number cells containing beneficial 8-mers and no 4-mers reached at least 90% of the population. The percentage of runs in which the beneficial 8-mer cells spread to high frequency is plotted against mutation probability
u in
Figure 8 for different values of the benefit parameter
β.
Only a small percentage of the runs lead to spread of the beneficial gene, and high values of
β are required in order to get appreciable probabilities of spread. For
β = 5, the maximum probability of spread is only about 2.3%. In comparison with
Figure 6B, we see that the reproduction rate of 8-mer cells with
β = 5 and
= 0.3 is approximately 8 times higher than for a 4-mer cell. A beneficial gene that gave an immediate 8-fold increase in fitness would have a very high probability of fixation in the usual approximation for the fixation rate used in population genetics [
43]. However, the usual theory for the fixation probability does not apply in our case, because there are multiple strands in each cell. The reproduction rate of a cell depends on the mixture of genetic strands that it contains and also on the number of copies of folded ribozymes, which is variable. Mixed cells containing both 4-mers and 8-mers with the
B gene do not spread to high frequencies. The spread of the
B gene only occurs if a cell is established that contains the beneficial gene but no 4-mers (as shown in
Figure 7). For this reason, the probability of spread is quite small, even when the benefit is large. To check that the
B gene cannot spread due to spread of mixed cells, we ran additional simulations in which the run was stopped on three separate conditions: either (i) the beneficial gene dies out; (ii) cells containing the
B gene and no 4-mers reach 90% of total; or (iii) mixed cells with both the
B gene and 4-mers reach 90%. It was found that the runs never stopped due to criterion (iii). Thus, it was never observed that mixed cells reach high proportion.
For each value of
β in
Figure 8, a peak occurs in the probability of spread at around
u = 0.15. The peak occurs because when
u is very low, the 4-mers replicate accurately. There are few 8-mer (or longer) circles created. If a
B gene arises on an 8-mer, it is in a cell that contains almost entirely 4-mers. It therefore has a low chance of spread. As
u increases, replication of the 4-mers is less accurate, and most of the cells also contain significant numbers of 8-mers and longer circles. If a
B gene arises on an 8-mer in this case, it has fewer 4-mers to compete with, and it is less likely to die out before the creation of a cell that contains only beneficial 8-mers. Thus the spread probability of the
B gene is larger. If
u is too high, however,
B genes disappear due to deleterious mutations. We assumed that the mutation probability
u of
B to
* is the same as that of
A and
Z.
The results in
Figure 7 and
Figure 8 have zero indel rate,
= 0. We supposed that the
B gene arises on an 8-mer because 8-mers are the most common type of longer sequence. However, if
is not zero, then shorter circles containing the beneficial gene can also arise by deletion. If the initial beneficial 8-mer is
YzaB***A, as before, then deletions of the
* characters can occur, giving 7-mers and 6-mers and, eventually, the 5-mer
YzaBA. The
B gene can also be deleted, giving the original minimal 4-mer
YzaA. Thus, the long-term survival of the
B gene depends on competition of the 5-mer and the 4-mer. We investigated this case by beginning with a population of cells each containing the 8-mer
YzaB***A, and allowing the simulation to reach a steady state. When the indel rate is small (
= 0.015) and the benefit is fairly large (
β = 5), 5-mers
YzaBA become dominant, alongside multiples of the 5-mer which also contain
B genes (shown in
Figure 9A). If the indel rate is too large, however, or if the benefit is too small, the minimal 4-mers arise.
Figure 9B shows the final steady-state distribution when
uindel = 0.015 and
β = 1. The
B gene has been lost, and we have a distribution of the 4-mer and its multiples.
In summary—in order for the B gene to survive, it has to maintain itself against deleterious mutations, deletions and selection favoring shorter sequences. These results show that this is fairly difficult, and it by no means occurs every time. However, at least sometimes, a beneficial gene becomes established. Thus, there is a route that leads from minimal, non-functional replicators towards replicating strands that encode beneficial functions.